Abstract
Transposition of Hoxd genes to a more posterior (5′) location within the HoxD complex suggested that colinearity in the expression of these genes was due, in part, to the existence of a silencing mechanism originating at the 5′ end of the cluster and extending towards the 3′ direction. To assess the strength and specificity of this repression, as well as to challenge available models on colinearity, we inserted a Hoxb1/lacZ transgene within the posterior HoxD complex, thereby reconstructing a cluster with a copy of the most anterior gene inserted at the most posterior position. Analysis of Hoxb1 expression after ectopic relocation revealed that Hoxb1-specific activity in the fourth rhombomere was totally abolished. Treatment with retinoic acid, or subsequent relocations toward more 3′ positions in the HoxD complex, did not release this silencing in hindbrain cells. In contrast, however, early and anterior transgene expression in the mesoderm was unexpectedly not suppressed. Furthermore, the transgene induced a transient ectopic activation of the neighboring Hoxd13 gene, without affecting other genes of the complex. Such a local and transient break in colinearity was also observed after transposition of the Hoxd9/lacZ reporter gene, indicating that it may be a general property of these transgenes when transposed at an ectopic location. These results are discussed in the context of existing models, which account for colinear activation of vertebrate Hox genes.
Keywords: Hox gene, transgenes, transposition, expression
Vertebrate Hox genes are key elements among the genetic determinants that organize positional information along the rostral-caudal axis. During development, their sequential activation, in time and space, results in the distribution of various combinations of proteins at each newly produced metameric level (e.g., Hunt et al. 1991; Kessel and Gruss 1991). For example, their successive transcription in presomitic mesoderm, in the course of gastrulation, will assign molecular addresses to emergent somitic condensations, thereby instructing these cells about their morphogenetic fates (Deschamps and Wijgerde 1993). Gene-targeting modifications to the complement of HOX protein have largely verified this proposal as they usually lead to corresponding and predictable alterations in the body plan, often referred to as homeotic transformations (see Krumlauf 1994). In this context, the molecular mechanism(s) controlling both the time- and level-specific activation of Hox genes play a crucial role in the proper organization and topology of structures. Hence, understanding these processes will be an important step in our analysis of vertebrate development. Interestingly, the spatial and temporal sequences of gene activation are colinear with the physical order of the genes along their respective clusters (Gaunt et al. 1988; Duboule and Dollé 1989; Graham et al. 1989; Izpisua-Belmonte et al. 1991). Although this correlation is likely to facilitate the control and coordination of the precise sequence of activation, the mechanism translating this genomic topological information into transcriptional outputs is elusive.
In recent years, this question has been investigated by use of mainly two sets of experimental designs. On the one hand, conventional transgenic approaches have revealed that isolated Hox genomic loci were able, in many instances, to drive expression of a reporter transgene in a way clearly reminiscent of the endogenous gene (e.g., Püschel et al. 1990; Whiting et al. 1991; Marshall et al. 1992; Behringer et al. 1993; Gérard et al. 1993; Becker et al. 1996). Although the transgenes generally did not faithfully recapitulate all the specificities of the locus, they nevertheless demonstrated that important regulatory elements necessary for some spatial and temporal Hox gene activation are located near the transcription units and can function outside the context of the Hox complex (Krumlauf 1994). On the other hand, experiments involving large rearrangements within the HoxD cluster in vivo, for example, by transferring genes from one position to another, have indicated that part of the regulation depends on the position of a given gene in the complex, regardless of its proximate flanking sequences (van der Hoeven et al. 1996). It was thus shown that when Hoxd9 or Hoxd11 transgenes were recombined upstream Hoxd13, that is, at a more posterior position in the complex, their expression was substantially delayed as compared with random integration sites. Consequently, it was proposed that whereas Hox genes can carry in cis elements capable of regulating their transcription in space and time, this conventional gene regulatory circuitry was subject to a silencing mechanism preventing posterior genes from being activated at an early stage (Dollé et al. 1989; van der Hoeven et al. 1996; Kondo et al. 1998).
Subsequent experiments in which deletions were engineered near the posterior side of the HoxD complex gave further support to this view, as deletion of an upstream fragment of DNA resulted in the deregulation of endogenous Hox gene expression (Kondo and Duboule 1999). This indicated that sequences outside of the complex were necessary to properly organize the silencing mechanism. A model was proposed to account for these results in which such sequences would be required to initiate a repressive chromatin configuration over the HoxD cluster. This proposal, however, failed to explain some observations: for example, a cis deletion of several genes in the posterior HoxD complex did not drastically change the activation timing of the resident genes, even though these latter transcription units were brought to a more posterior position, that is, near the potential upstream sequence required for organizing a presumptive high order structure (Zákány and Duboule 1996).
In these approaches, the interpretation of the results as well as their integration into a conceptual framework, were subject to an additional difficulty related to the multiphasic aspect of Hox gene expression. Experimental evidence indicates that the early phase of activation needs to be subsequently maintained by a process not necessarily related at the mechanistic level (Belting et al. 1998; Gould et al. 1998; Stern and Foley 1998; Kondo and Duboule 1999). For instance, ectopic activation of posterior genes in anterior regions is usually maintained only in those domains in which other (or the same) endogenous posterior genes are expressed, suggesting that cross-regulation via paralogous genes or autoregulation is an important factor in the refinement of the expression specificities. This particular point must be carefully considered when interpreting transgenic mice experiments to examine regulatory components, as all of the potential cross-regulatory Hox proteins may need to be removed, and this is often not possible or very difficult. An example of this cross-regulation is the rhombomere 4 (r4) specific expression domain of the Hoxb1 transgene, which was completely abolished only when endogenous Hoxb1 and Hoxa1 functions were removed (Studer et al. 1998).
To gain insight into the nature of the down-regulation observed upon transgene relocation in the posterior HoxD complex and to further understand the mechanisms underlying colinearity, we wanted to transpose the anterior Hoxb1 transgene to the posterior end of HoxD. The reasons why we selected this particular transgene are as follows: first, this is one of the earliest and more anteriorly expressed Hox genes (Hunt et al. 1991; Murphy and Hill 1991); that is, it is expressed at a time and in body regions in which all 5′ Hoxd genes (from Hoxd9 to Hoxd13) are still silent. Second, the regulatory potential of this transgene has been thoroughly studied by conventional transgenesis leading to a precise characterization of the various enhancers present therein (Marshall et al. 1994; Studer et al. 1994; Popperl et al. 1995) as well as several of the factors interacting with them. Third, Hoxb genes, unlike Hoxd's, are not involved in limb or genital patterning and the transgene does not carry any Hoxd-specific regulatory element, allowing us to clearly distinguish between a Hoxd versus a Hoxb-type of regulation when inserted within the HoxD cluster. Fourth, r4-specific expression of the transgene only requires Hoxb1, Hoxa1, and Pbx/Prep 1 functions (Popperl et al. 1995; Di Rocco et al. 1997; Berthelsen et al. 1998; Studer et al. 1998) in an autoregulatory loop and, finally, the transgene was shown to be responsive to retinoic acid (RA) treatment in vivo (Marshall et al. 1992). These latter two criteria were important as they would, in principle, allow us to challenge the accessibility of the transgene, for example, after exposure to RA, when transposed posteriorly.
Here, we report the results of such a Hoxb1 transgene relocation experiment. In the developing nervous system, expression of Hoxb1 in r4 was totally abolished when placed near Hoxd13, either in the presence or absence of exogenous retinoic acid. Surprisingly however, such a strong repression was not observed in the early mesoderm domain in which the transgene was unexpectedly transcribed in a manner similar to its endogenous counterpart. Furthermore, the presence of the transgene was able to trigger ectopic and transient expression of the neighboring Hoxd13 gene (but not Hoxd12) in a related domain, that is, at a time and places in which Hoxd13 is normally repressed. These results suggest that a potential repressive mechanism may not be equally implemented in all tissues. They also indicate that the position of a given Hox gene within a complex is not solely responsible for its timing of activation. The significance of these observations for our understanding of colinearity is discussed in the context of the different models that have been proposed.
Results
Transposition of Hoxb1 in the HoxD complex
To study the effect of the posterior HoxD complex on the regulation of an anterior Hox gene, we selected the well-characterized Hoxb1/lacZ transgene. This reporter construct is known to mimic the endogenous Hoxb1 expression pattern (Marshall et al. 1992, 1994; Studer et al. 1994) and carries most of the cis regulatory sequences sufficient for the early expression in mesoderm and neuroectoderm. In particular, the early neural and mesodermal enhancer, containing the 3′ retinoic acid responsive element (RARE; Marshall et al. 1994) was included in this transgene as well as the r4 autoregulatory enhancer with the labial-PBX binding sites (Popperl et al. 1995; Fig. 1). A targeting vector was engineered by introducing a PGKneo selection cassette, flanked by loxP sites, at the 5′ end of the Hoxb1/lacZ transgene. The recombination site in the HoxD cluster (Fig. 1; IS) was as reported previously for the targeted insertions of Hoxd9 and Hoxd11 (van der Hoeven et al. 1996), that is, in between Evx2 and Hoxd13, to facilitate subsequent comparisons. To this aim, the Hoxb1/lacZloxPneoloxP construct was flanked by both halves of the Evx2/Hoxd13 intergenic region (Fig. 1, bold lines) to produce the final targeting vector. After electroporation and selection, ES cell clones were identified in which either homologous recombination (Fig. 1, TgH[b1/lacZ/neo]GE) or random integration (Fig. 1, TgN[b1/lacZ/neo]GE) had occurred. After injection into blastocysts and germ-line transmission, homozygous lines of mice were established for both configurations. Potential interference on the regulation of the surrounding loci induced by the PGK-neo cassette was prevented by deleting the cassette with the Cre/loxP system. Both TgH[b1/lacZ/neo]GE and TgN[b1/lacZ/neo]GE mice were crossed with CMV/Cre deleter mice (Dupé et al. 1997) to produce the TgHb1 (targeted) and TgNb1 (random) mice, respectively (Fig. 1). Both TgHb1 and TgNb1 mice were used for expression studies. The TgNb1 mice contained a single-copy integration and serve as control for transgene expression in TgHb1 mice.
Figure 1.
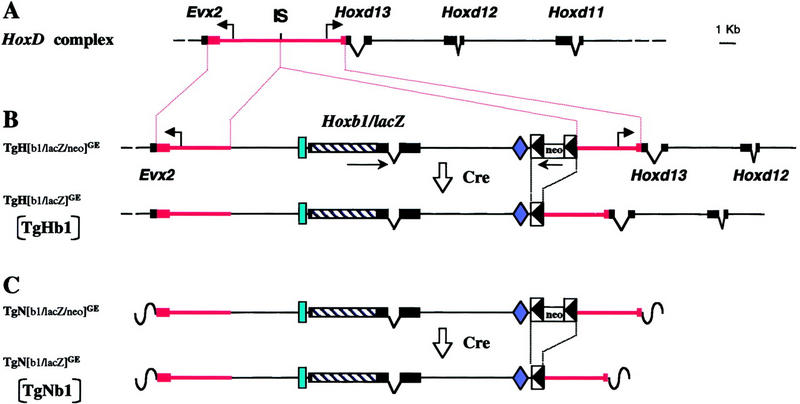
Transposition of the Hoxb1/lacZ reporter transgene in the posterior part of the HoxD complex. (A) Scheme of the upstream HoxD complex. The position of the insertion site (IS) is shown between Evx2 and Hoxd13. The red lines indicate the extent of the homologous arms used for recombination. (B) Drawing of the targeting vector after integration at the HoxD locus by homologous recombination. The Hoxb1/lacZ transgene, together with flanking genomic sequences, was recombined in the same transcriptional orientation as the neighboring Hoxd genes (arrow). The PGK-neo selection cassette, flanked by loxP sites (boxed arrowheads), was introduced in the opposite orientation. The position of the r4 enhancer is shown by a vertical rectangle, whereas the early neural-mesodermal enhancer is shown as a losange. After crossing these mice with a Cre deleter strain, the cassette was excised to generate the TgHb1 allele (bottom). (C) The same targeting vector after random integration (top). Mice carrying this allele were also crossed over a deleter strain to remove the selection cassette and produce the TgNb1 allele (bottom), in which a single copy transgene, together with homologous arms, was integrated randomly.
Expression of Hoxb1 inside and outside of HoxD
We first tested whether the posterior HoxD environment would interfere with the early activation of the Hoxb1 transgene, as suspected from previous work. For this purpose, we analyzed transgene expression at 8.5 d.p.c., a stage at which 5′ Hoxd genes are transcriptionally silent (Dollé et al. 1989). At this early stage, the randomly integrated single-copy transgene was expressed as described previously (Fig. 2, left), with a pattern reminiscent of both the endogenous Hoxb1 and the conventional transgene (Marshall et al. 1992, 1994; Studer et al. 1994). This demonstrated that the presence of both Hoxd homologous arms (covering the Evx2/Hoxd13 intergenic region; bold lines in Fig. 1) flanking the original transgene did not modify the regulation of its expression when integrated randomly. LacZ staining was strong in presomitic and somitic mesoderm as well as in r4 (Fig. 2, left). We next looked at the expression of the same construct when integrated upstream Hoxd13 and observed surprisingly a similar early onset of expression (Fig. 2, right). This showed that the Hoxb1 transgene could be active in the posterior HoxD complex. However, the pattern was markedly different from that seen with the randomly integrated version as the targeted Hoxb1 was not expressed in the hindbrain (Fig. 2, arrowhead). In addition, the anterior limit of expression within the somitic mesoderm was clearly more rostral than seen with the random integrant (Fig. 2, red arrows). Hence, repression varied between tissues and the transgene was unable to respond in the hindbrain, even though r4 expression depends on autoregulation and endogenous labial members that were present.
Figure 2.
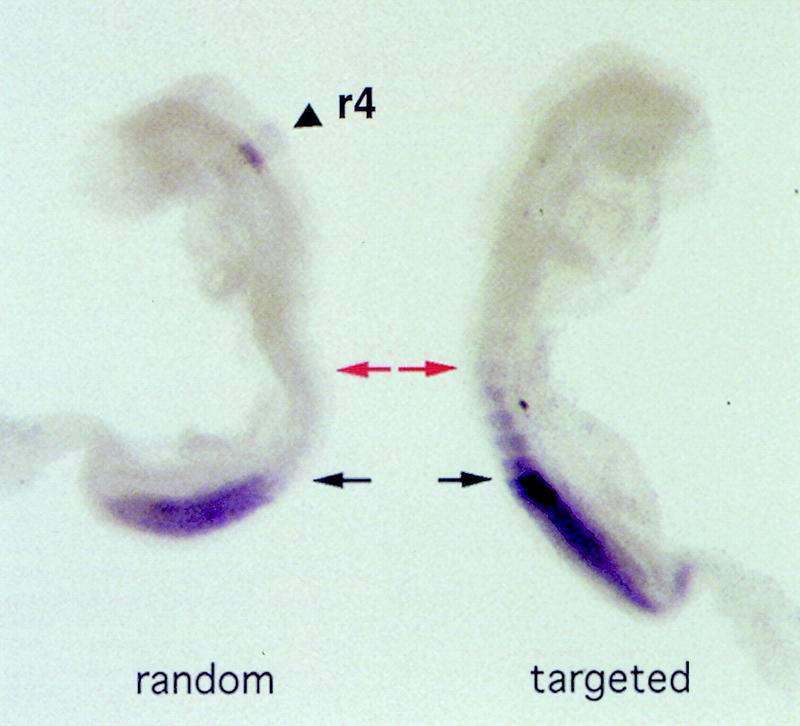
Expression of the targeted and randomly integrated versions of the Hoxb1/lacZ transgene during early development. Two 8.5-d.p.c. fetuses expressing either the randomly integrated (left), or the HoxD targeted (right) Hoxb1 transgene. (Left) The expected expression domains were scored, both in the fourth rhombomere of the hindbrain (r4, arrowhead) and in the posterior part of the developing trunk. (Right) Expression in r4 was no longer detected when the transgene was recombined upstream Hoxd13. However, early expression in the trunk was maintained, extending more anteriorly in somitic mesoderm than for the randomly integrated version (arrows). Black and red arrows indicate the anterior limits of transgene expression in TgNb1 (black) and TgHb1 (red) embryos, respectively.
We followed the evolution of this unexpected expression pattern during later stages of fetal development, that is, when both the endogenous Hoxb1 and 5′ Hoxd genes are known to be transcriptionally active. At day 10.5, the relocated version of Hoxb1, unlike the randomly integrated copy, was still silent in r4 (Fig. 3A). In contrast, expression of the relocated transgene was now detected in the most distal part of the growing limb buds as well as in the genital bud (Fig. 3A, arrows), whereas these structures were negative in the randomly integrated counterpart (Fig. 3A, left). Expression in both limb and genital buds was reminiscent of posterior Hoxd gene expression pattern, indicating Hoxb1 had come under the influence of Hoxd regulatory controls. From 11.5 d.p.c. onward, the relocated transgene continued to be strongly transcribed in developing limbs and genitalia (Fig. 3B) in a way similar to the neighboring Hoxd genes, but in marked contrast to the endogenous Hoxb1. At this stage, neither the endogenous Hoxb1 gene, nor the randomly integrated transgene, were expressed any longer.
Figure 3.
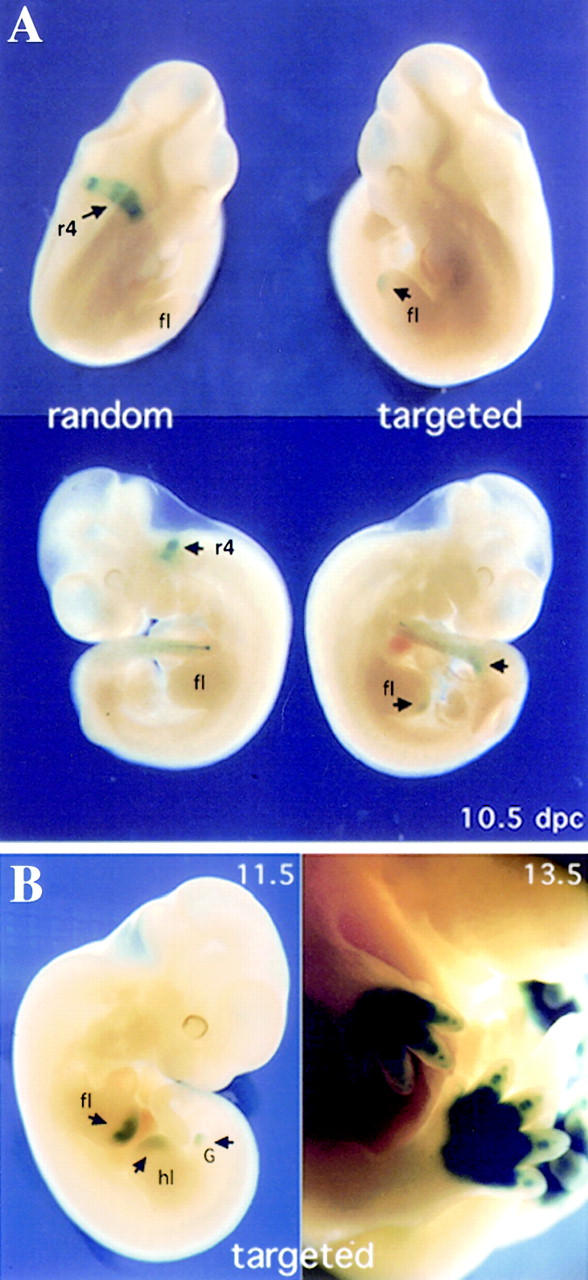
Expression of the targeted and randomly integrated versions of the Hoxb1 transgene in fetal stages. (A) Comparison between targeted (right) and randomly integrated (left) Hoxb1/lacZ expression in 10.5-d.p.c. embryos. The randomly integrated transgene was expressed in the r4 domain (arrow), whereas the activity of the targeted transgene was suppressed in this domain. In addition, the recombined version was expressed in the tip of the developing limb bud (short arrow) as well as in the emerging genital bud (arrow). Note that expression in the tail bud was similar in both configurations. (B) Expression of targeted Hoxb1/lacZ in the distal part of the limb and genital buds. At 11.5 and 13.5 d.p.c., expression of the recombined transgene was like that of posterior Hoxd genes (e.g., Hoxd13), with a conspicuous domain in the developing digits and external genitalia. (fl) forelimb; (hl) hindlimb; (G) genital bud.
Repression of Hoxb1 expression in r4
Expression analysis of developing TgHb1 animals revealed that the r4 enhancer, located on the transposed Hoxb1 locus, was no longer functional when integrated at the 5′ end of the HoxD complex. This is an auto-/cross-regulatory element (Popperl et al. 1995; Studer et al. 1998), and as endogenous Hoxb1 was normally transcribed in these animals (not shown), the absence of lacZ staining could not be explained by a lack of upstream regulators and/or cofactors. As the relocated transgene might have required higher levels of endogenous Hoxb1 for proper expression in r4, we stimulated Hoxb1/lacZ expression by treatment with RA. This was shown previously to act positively on the autoregulatory loop involved in Hoxb1 activation in this particular rhombomere (Marshall et al. 1992; Popperl et al. 1995).
We challenged both TgHb1 and TgNb1 7.5-d.p.c. embryos in utero with RA treatment and the response of the transgenes to this exogenous RA delivery was monitored at 8.5 d.p.c. RA-induced lacZ staining of the randomly integrated transgene in a broad domain extending anterior to r4 (Fig. 4, arrow). This anterior shift was similar to those reported previously, for either the endogenous Hoxb1 gene (Conlon and Rossant 1992), or the conventional Hoxb1 transgenes (Marshall et al. 1992; Popperl et al. 1995), and was associated with severe alterations in the rostral part of the embryo. In contrast, whereas RA-treated Hoxb1-relocated embryos did show the expected morphological defects, no lacZ staining was ever detected in their hindbrain, thus corroborating the absence of lacZ staining in r4 of untreated TgHb1 embryos. We concluded that even in the presence of a substantially elevated amount of endogenous Hoxb1 protein obtained after stimulation by RA, the Hoxb1/lacZ transgene, unlike its endogenous counterpart, was unable to respond. This indicates that the Hoxd location differentially influences neural and mesodermal expression of the relocated Hoxb1 transgene.
Figure 4.
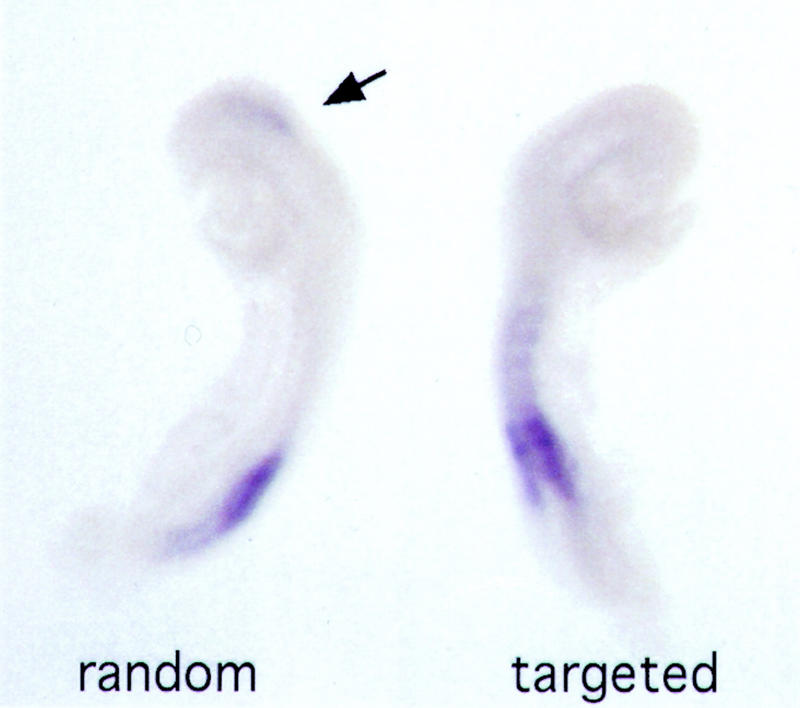
Treatment of both random and targeted lines with RA. LacZ expression is shown in two 8.5-d.p.c. fetuses derived from females treated with RA. Both fetuses showed severe alterations in their anterior morphologies, indicating that RA treatment was efficient. At left, an embryo expressing the randomly integrated transgene is shown. This TgNb1 embryo displayed a clear anterior extension of the lacZ reporter gene activity, which was no longer restricted to a putative r4 domain (arrow). In contrast, RA treatment of an embryo carrying the recombined transgene (right) was not able to trigger lacZ expression, even though the endogenous Hoxb1 gene was deregulated and expressed anteriorly (not shown).
Misregulation of HoxD genes
Because the Hoxb1/lacZ transgene was activated unexpectedly early in mesoderm when transposed at the 5′ end of the HoxD complex, we investigated whether expression of the neighboring Hox genes were deregulated by this insertional event. We first examined expression of the immediately adjacent gene Hoxd13 by whole-mount in situ hybridization and detected strong expression in 7.5-d.p.c. TgHb1 embryos (Fig. 5B). At this early developmental stage, expression was similar to that of both the endogenous Hoxb1 gene and the Hoxb1 transgene (Fig. 5A), with a strong signal in mesoderm emerging from the primitive streak. This represented a severe deregulation of Hoxd13, as transcription of this gene normally starts at day 9 of development and appears in posterior mesoderm at the base of the allantois (Fig. 5F, arrow). By day 8.5 in TgHb1 embryos, an ectopic Hoxd13 signal was detected in the node and presomitic and somitic mesoderm with an anterior limit located at the level of the third somite (Fig. 5C). Hoxd13 expression was strongly detected in the posterior part of the embryo at day 9.5, as if its normal transcript domain was reinforced by the presence of the ectopic domain induced by the Hoxb1 transgene (Fig. 5, cf. D and F). Consistent with this idea, and unlike the wild-type specimen, expression was also observed in somites from the caudal end to the level of the future forelimb (Fig. 5D, arrow). As with the relocated Hoxb1 gene, we never observed r4 expression during any of these stages. One day later, expression in somitic mesoderm was down-regulated and became restricted to the posterior end of the embryo. The patterns at 10.5 d.p.c. appeared essentially as the wild-type Hoxd13, although a slight anteriorization was maintained (Fig. 6A, arrow). With the exception of Hoxd13, none of the other 5′ Hoxd expression domains were affected by the Hoxb1 relocation, neither in terms of activation time, nor with respect to their expression domains (not shown).
Figure 5.
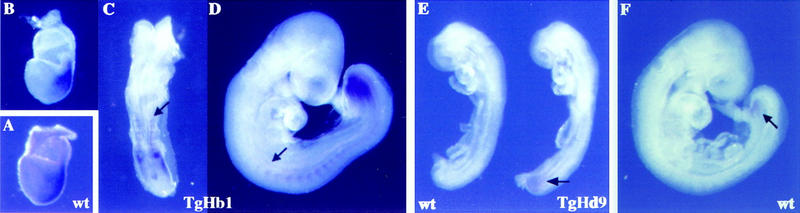
Ectopic expression of Hoxd13 after transgene relocation. (A) Endogenous Hoxb1 expression in a 7.5-d.p.c. wild-type embryo used as a control. (B–D) Hoxd13 expression in Hoxb1/lacZ relocated (TgHb1) embryo. Expression of Hoxd13 at 7.5 d.p.c. (B) was strongly reminiscent of that of Hoxb1 (A). One day later (C), expression was strong in the node, the somitic and presomitic mesoderm. As for the relocated transgene, staining was not detected in the hindbrain. At 9.5 d.p.c. (D), a strong expression domain was scored in posterior mesoderm, resembling that of posterior Hoxd genes, although of much stronger intensity (cf. with F). However, ectopic expression was still present in somitic mesoderm up to the level of the emerging forelimb bud (arrow). Comparison between C and D clearly shows the posterior regression of the ectopic Hoxd13 domain. (E) Hoxd13 ectopic expression in an 8.5 d.p.c. Hoxd9/lacZ (TgHd9) embryo. Whereas Hoxd13 was not detected in the wild-type control embryo (left), expression was visible in the posterior part of an aged-matched TgHd9 embryo (arrow). (F) Onset of Hoxd13 expression in a wild-type foots of 9.5 d.p.c.. Expression can hardly be documented before this stage.
Figure 6.
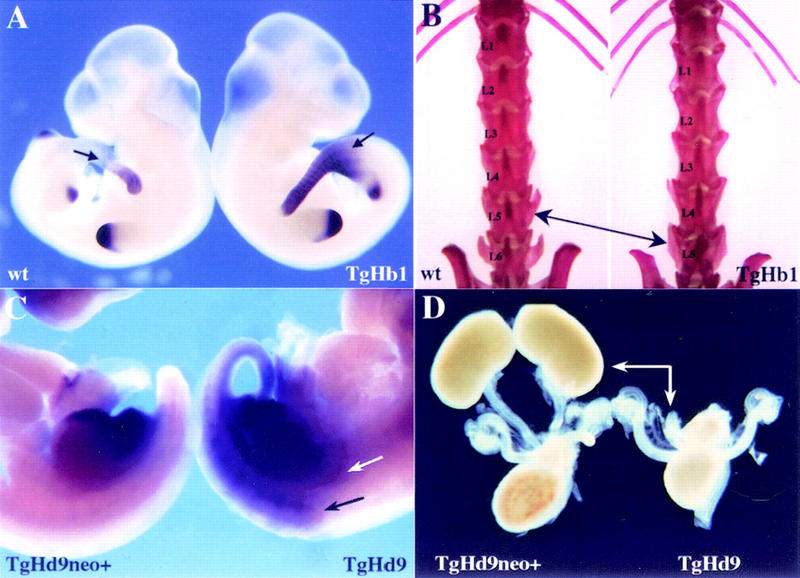
Ectopic Hoxd13 expression and concurrent phenotypic alterations. (A) Hoxd13 expression in wild-type (left) and TgHb1 (right) 10.5-d.p.c. fetuses. Expression patterns were identical to each other in limb buds. In addition, the signal was restricted to the posterior part of the trunk for both wild-type and TgHb1 embryos. However, expression in the latter embryo was slightly anteriorized (arrows). (B) Adult skeletal preparations showed that all animals carrying the relocated transgene (TgHb1) displayed five lumbar vertebrae (left), instead of the six found among control littermates (right). (C) Expression of Hoxd13 in 11.5-d.p.c. fetuses carrying the relocated Hoxd9/lacZ transgene, either in the presence (TgHd9neo+, left) or in the absence (TgHd9, right) of the PGK-neo selection cassette. No obvious ectopic Hoxd13 expression was detected in the left fetus, which showed signals in the tip of developing limb buds and in the emerging genital eminence. After excision of the neo cassette, however, strong expression was de tected throughout the posterior part, including in somitic (black arrow) and intermediate (white arrow) mesoderm. This latter domain included the metanephric blastema, as verified by histological sections (not shown). (D) Dissected urogenital system of newborn animals carrying the Hoxd9/lacZ transgene relocated upstream Hoxd13, with (left) or without (right) the PGK-neo selection cassette. In the absence of the neo gene, kidneys were not formed (arrows), whereas truncated ureteres of variable lengths could be observed at the expected position.
This unexpected activation of Hoxd13 in TgHb1 embryos lead us to test whether this was specific for Hoxb1 or whether it could also be observed with other Hoxd/lacZ transgenes that we relocated previously to the same genomic position. Therefore, we assayed for changes in Hoxd13 expression during early development of embryos carrying the Hoxd9/lacZ transgene upstream of Hoxd13 (van der Hoeven et al. 1996). In those embryos in which the PGK-neo selection cassette was present between the transgene and Hoxd13, ectopic expression of Hoxd13 was not detected. In contrast, after this allele was crossed with a Cre deleter mouse, Hoxd13 was expressed prematurely (Fig. 5E, arrow). Interestingly, this stage (8.5 d.p.c., about four somites) was older than the time of premature transcription observed in the presence of the Hoxb1/lacZ transgene (7.5 d.p.c.). Conversely, the Hoxd9/lacZ transgene reporter itself was not transcribed earlier than expected for its genomic position. Instead, it became active at the expected time for the most posterior Hoxd gene (van der Hoeven et al. 1996). Hence, the types of changes observed by transposition varied with the particular transgene inserted.
Phenotypes
The above Hoxd13 expression analyses revealed that in at least two cases in which a transgene had been recombined between Evx2 and Hoxd13, transcription of Hoxd13 was deregulated, either in time or in space. Such misregulation of a posterior Hox gene would be expected to generate concurrent gain-of-function phenotypes, as shown previously in related circumstances in both limbs and trunk (van der Hoeven et al. 1996). Consequently, we searched for phenotypic alterations in animals carrying either the Hoxb1/lacZ or the Hoxd9/lacZ transgenes, without the PGK-neomycin selection cassette.
In the case of the Hoxb1/lacZ relocation, we suspected that the early and widespread Hoxd13 ectopic expression might be detrimental to the developing embryo. However, animals carrying this configuration didn't show any particularly visible alteration and survived as well as their wild-type littermates. The only significant phenotype was a completely penetrant transformation of the sixth lumbar vertebra (L6) into the morphology of the first sacral (S1), leading to a total of five lumbar vertebrae instead of six (Fig. 6B). The sacrum, however, remained unaffected, suggesting that the entire posterior axial skeleton had shifted anteriorly or, alternatively, and that the sixth lumbar vertebra had disappeared. This weak phenotype was also found in Hoxd11/lacZ relocated animals (Zákány and Duboule 1996) in which ectopic Hoxd13 expression in the main body axis was similar to that reported here for 10.5-d.p.c. TgHb1 embryos. Surprisingly however, no phenotype was found associated with the early and widespread misexpression of Hoxd13 in the Hoxb1/lacZ configuration.
We next checked whether ectopic expression of Hoxd13 in the Hoxd9/lacZ allele would affect development. Mice carrying the relocated Hoxd9/lacZ transgene with the selection cassette were thus crossed with a Cre deleter strain to obtain adult animals without the PGK-neo gene. At birth, animals of this genotype were indistinguishable from their wild-type littermates. However, all of them died the next day and the autopsies revealed that they had no kidneys (Fig. 6D). The rest of the urogenital system nevertheless appeared normal with the exception of the ureteres, which were truncated at various distances from the bladder (Fig. 6D). We verified that this kidney agenesis correlated with an ectopic expression of Hoxd13 in developing metanephric blastema and found expression of Hoxd13 therein (Fig. 6C, white arrow), following a pattern that had been described previously for the Hoxd9/lacZ transgene when integrated randomly (Renucci et al. 1992). Interestingly, this kidney defect phenocopied the effect of a Hoxd11/Hoxa11 double loss of function (Davis et al. 1995), suggesting that ectopic Hoxd13 may negatively affect the function of group 11 genes, as described for limb development (van der Hoeven et al. 1996; Hérault et al. 1997; Peichel et al. 1997).
TAMERE-dependent anterior relocations of Hoxb1/lacZ
The failure of relocated Hoxb1/lacZ to be expressed in r4 suggested that the transgene was the target of a negative effect exerted by the posterior part of the complex in neuroectodermal cells. To further investigate this possibility, we generated two different deletions whereby the Hoxb1/lacZ transgene was brought to more anterior positions within the cluster, while keeping the same position with respect to Evx2 (Fig. 7). To achieve this result, we made use of the TAMERE (targeted meiotic recombination) method, allowing for targeted recombination to be induced during meiosis between homologous chromosomes carrying loxP sites (Hérault et al. 1998). We used two additional lines of mice (Fig. 7A); the first one carried a loxP site between Hoxd11 and Hoxd10 (the C6 line, Gérard et al. 1996; allele Ch.2 in Fig. 7A); the other one carried a deletion of the posterior HoxD complex until Hoxd3, in which a loxP site was located (Zákány and Duboule 1999; Ch.3 in Fig. 7A). Males were produced in which the relocated Hoxb1 allele (Fig. 7A, Ch.1) was complemented with either one of the two other alleles (Fig. 7A, top or bottom), together with the transgene expressing the Cre recombinase in meiotic prophase. In the progeny of these males, animals were recovered that derived from meiotic cells wherein interallelic (trans-chromosomal) recombination had occurred between loxP sites (broken red line in Fig. 7A). In these animals, the Hoxb1/lacZ transgene was therefore located at different positions, either upstream of Hoxd13 (Fig. 7B, TgHb1), upstream of Hoxd10 (Fig. 7B, TgHb1Del3), or upstream of Hoxd3 (Fig. 7B, TgHb1Del7).
Figure 7.
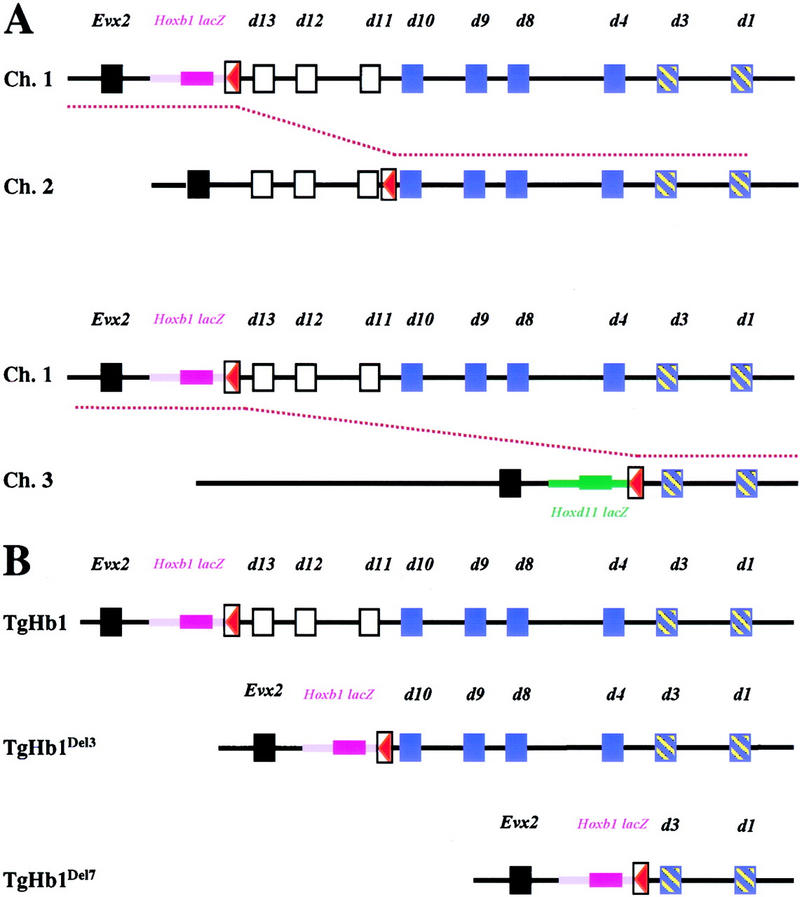
Strategy used to produce anterior relocations of the Hoxb1/lacZ transgene with TAMERE. (A) Description of the different alleles used for the interallelic recombination. (Top) The TgHb1 chromosome with the transgene in magenta and the loxP site in red (referred to as Ch.1). Below, the C6 chromosome (Gérard et al. 1996), containing a loxP site between Hoxd11 and Hoxd10 (or Ch.2). The broken red line between Ch.1 and Ch.2 indicates the unequal targeted recombination event occurring at meiosis to generate the TgHb1Del3 chromosome (B). The other recombination event involved Ch.1 as well as a chromosome containing the Hoxd11/lacZ transgene (green) and a loxP site (red), located between Hoxd3 and Hoxd4 along with a deletion of the seven Hoxd genes posterior to the insertion site (the HoxDDel7 allele in Zákány and Duboule 1999; depicted here as Ch.3). Again, the red broken line shows the meiotic recombination event leading to the production of the TgHb1Del7 allele shown in B. (B) Description of the two alleles produced by TAMERE together with the starting chromosome (Ch.1). In all three cases, the Hoxb1/lacZ reporter transgene maintained its posterior (5′) neighborhood, while moving to more anterior locations. (Black squares) The Evx2 gene; (white squares) Hoxd genes deleted in the first recombination event (in the TgHb1Del3 allele); (blue squares) Hoxd genes subsequently deleted in the second recombination event (the TgHb1Del7 allele). The remaining Hoxd1 and Hoxd3 genes are present in all configurations. In magenta, the Hoxb1/lacZ transgene; in green, the Hoxd11/lacZ transgene; in red, the loxP sites.
We analyzed β-gal staining in embryos from these lines at development stages in which the endogenous Hoxb1 r4 enhancer was known to be active (9–11 d.p.c.; Fig. 8, r4 arrow). In the case in which the transgene was upstream Hoxd10, no staining was detected within the hindbrain, as for the control recombination upstream Hoxd13 (Figs. 7 and 8, TgHb1Del3 and TgHb1, respectively). In the TgHb1Del3 configuration, the anterior limit of expression was somewhat reminiscent of the Hoxd10 pattern. Interestingly, when the transgene was relocated upstream of Hoxd3, lacZ expression did appear in the neuroectoderm, from the posterior end until the limit between the presumptive r5/r6 (Fig. 8, TgHb1Del7, arrow), in a way related to the Hoxd3 expression pattern (Tan et al. 1996). However, in all cases, staining was never detected in r4. These results showed that although there was not a complete block in the ability of the transposed Hoxb1 transgene to be expressed in neural tissue, as evidenced by the expression up to r5/r6, the specific Hoxb1 domain in r4 was still negatively regulated (Fig. 8). Furthermore, in both cases, expression was detected in developing limb and genital buds, likely due to the presence of the genomic region located upstream the complex (Hérault et al. 1999).
Figure 8.
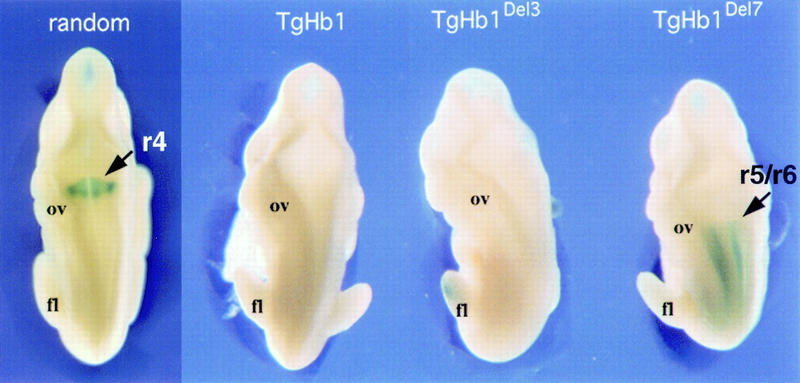
Comparison between the expression patterns of the Hoxb1/lacZ transgene in the randomly integrated version and at the three relocated positions (TgHb1, TgHb1Del3, and TgHb1Del7, from posterior to anterior, respectively, see Fig. 7). X-gal Staining of 10.5-d.p.c. fetuses are shown from a dorsal-anterior view. Expression in r4 was clearly detected in the TgNb1 (random) embryo (arrow), although absent from the relocated version (TgHb1; see Figs. 2 and 3). Moving the Hoxb1 transgene between Hoxd11 and Hoxd10 (TgHb1Del3) did not modify this negative regulation as no staining was scored. However, moving the transgene more anteriorly (TgHb1Del7) resulted in the appearance of a lacZ pattern in the hindbrain that was nevertheless restricted to a domain posterior to the r5/r6 rhombomeric boundary (arrow). Here again, staining in r4 was not observed. (ov) Otic vesicle; (fl) forelimb bud.
Discussion
The regulation of vertebrate Hox gene expression presents an interesting paradox; whereas their tight clustered organization has been maintained and reflects the temporal and spatial organization of their expression domains, some genes appear to be properly regulated outside of this clustered environment, as illustrated by many transgenic studies (e.g., Püschel et al. 1990; Whiting et al. 1991; Marshall et al. 1992; Behringer et al. 1993; Gérard et al. 1993; Becker et al. 1996). Although shared local regulatory regions could be important for directing the proper balance of spatial and temporal expression, Hoxd gene transpositions have provided evidence for a progressive and linear release of a potential repressive configuration as a possible mechanism to sequentially activate these genes (van der Hoeven et al. 1996; Kondo et al. 1998). Subsequent targeted modifications at the HoxD locus supported this hypothesis and indicated that DNA sequences required for organizing this silencing mechanism were located upstream of the complex, near its 5′ end. Further deletion of these sequences led to ectopic expression of the remaining Hox genes, as if silencing had been released (Kondo and Duboule 1999). The experiment reported in this paper, in which an anterior Hoxb1/lacZ gene was transposed into the posterior HoxD complex, reveal a surprising and dynamic interplay between regulatory influences of the endogenous complex and the transposed gene. This raises several important questions on models and mechanisms of colinearity.
Relocation of Hoxb1: expression in hindbrain
An apparent example consistent with the posterior repression model is the inhibition of r4 expression in TgHb1 mice. During hindbrain development of these mice, the transgene-specific Hoxb1 r4 enhancer was totally inactive. This was somewhat unexpected as the upstream factors required for its activity are known and present in these animals. The r4 enhancer was identified as a 148-bp DNA fragment located immediately 5′ to the Hoxb1 start site (Popperl et al. 1995). This fragment alone, fused to a lacZ reporter transgene, was able to drive expression in r4 with a high penetrance (Popperl et al. 1995). It was also shown that this element is the target of an autoregulatory loop from paralogy group I proteins acting in concert with cofactors of the Pbx/exd type (Di Rocco et al. 1997). In the absence of both Hoxb1 and Hoxa1 functions, Hoxb1/lacZ transgene expression in r4 was abolished (Studer et al. 1998). In addition, this autoregulatory loop mediates the action of RA which, at teratological doses, was able to extend Hoxb1/lacZ expression up into the r2 domain (Popperl et al. 1995).
The present results support the existence, in r4 cells, of a silencing mechanism operating over the posterior HoxD complex. In TgHb1 mice, cells expressing the endogenous Hoxb1 gene in r4, that is, cells that did contain all the factors necessary for this expression, were nevertheless unable to transcribe the targeted Hoxb1/lacZ transgene, even though the r4 enhancer was present in its native form, as judged by sequencing the targeted mouse DNA. The same transgene, however, could fully respond to these factors when integrated randomly, even as a one-copy insert. In the randomly integrated locus, the presence of both HoxD-specific homologous arms indicated that silencing was not due to a local neighborhood effect exerted by surrounding sequences over the r4 enhancer. Therefore, we interpret this result as an illustration of a broadly acting silencing mechanism implemented over this DNA region in this cell type. Consistent with this idea, we were unable to compete out this repression through a transcriptional stimulation of the relocated transgene by application of teratological doses of RA. Whereas the randomly integrated Hoxb1/lacZ copy readily responded to RA stimulation, the Hoxd relocated Hoxb1/lacZ transgene remained totally silent in the hindbrain. This indicates that RA stimulation was not sufficient to overcome silencing of the reporter gene when placed upstream Hoxd13.
Expression of the targeted transgene in r4 was not recovered even after it was positioned at two more anterior locations in the HoxD complex. Internal deletions that concurrently moved the Hoxb1 reporter gene either upstream of Hoxd10 or adjacent to Hoxd3, did not lead to the recovery of a signal in r4. When placed at these anterior positions, however, the transgene behaved as if it would respond to Hoxd10 or Hoxd3 regulatory controls, respectively. For example, transcripts in the CNS appeared in the hindbrain up to the r5/r6 boundary and the transgene was expressed at the time and place where its novel 3′ neighbor gene (Hoxd3) was activated. However, none of these configurations allowed for Hoxb1/lacZ to be expressed in r4. In the case of the Hoxd3 locus, even though the r4 element was still inactive, silencing did not prevent neural expression of the transgene in response to endogenous Hoxd3 regulatory controls. This early CNS expression of the transgene when relocated to the Hoxd3 locus should nevertheless be considered in the context of the accompanying large deletion (from Hoxd13 to Hoxd4 included). It is likely that such a deficiency may affect the proper implementation of the silencing mechanism through the removal of important elements (Kondo et al. 1998).
A possible explanation for this difference in CNS expression of the Hoxb1/lacZ transgene is that a local chromatin configuration may be destabilized by protein complexes triggering transcriptional activation, such as in the Hoxd3 locus, whereas subsequent controls based on autoregulatory loops would not have this capacity. This may explain why the transgene was expressed in early mesoderm as well as in posterior hindbrain. However, expression in r4 essentially depends on a maintenance mechanism on the basis of auto-regulatory loops, and this mechanism may not have the capacity to reconfigure local chromatin. In the former cases, activating complexes may locally access the locus, even in the presence of a repressive configuration, whereas the maintenance complexes may not have the required properties to overcome silencing, even when present in increased amounts such as after treatment with retinoic acid. Expression of the Hoxb1 reporter gene in r6, when placed near the Hoxd3, demonstrates that it had conserved its capacity to be activated in hindbrain neural cells under certain conditions, suggesting that colinearity could be broken not only in mesoderm cells but also during CNS development.
Relocation of Hoxb1: expression in mesoderm
A surprising finding in this study, which contrasts with the suppression of r4-specific transcription, is that expression of the relocated Hoxb1 transgene in presomitic and somitic mesoderm was not dramatically affected by its new genomic location, as if the surrounding HoxD environment had little impact on its regulation in this tissue. Furthermore, the transposed Hoxb1, whose expression was anteriorized, caused changes in Hoxd13 as it was ectopically expressed, in a manner following a Hoxb1-like pattern and well before 5′ Hoxd genes are normally activated. This was not seen if the selection cassette was left in place, indicating that this cassette can behave as a potent enhancer block. Interestingly, premature and anterior activation was restricted to Hoxd13 and did not affect other Hoxd genes. Therefore, Hoxb1 induced a local break in both spatial and temporal colinearities rather than a global change in expression.
Hoxd9/lacZ recombined at the same position, after the PGK-neo cassette had been removed, also induced the neighboring Hoxd13 gene to be expressed too early and anteriorly, in a way somewhat reminiscent of endogenous Hoxd9 expression. However, even though Hoxd13 regulation was clearly perturbed, the Hoxd9 transgene itself, unlike Hoxb1, was not expressed until all posterior Hoxd genes had been activated (van der Hoeven et al. 1996). This suggests that the integration of this foreign locus near Hoxd13 lead to the deregulation of this latter gene before the transgene itself was activated. In both cases, premature activation of Hoxd13 as well as the observed early expression of Hoxb1/lacZ are at odds with a model whereby a repression over the posterior HoxD complex would prevent any 5′-located genes to be activated early on (van der Hoeven et al. 1996; Kondo et al. 1998; Kondo and Duboule 1999). Therefore, whereas the block in r4 expression is consistent with this view, an extreme interpretation of this model whereby repression occurs in all tissues and genes through the existence of a tight global silencing mechanism seems unlikely, and a more dynamic mechanism, capable of some discrimination between tissues and, perhaps, activating and maintenance complexes must be operating.
One possibility is that factors necessary for Hoxb1 activation can recognize the ectopic locus and access the Hoxb1 promoter by inducing a local opening within an otherwise refractory structure. Because of the proximity of the Hoxd13 promoter, this latter gene would also be activated, either through the Hoxb1 machinery itself or as a response to posterior factors that would normally not have access to the locus but which, in this particular context, would take advantage of a local disorganization (Fig. 9). In this model (Model I; Kondo et al. 1998), cells currently engaged in Hox gene activation would make posterior genes progressively accessible to transcription along with time (Fig. 9, A–A2). Cells with a given state of Hox gene activation, that is, cells that have reached a particular AP level as a result of gastrulation, would leave this mode of activation and maintained their current state by the same silencing system, preventing posterior genes to be activated (A–A0). In the case in which a transgene is introduced (Fig. 9, Model I, bottom), factors normally required for the transcription of the endogenous copies could locally unwind the repressive structure and allow for transcription to occur for both the transgene and the nearby located Hox gene (A′–A2′, red arrows).
Figure 9.
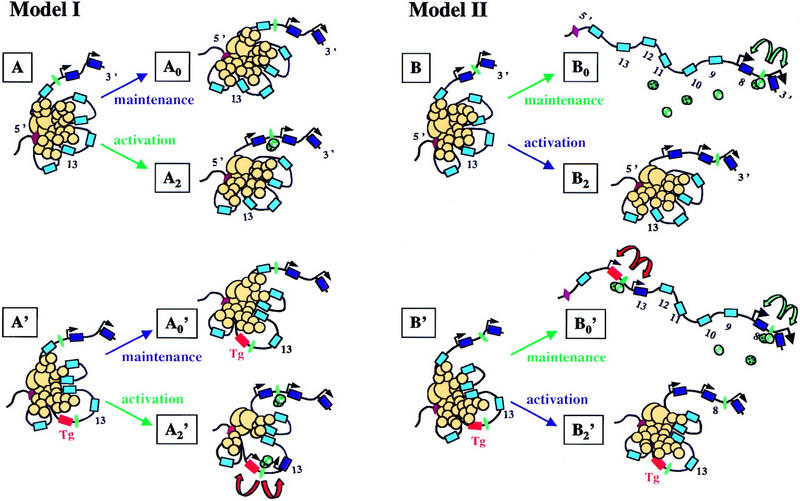
Models for Hoxd gene colinear activation and maintenance. (Model I) Hox genes (blue) are activated (dark blue), during gastrulation, one after the other, as a result of the linear release of a silencing mechanism (A–A2). This mechanism is illustrated here by the aggregation of protein complexes (yellow circles) triggered by a major nucleation sequence located upstream the complex (brown, Kondo and Duboule 1999). Genes will become progressively accessible for transcription (black arrows), in conjunction with the presence of the appropriate factors (green). Thus, whereas only two genes are active in A, three genes will be transcribed at A2 (black arrows). In the meantime, cells can escape activation and maintain their current status of Hox gene activation (A–A0). In the case in which an anterior transgene is integrated posteriorly (Tg, red box), those factors normally acting on anterior genes (green) can access the ectopic locus through recognition of early enhancer elements (green). This may destabilize locally the silencing mechanism and allow for both the transgene and the immediate neighboring gene to be expressed (A′–A2′, red arrows). In this view, maintenance of the expressed state, in a given cell, will depend on the high-order structure of the complex, i.e., the extent of silencing will be memorized once the cell will quit the population that implement the opening process. (Model II) Early colinear activation is as for Model I (B–B2). However, cells leaving this environment will remember their state of activation mainly through trans-acting regulation, without the need for the silencing mechanism to be pertained (B–B0). In this case, posterior genes will be silent anteriorly because of the absence of the necessary trans-acting factors, for example auto- and trans-regulatory complexes. If an anterior transgene is inserted posteriorly, early activation will not occur. However, those cells derived from this early stage will activate this transgene because of the presence of factors maintaining expression of the endogenous anterior counterpart (B0′, green arrows). In this view, ectopic expression of anterior transgenes described in this work would not result from their normal activation but, instead, may illustrate a maintenance mechanism whereby all genes would be accessible, e.g., to cross- or auto-regulatory controls, unlike in Model I, in which only properly activated genes can be the target of such controls.
In this scheme, transcriptional activation is tightly linked to the linear retraction of the repressive mechanism, and silencing over the ectopic transgene would be competed by transgene-specific activating complexes. Under this model, expression mediated by the r4 enhancer did not occur, either due to the lack of access, or the weak remodeling capacity of the Hox/Pbx complex. Another potential explanation is that Hox gene activation in mesoderm strictly depends on combinations or gradients of upstream regulators acting in cis, without any contribution of a higher type of regulation. In such a view, clustering would have no critical function in the time course of activation. Whereas this simple scheme explains several results obtained by conventional transgenesis, it fails to account for observations made on recombining the HoxD complex (van der Hoeven et al. 1996; Kondo and Duboule 1999). We therefore consider this possibility as unlikely.
Alternatively, silencing may occur transiently and exclusively in those cells in which Hox gene activation occurs, that is, in a restricted cellular population during gastrulation. In this view, although cells activate their Hox genes through a linear release of silencing (Fig. 9, Model I, B–B2), subsequent maintenance would rely on the interplay between gene-specific enhancer sequences (Fig. 9, Model II). Accordingly, particular states of activation would be maintained without the requirement for a high-order type of regulation (B–B0), for example, by relying on cross- and auto-regulation as well as the action of local enhancers (Model II, green arrows). In this context, it is possible that the Hoxb1 transgene, even though expressed early on, was properly silenced during the activation phase (Fig. 9, B′). However, it might subsequently be able to respond to a maintenance phase before posterior Hoxd's were activated, through the early enhancers that were transferred together with the relocated locus (B′–B0′, red arrows). The difference between Models I and II (Fig. 9) is that the transgene would respond either to genuine activating signals (Model I) or to the maintenance machinery (Model II). Nevertheless, both models imply that Hox transgenes carry with them regulatory information regarding their expression in a defined domain and at a defined developmental stage. As it is difficult to discriminate between regulatory elements responding to either cross- or auto-regulatory loops (directly or indirectly) and elements responding to the original activating signal, the activation versus maintenance question is difficult to address. A genetic approach should help in investigating this problem, but the redundancy of the system requires that multiple inactivation strategies be implemented.
Phenotypes
On relocation near Hoxd13, Hoxb1 ectopic expression was induced in mesoderm. However, no related morphological alteration was scored. This may be due to the rapid disappearance of Hoxd13 transcripts, which were detected from day 7 to late day 9. The rapid turnover in ectopic Hox gene transcription was already noticed in another context (Kondo and Duboule 1999) and may highlight the necessity for cross-regulatory interactions among posterior Hox genes to occur to maintain a previously established pattern. Consequently, ectopic Hoxd13 was maintained only in those domains in which posterior Hoxd genes were normally active. The requirement for a prolonged and precisely localized ectopic expression to induce developmental defects was exemplified by the relocation of Hoxd9/lacZ. In this case, all newborns died from complete kidney agenesis. This severe defect coincided with a sustained ectopic expression of Hoxd13 in a broad region, including the metanephric blastema, a structure in which group 11 genes are required and from which group 13 genes are normally excluded. Interestingly, kidney agenesis, together with the loss of the intermediate pieces of both fore- and hindlimbs, was also obtained in mice lacking the functions of both Hoxd11 and Hoxa11 (Davis et al. 1995). It is therefore probable that the presence of the HOXD13 protein in developing kidneys functionally inactivated group 11 function, much in the same way as in the limbs (van der Hoeven et al. 1996; Hérault et al. 1997; Peichel et al. 1997). This negative effect of group 13 proteins over group 11 function may not necessarily be triggered by a transcriptional down-regulation and may involve other mechanisms (posterior prevalence; Duboule and Morata 1994). The fact that kidney agenesis was directly due to ectopic Hoxd13 expression is demonstrated by the production of healthy mice carrying the Hoxd9/lacZ transgene at the same position, but together with a deletion of Hoxd13 (Kondo and Duboule 1999).
Hoxb1 in digits and genitalia
Posterior Hoxd genes share similar expression domains in the distal part of the limb and genital buds (Dollé et al. 1991; Nelson et al. 1996; Sordino et al. 1996; Kondo et al. 1997). Analysis of a variety of engineered alleles suggested that these two sites of expression were controlled by one and the same enhancer sequence located upstream of the HoxD complex (see references in Hérault et al. 1999). Here, we show that Hoxb1, an early and anterior Hox gene that is neither normally expressed in developing limbs, nor in genitalia, readily adopted the conventional posterior Hoxd pattern when relocated in this part of the complex. This observation emphasizes the versatility of Hox gene promoters as well as the importance of the topological context for the expression specificity of a given gene within these clusters. In the case of relocated Hoxb1, limb expression was not likely controlled by nearby located sequences, as the randomly integrated copy was unable to express this specificity. This observation reinforces the idea that a remote enhancer or regulatory sequence is able to globally regulate Hox genes in limbs and genitalia and illustrates that cluster-specific traits may be derived from the evolution of enhancers acting over several genes at once. This suggests that the existence of gene complexes may have facilitated the design of shared enhancer elements, thereby leading to an increase in the level of regulatory complexity, further constraining this particular genomic organization.
Materials and methods
Constructs, ES cells, and mice
For both TgHb1 and TgNb1, a loxP-PGKneo-loxP selection cassette was cloned immediately 3′ to the Hoxb1/lacZ transgene (Marshall et al. 1994). The resulting Hoxb1lacZ-loxP-PGK-neo-loxP construct was further introduced into the NsiI site located in the middle of the 9.5-kb DNA fragment spanning the entire intergenic region between Evx2 and Hoxd13. This 23-kb large construct was electroporated into D3 ES cells (Doetschmann et al. 1985) that were selected and amplified according to Joyner (1993). Both homologous recombination events and random chromosomal integrations were identified by Southern blot analysis with external and internal probes. Two distinct ES cell clones were injected into C57BL/6 mouse blastocysts, either for the random integration (TgNb1), or for the homologous recombination (TgHb1). Chimeric animals were either sacrificed at day 11.5 d.p.c. to assay for transgene expression, or crossed with wild-type females to generate heterozygous animals. Deletion of the PGK-neo selection cassette was carried out in vivo by crossing mice heterozygous for either the relocated or the randomly integrated allele, with mice expressing the Cre recombinase under the control of the cytomegalovirus (CMV) promoter (Dupé et al. 1997). Homozygous lines were derived for both alleles. Mice were genotyped by Southern blotting with tail DNA. The derivation of mouse stocks carrying the Hoxd11/lacZ and Hoxd9/lacZ transgenes, either randomly integrated or recombined at the same NsiI site, was reported previously (van der Hoeven et al. 1996). To delete the PGK-neo cassette from Hoxd9/lacZ mice, heterozygous females were crossed with males carrying the same Cre-expressing transgene (Dupé et al. 1997).
X-gal staining, in situ hybridization and skeletal preparations
Expression of the reporter transgene in embryos older than 9 d.p.c. was monitored by detection of β-gal activity (Zákány et al. 1988). For younger embryos, expression of the lacZ fusion gene was detected by whole-mount in situ hybridization with a lacZ RNA probe. Expression of 5′ Hoxd genes was performed by whole-mount in situ hybridization (Gérard et al. 1996) with conventional probes (Hoxd12, Izpisua-Belmonte et al. 1991; Hoxd13 and Evx2, Dollé et al. 1994; Hoxd11, Gérard et al. 1996). For skeletal preparations, adult animals were sacrificed, dissected, eviscerated, and stained according to standard alizarin red staining protocol (Inouye 1976).
TAMERE
Subsequent relocations of the Hoxb1/lacZ transgene at various anterior locations within the HoxD complex were generated through the TAMERE approach (Hérault et al. 1998). We generated mice that carried both the TgHb1 allele and an allele containing a loxP site between Hoxd11 and Hoxd10 (Ch. 2 in Fig. 7a; Gérard et al. 1996). In addition, these males were hemizygous for a Sycp1 promoter-driven Cre transgene (Vidal et al. 1998). Such transloxer males were crossed with wild-type females to recover in the progeny individuals carrying the transloxed allele containing the Hoxb1 transgene upstream Hoxd10, along with a deletion of the Hoxd13 to Hoxd11 region (TgHb1Del3). Likewise, the relocation upstream of Hoxd3 was obtained by combining an allele containing a loxP site upstream Hoxd3 (Ch. 3 in Fig. 7a; Zákány and Duboule 1999) and the TgHb1 allele. The resulting novel Hoxb1/lacZ transgene relocation was associated with a deletion encompassing genomic DNA from Hoxd13 to Hoxd4 (TgHb1Del7). The frequencies of such interallelic recombination were close to those reported previously, that is, ∼10% (Hérault et al. 1998; Kondo and Duboule 1999). Mice heterozygous for the two novel relocations were genotyped by Southern blotting.
Acknowledgments
We thank M. Friedli for technical assistance, H. Popperl for the Hoxb1 construct and P. Chambon and F. Cuzin for mice. We also thank members of the laboratory for their comments on the manuscript and for sharing reagents. This work was supported by funds from the Canton de Genève, the Swiss National Research Fund, the Claraz, Latsis, Cloetta and Louis-Jeantet foundations.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Denis.Duboule@zoo.unige.ch; FAX 41 22 702 6795.
References
- Becker D, Jiang Z, Knodler P, Deinard AS, Eid R, Kidd KK, Shashikant CS, Ruddle FH, Schughart K. Conserved regulatory element involved in the early onset of Hoxb6 gene expression. Dev Dyn. 1996;205:73–81. doi: 10.1002/(SICI)1097-0177(199601)205:1<73::AID-AJA7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Behringer RR, Crotty DA, Tennyson VM, Brinster RL, Palmiter RD, Wolgemuth DJ. Sequences 5′ of the homeobox of the Hox-1.4 gene direct tissue-specific expression of lacZ during mouse development. Development. 1993;117:823–833. doi: 10.1242/dev.117.3.823. [DOI] [PubMed] [Google Scholar]
- Belting HG, Shashikant CS, Ruddle FH. Multiple phases of expression and regulation of mouse Hoxc8 during early embryogenesis. J Exp Zool. 1998;282:196–222. [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep 1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon RA, Rossant J. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development. 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Deschamps J, Wijgerde M. Two phases in the establishment of HOX expression domains. Dev Biol. 1993;156:473–480. doi: 10.1006/dbio.1993.1093. [DOI] [PubMed] [Google Scholar]
- Di Rocco G, Mavilio F, Zappavigna V. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J. 1997;16:3644–3654. doi: 10.1093/emboj/16.12.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: Formation of viceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- Dollé P, Izpisua-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- Dollé P, Izpisua-Belmonte JC, Tickle JM, Duboule D. Hox-4 genes and the morphogenesis of mammalian genitalia. Genes Dev. 1991;5:1767–1776. doi: 10.1101/gad.5.10.1767. [DOI] [PubMed] [Google Scholar]
- Dollé P, Fraulob V, Duboule D. Developmental expression of the mouse Evx-2 gene: Relationship with the evolution of the HOM/Hox complex. Dev Suppl. 1994;45:143–153. [PubMed] [Google Scholar]
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Dupé V, Davenne M, Brocard J, Dollé P, Mark M, Dierich A, Chambon P, Rijli FM. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE) Development. 1997;124:399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ. Mouse homeobox gene transcripts occupy different but overlapping domains in embryonic germ layers and organs: A comparison of Hox-3.1 and Hox-1.5. Development. 1988;103:135–144. doi: 10.1242/dev.103.1.135. [DOI] [PubMed] [Google Scholar]
- Gérard M, Duboule D, Zákány J. Structure and activity of regulatory elements involved in the activation of the Hoxd-11 gene during late gastrulation. EMBO J. 1993;12:3539–3550. doi: 10.1002/j.1460-2075.1993.tb06028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard M, Chen JY, Gronemeyer H, Chambon P, Duboule D, Zákány J. In vivo targeted mutagenesis of a regulatory element required for positioning the Hoxd-11 and Hoxd-10 expression boundaries. Genes & Dev. 1996;10:2326–2334. doi: 10.1101/gad.10.18.2326. [DOI] [PubMed] [Google Scholar]
- Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:19–51. doi: 10.1016/s0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Hérault Y, Fraudeau N, Zákány J, Duboule D. Ulnaless (Ul), a regulatory mutation inducing both loss-of-function and gain-of-function of posterior Hoxd genes. Development. 1997;124:3493–3500. doi: 10.1242/dev.124.18.3493. [DOI] [PubMed] [Google Scholar]
- Hérault Y, Rassoulzadegan M, Cuzin F, Duboule D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE) Nat Genet. 1998;20:381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- Hérault Y, Beckers J, Gérard M, Duboule D. Hox gene expression in limbs: Colinearity by opposite regulatory controls. Dev Biol. 1999;208:157–165. doi: 10.1006/dbio.1998.9179. [DOI] [PubMed] [Google Scholar]
- Hunt, P., M. Gulisano, M. Cook, M.H. Sham, A. Faiella, D. Wilkinson, E. Boncinelli and R. Krumlauf. 1991. A distinct Hox code for the branchial region of the vertebrate head. Nature : 861–864. [DOI] [PubMed]
- Inouye M. Differential staining of cartilage and bone in fetal mouse skeleton by alcian blue and alizarin red. S Cong Anom. 1976;16:171–173. [Google Scholar]
- Izpisua-Belmonte JC, Falkenstein H, Dollé P, Renucci A, Duboule D. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991;10:2279–2289. doi: 10.1002/j.1460-2075.1991.tb07764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL. Gene targeting. A practical approach. Oxford, UK: Oxford University Press; 1993. [Google Scholar]
- Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- Kondo T, Duboule D. Breaking colinearity in the mouse HoxD complex. Cell. 1999;97:407–417. doi: 10.1016/s0092-8674(00)80749-7. [DOI] [PubMed] [Google Scholar]
- Kondo T, Zákány J, Innis JW, Duboule D. Of fingers, toes and penises. Nature. 1997;390:29. doi: 10.1038/36234. [DOI] [PubMed] [Google Scholar]
- Kondo T, Zákány J, Duboule D. Control of colinearity in AbdB genes of the mouse HoxD complex. Mol Cell. 1998;1:289–300. doi: 10.1016/s1097-2765(00)80029-5. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Marshall H, Nonchev S, Sham MH, Muchamore I, Lumsden A, Krumlauf R. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature. 1992;360:737–741. doi: 10.1038/360737a0. [DOI] [PubMed] [Google Scholar]
- Marshall H, Studer M, Popperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- Murphy P, Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991;111:61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, Gonzales E, Tessarollo L, Parada LF, Tabin C. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Prabhakaran B, Vogt TF. The mouse Ulnaless mutation deregulates posterior HoxD gene expression and alters appendicular patterning. Development. 1997;124:3481–3492. doi: 10.1242/dev.124.18.3481. [DOI] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Puschel AW, Balling R, Gruss P. Position-specific activity of the Hox1.1 promoter in transgenic mice. Development. 1990;108:435–442. doi: 10.1242/dev.108.3.435. [DOI] [PubMed] [Google Scholar]
- Renucci A, Zappavigna V, Zákány J, Izpisua-Belmonte JC, Burki K, Duboule D. Comparison of mouse and human HOX-4 complexes defines conserved sequences involved in the regulation of Hox-4.4. EMBO J. 1992;11:1459–1468. doi: 10.1002/j.1460-2075.1992.tb05190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordino P, Duboule D, Kondo T. Zebrafish Hoxa and Evx-2 genes: Cloning, developmental expression and implications for the functional evolution of posterior Hox genes. Mech Dev. 1996;59:165–175. doi: 10.1016/0925-4773(96)00587-4. [DOI] [PubMed] [Google Scholar]
- Stern CD, Foley AC. Molecular dissection of Hox gene induction and maintenance in the hindbrain. Cell. 1998;94:143–145. doi: 10.1016/s0092-8674(00)81412-9. [DOI] [PubMed] [Google Scholar]
- Studer M, Popperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728–1732. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli F, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- Tan DP, Shao X, Pu L, Guo V, Nirenberg M. Sequence and expression of the murine Hoxd-3 homeobox gene. Proc Natl Acad Sci. 1996;93:8247–8252. doi: 10.1073/pnas.93.16.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven F, Zákány J, Duboule D. Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell. 1996;85:1025–1035. doi: 10.1016/s0092-8674(00)81303-3. [DOI] [PubMed] [Google Scholar]
- Vidal F, Sage J, Cuzin F, Rassoulzadegan M. Cre expression in primary spermatocytes: A tool for genetic engineering of the germ line. Mol Reprod Dev. 1998;51:274–280. doi: 10.1002/(SICI)1098-2795(199811)51:3<274::AID-MRD6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Whiting J, Marshall H, Cook M, Krumlauf R, Rigby PW, Stott D, Allemann RK. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes & Dev. 1991;5:2048–2059. doi: 10.1101/gad.5.11.2048. [DOI] [PubMed] [Google Scholar]
- Zákány J, Duboule D. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature. 1996;384:69–71. doi: 10.1038/384069a0. [DOI] [PubMed] [Google Scholar]
- ————— Hox genes and the making of sphincters. Nature. 1999;401:761–762. doi: 10.1038/44511. [DOI] [PubMed] [Google Scholar]
- Zákány J, Tuggle CK, Patel MD, Nguyen-Huu MC. Spatial regulation of homeobox gene fusions in the embryonic central nervous system of transgenic mice. Neuron. 1988;1:679–691. doi: 10.1016/0896-6273(88)90167-5. [DOI] [PubMed] [Google Scholar]


