Abstract
Surfaces that resist protein adsorption are important for many bioanalytical applications. Bovine serum albumin (BSA) coatings and multi-arm poly(ethylene glycol) (PEG) coatings display low levels of non-specific protein adsorption and have enabled highly quantitative single-molecule (SM) protein studies. Recently, a method was developed for coating a glass with PEG–BSA nanogels, a promising hybrid of these two low-background coatings. We characterized the nanogel coating to determine its suitability for SM protein experiments. SM adsorption counting revealed that nanogel-coated surfaces exhibit lower protein adsorption than covalently coupled BSA surfaces and monolayers of multi-arm PEG, so this surface displays one of the lowest degrees of protein adsorption yet observed. Additionally, the nanogel coating was resistant to DNA adsorption, underscoring the utility of the coating across a variety of SM experiments. The nanogel coating was found to be compatible with surfactants, whereas the BSA coating was not. Finally, applying the coating to a real-world study, we found that single ligand molecules could be tethered to this surface and detected with high sensitivity and specificity by a digital immunoassay. These results suggest that PEG–BSA nanogel coatings will be highly useful for the SM analysis of proteins.
Keywords: adsorption, total internal reflection fluorescence, antibody binding, protein detection, digital immunoassay, surfactant
1. Introduction
Single-molecule (SM) fluorescence microscopy studies hold great promise for elucidating biological systems [1], but the non-specific surface adsorption of fluorescently labelled proteins [2,3], antibodies [4] and bioconjugated nanoparticles [5] is often a significant source of experimental noise. Recently, low-background surface coatings have been developed that reduce protein adsorption to SM levels—levels at which a digital signal from individual target molecules can be reliably quantified above the background of non-specifically adsorbed molecules. For example, Tessler et al. [4] surveyed 12 different surface coatings and found that the best performing of these, a covalently coupled bovine serum albumin (BSA) coating, allowed accurate protein quantification of as few as 55 molecules per 1000 µm2 by SM antibody binding. The low-background surface enabled target protein molecules to be tethered to the surface and digitally detected with total internal reflection fluorescence (TIRF) microscopy and without the need for fluorescence resonance energy transfer or two-colour coincidence detection [4]. Poly(ethylene glycol) (PEG) coatings that are highly resistant to protein adsorption have also been developed [6]. Groll et al. [7] demonstrated that a monolayer of multi-arm PEG covalently coupled to a surface reduced protein adsorption to SM levels, allowing quantitative monitoring of protein folding by TIRF .
Recently, Scott et al. [8] developed a method for coating surfaces with nanoscale hydrogels (nanogels) formed by cross-linking multi-arm PEG to albumin. Surfaces treated with these nanogels displayed very low levels of protein adsorption, as measured by optical waveguide lightmode spectroscopy (OWLS) and cell adhesion. The nanogel coating holds great potential for SM applications because it is a hybrid of two good SM surface coatings and is thin enough (≅ 75 nm) to perform total internal reflection imaging.
Here, we characterized nanogel-coated surfaces for use in SM protein studies. We measured protein adsorption onto PEG–BSA nanogel coatings using a variety of proteins, fluorophores and labelling methods, as well as with DNA. We examined the resilience of the nanogel coating to sodium dodecyl sulphate (SDS), a reagent commonly used in binding studies. To assess the utility of nanogel coatings in the context of a real-world SM experiment, we adapted the surface to perform digital measurements of antibody binding. Finally, we examined the roles of capping steps, cross-linker molecules and substrate coupling methods via adsorption studies and atomic force microscopy (AFM). Our studies reveal that PEG–BSA nanogel surface coatings show substantially higher performance than previously characterized SM coatings and should be of benefit to SM protein studies.
2. Material and methods
2.1. Synthesizing solutions of nanogels
PEG–BSA nanogel coatings were prepared using partially cross-linked solutions of eight-arm PEG and BSA. First, eight-arm PEG-OH (PEG8-OH; MW 10 000; Creative PEGWorks, Winston Salem, NC, USA) was used to synthesize PEG-octovinylsulfone (PEGOVS) in a four-step reaction, as described previously [9,10]. PEGOVS and fatty acid-free BSA (Sigma-Aldrich, St Louis, MO, USA) solutions were prepared at 20 per cent (w/v) in phosphate-buffered saline at pH 7.4 (PBS) and sterile filtered with 0.22 µm syringe filters (Millipore, Billerica, MA, USA). Assuming 36 reactive amines per BSA (obtained from crystal structures), PEGOVS and BSA solutions were combined at a 1 : 1 ratio of vinylsulfone to amine groups and reacted by incubation at 37°C, with rotation. The progress of the reactions was followed by dynamic light scattering (DLS) until a mean effect diameter (dPCS) of about 100 nm was achieved—generally approximately 6 h. (At this step, the DLS polydispersity index for the nanogel solution was 6.2, and the complexes in the nanogel solutions had a standard deviation of approx. 20 nm.) The nanogel-containing solution was then diluted 1 : 1 with PBS to 10 per cent (w/v) and stored until use at −80°C. For long-term storage, nanogel solutions were stored at −140°C. To generate PEG–PEGOA nanogels, the same protocol as above was used with the exception that BSA was substituted by PEG-octoamine (PEGOA, MW 10 000). (For details, see the electronic supplementary material.)
2.2. Coating substrates with nanogels
Nanogel solutions were either thiol-reacted or epoxy-reacted to the glass substrates. The glass substrates (40 mm diameter circles, no. 1.5) were obtained from Erie Scientific (Waltham, MA, USA). For the epoxide-reacted coatings, the substrates were epoxysilanated by the vendor. For the thiol-reacted coatings, the coverslips were mercaptosilanated as follows. The coverslips were washed 3× in diH2O and 3× in ethanol. Then they were etched with oxygen plasma at 100 W for 10 min (Femto, Diener Electronic, Royal Oak, MI, USA). Coverslips were then washed 3× in acetone. Mercaptosilanation was achieved by treating the glass for 1 h at 25°C with a 5 per cent (v/v) solution of mercaptopropyltrimethoxysilane (MPTS, Sigma-Aldrich) in acetone. Surfaces were washed 3× in acetone and cured at 100°C for 25 min.
Substrates were coated with PEG–BSA nanogel-containing solutions in either a flow cell (FSC2, Bioptechs, Butler, PA, USA) or in a 60 mm diameter Petri dish (for cell seeding experiments). Substrates were incubated in 10 per cent nanogel-containing solutions for 1 h at 37°C. Substrates were washed with 600 µl PBS and capped for 1 h with BSA 50 mg ml−1 in PBS, 37°C. Unreacted vinylsulfone or epoxide groups were then quenched with 1 M Tris at pH 8.0 for 15 min at room temperature. (A time course showed that quenching went to completion.) Coverslips were then washed with 600 µl PBS.
2.3. Covalently coupled bovine serum albumin-coated surfaces
The epoxide-reacted BSA-(coated) surfaces were generated by reacting 50 mg ml−1 BSA, in PBS, to epoxysilanated glass coverslips within a flow cell for 1 h at 37°C. Unbound BSA was washed away with PBS, and unreacted epoxides were quenched with 1 M Tris at pH 8.0 for 15 min at room temperature. Coverslips were then washed with 600 µl PBS.
2.4. Multi-arm poly ethylene glycol monolayer-coated surfaces
The multi-arm PEG monolayers were generated by reacting 200 mg ml−1 PEGOVS, in PBS, to mercaptosilanated glass coverslips within a flow cell for 1 h at 37°C. Coverslips were washed with 600 µl PBS and incubated 1 h with 50 mg ml−1 BSA in PBS at 37°C. Unreacted vinylsulfone groups were quenched with 1 M Tris at pH 8.0 for 15 min at room temperature. Coverslips were then washed with 600 µl PBS.
2.5. Single-molecule adsorption measurements
Three different proteins and one DNA were used in the SM adsorption experiments. These include polyclonal goat IgG labelled with multiple Cy5 fluorophores (Abcam, Cambridge, MA, USA), streptavidin labelled with multiple AlexaFluor 647 fluorophores (Invitrogen, Carlsbad, VA, USA), Escherichia coli methionine aminopeptidase fused to mCherry fluorescent protein, and DNA thrombin binding aptamer labelled with a single Cy3 fluorophore (Integrated DNA Technologies, Coralville, IA, USA). Each of the surfaces under investigation was prepared within a flow cell (FSC2, Bioptechs). An uncoated control surface was generated by quenching an epoxysilanated glass coverslip with 1 M ethanolamine-HCl at pH 8.0 for 30 min. Flow cells were fitted with perfusion ports to allow for reagents to be passed over the surface by a custom vacuum pump. The flow cells were washed with 600 µl PBS and loaded with 200 µl of 1 nM fluorescent protein or DNA. The fluorescent molecules were incubated for 25 min in the dark at room temperature, and unbound protein or DNA was washed off with 600 µl PBS. Images were acquired and processed as described above. Standard deviations were obtained from triplicate (for antibody) or duplicate (for all other molecules) surfaces.
2.6. Measuring detergent resistance
Each of the surfaces under investigation was prepared within a flow cell. Surfaces were exposed to 100 ng ml−1 Cy5-labelled antibody for 25 min in the dark at room temperature to assess initial levels of non-specific protein adsorption. Unbound antibody was washed out of the flow cells with 600 µl PBS, and the flow cells were imaged. The flow cells were then exposed to 0.1 per cent SDS in PBS for 5 min at room temperature, washed with 600 µl PBS and imaged. The flow cells were exposed for the second time to antibody for 25 min, to measure adsorption after SDS treatment. Surfaces were washed with 600 µl PBS, and imaged. Finally, the flow cells were washed in 600 µl 0.1 per cent SDS in PBS for the second time, washed in 600 µl PBS and imaged. Images were processed as described above. Standard deviations were obtained by replicates on two separate surfaces.
2.7. Digital immunoassays
Nanogel-coated surfaces were generated in a flow cell as described above. The antibody binding experiment was performed as previously described [4]. First, the surface was activated by 0.2 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and 0.05 M N-hydroxysuccinimide (NHS) (Pierce, Rockford, IL, USA) in sodium phosphate buffer (SPB) at pH 5.8 for 10 min. The flow cell was washed with 600 µl of SPB, and Cy3-labelled target protein (IgG obtained from goat, Abcam, Cambridge, MA, USA) was tethered to the activated surface for 10 min at 100 ng ml−1 in PBS in the dark. Unreacted cross-linking groups were quenched with 1 M Tris at pH 8.0 for 5 min. Then the surface was probed with Cy5-labelled antibody (anti-Goat IgG, Abcam) for 2 h at 100 ng ml−1 in PBS in the dark. The flow cell was washed with 600 µl of PBS and imaged at 540 and 635 nm. Images of Cy3 and Cy5 channels were merged to determine the fraction of targets that were bound by antibody and the specificity of the antibody for the targets compared with random binding. (See the electronic supplementary material for details.)
3. Results
3.1. Nanogel coatings display lower protein adsorption than bovine serum albumin or polyethylene glycol
We first sought to quantify antibody adsorption onto PEG–BSA nanogel-coated surfaces. We generated covalently coated BSA surfaces, multi-arm PEG monolayer-coated surfaces and nanogel-coated surfaces within flow cells (figure 1a), exposed them to fluorescently labelled antibody, and quantified the adsorbed molecules by total internal reflection fluorescence (TIRF) imaging. For adequate sampling, images were acquired at five positions per surface, with independent surfaces analysed in triplicate. So, thousands of molecules were sampled to obtain each reported data point. (For details see the electronic supplementary material.)
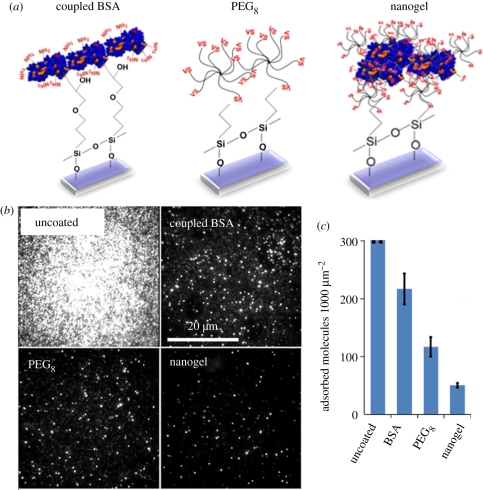
Figure 1.
(a) Schematic of three coating methodologies: bovine serum albumin (BSA) covalently coupled to epoxysilanated glass, multi-arm PEG (PEG8) coupled to mercaptosilanated glass and PEG–BSA nanogels coupled to mercaptosilanated glass. (b) Antibody adsorption onto uncoated, BSA-coated, multi-arm PEG-coated and PEG–BSA nanogel-coated surfaces was quantified by total internal reflection fluorescence (TIRF) imaging, and representative raw TIRF images are shown. (c) Molecule counts of antibody adsorption per unit area. On the uncoated surface, molecular density was so high that single-molecule counting was not possible. BSA-coated, multi-arm PEG-coated and nanogel-coated surfaces show decreased antibody adsorption compared to the uncoated surfaces, with the nanogel-coated surface performing the best. Error bars represent standard deviation of triplicate substrates.
We found that antibody adsorption onto uncoated control surfaces was too high to allow single antibodies to be reliably resolved. By contrast, the BSA-, multi-arm PEG- and nanogel-coated surfaces were highly resistant to antibody adsorption (figure 1b). The BSA-coated, multi-arm PEG-coated and nanogel-coated surfaces adsorbed antibody at densities of 217, 117 and 50 molecules per 1000 µm2 image, respectively (figure 1c). Thus, the nanogel-coated surfaces adsorbed over fourfold less antibody than did the BSA-coated surfaces (p = 5.5 × 10−5, t-test) and twofold less antibody than did the multi-arm PEG surfaces (p = 7.6 × 10−4, t-test). Cell adhesion studies agreed with this finding and additionally showed that nanogel-coated surfaces can perform well for 7–9 days compared with 1–3 days for covalently coupled BSA surfaces (electronic supplementary material, figure S1).
By converting the observed molecular density on the surface to absolute mass density, we determined that the nanogel-coated surfaces adsorbed 1.28 pg cm−2 of antibody. Thus, the PEG–BSA nanogel coating is highly resistant to antibody adsorption, outperforming both the BSA-only coating and the PEG-only coating (p = 4.0 × 10−6, ANOVA). Notably, the adsorption we measured was approximately 1000-fold lower than the limit of detection of standard protein adsorption measurement methods (e.g. OWLS and surface plasmon resonance).
3.2. Nanogel coatings resist adsorption of a variety of biomolecules
SM protein studies often involve labelled biomolecules other than antibodies, such as enzymes and DNA molecules. Also, studies employ proteins that are labelled by a variety of methodologies including single-dye labelling, multi-dye labelling and fusion to fluorescent proteins. We wanted to investigate the utility of the nanogel surface in a variety of contexts. To this end, we interrogated nanogel coatings with Cy5-streptavidin (multi-labelled protein), mCherry-E. coli methionine aminopeptidase (singly labelled enzyme) and Cy5-thrombin binding aptamer (singly labelled DNA).
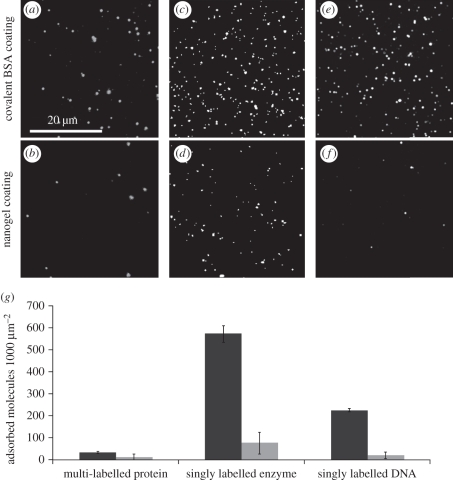
The protein, enzyme and DNA were adsorbed at levels of 33, 573 and 225 molecules per 1000 µm2 onto the BSA-coated surface and 11, 75 and 19 molecules per 1000 µm2 onto the nanogel-coated surface, respectively (figure 2a–f). For these three biomolecules, adsorption onto nanogel coatings was significantly improved over a covalently coupled BSA coating (respectively, p = 0.025, p = 0.0002, p = 0.0001, t-test); (figure 2g). Remarkably, the improvement by the nanogel surface ranged from threefold to 12-fold, depending on the biomolecule. So, the relative improvement by the nanogel coating is dependent on the size and/or the biochemistry of the fluorescent biomolecule. The fact that the nanogel coating showed improved performance across a variety of biomolecules underscores the potential utility of the nanogel coating in a broad variety of SM studies.
Figure 2.
TIRF adsorption measurements for three types of biomolecules show improved performance by nanogel coatings compared to covalently-coupled BSA coatings. The nanogel coating reduces adsorption of (a,b) the multi-labelled protein, Cy5-labelled streptavidin, (c,d) the singly labelled enzyme, mCherry-E. coli methionine aminopeptidase and (e,f) the singly labelled DNA, Cy5-labelled thrombin binding aptamer. (g) Molecule counts of biomolecule adsorption per unit area onto (black bars) covalently coupled BSA coatings and (grey bars) nanogel coatings. Error bars represent standard deviation of duplicate substrates.
3.3. Nanogel-coated surfaces are resilient to detergent
In solid phase in vitro binding studies, surfactants such as Triton X-100, Tween 20 and SDS are commonly used for performing wash steps or to regenerate ligands after protein binding [4,11]. Therefore, we measured the resilience of PEG–BSA nanogel coatings to surfactant exposure. This was done by quantifying protein adsorption onto coated surfaces before and after an SDS wash.
Surfaces were prepared, exposed to fluorescently labelled antibody, washed with 0.1 per cent SDS, exposed to antibody a second time, and washed with SDS a second time. Imaging was performed before and after each step to quantify the amount of attached antibody.
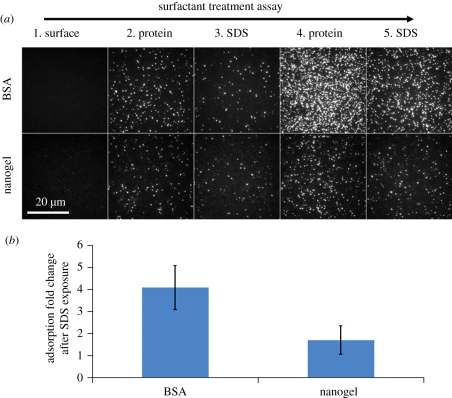
We found that the BSA coating was strongly affected by the SDS wash (figure 3a, row 1) while the nanogel coating was mostly resilient (figure 3a, row 2). The BSA coating displayed a 4.1-fold increase in antibody adsorption after SDS exposure when compared with before SDS exposure, whereas the nanogel coating displayed only a 1.7-fold increase in antibody adsorption (figure 3b). Thus, thin, PEG–BSA nanogel coatings should be superior to BSA coatings for experiments that use surfactants.
Figure 3.
Nanogel-coated surfaces show resilience to a harsh surfactant environment compared with BSA-coated surfaces. Coated surfaces were exposed to antibody, washed with sodium dodecyl sulfate (SDS), re-exposed to antibody and re-washed with SDS. Adsorption measurements were obtained in between each step. (a) Raw TIRF images of (top row) a covalently coupled BSA-coated surface and (bottom row) a nanogel-coated surface over the course of a five-step antibody adsorption experiment. (1) Surfaces prior to antibody exposure. (2) Surfaces after exposure to antibody. (3) Surfaces after SDS wash. (4) Surfaces after second exposure to antibody. (5) Surfaces after second SDS wash. (b) The bar chart depicts the ratio of adsorption change after SDS exposure to adsorption change before SDS exposure, i.e. (step 4 – step 3) : (step 2 – step 1). Error bars represent standard deviation of duplicate substrates.
3.4. Nanogel coatings are compatible with digital antibody binding experiments
Protein adsorption is an important parameter to consider when evaluating a surface for SM protein experiments, but it is not the only one. If target molecules cannot be tethered to the surface, or if the tethered molecules are not accessible for in vitro binding, then the method will lack sensitivity of detection. Conveniently, nanogel coatings and BSA coatings contain surface-exposed carboxyl groups (provided by the albumins), which allow for the near-universal tethering of protein analytes via the use of a cross-linker (see §2) [4]. Making use of this tethering method, we evaluated the compatibility of PEG–BSA nanogel-coated surfaces with digital antibody binding experiments.
We generated nanogel-coated surfaces and tethered target protein molecules labelled with Cy3 to the surfaces. Then we bound the targets with Cy5-labelled antibody, washed away unbound antibody, and used SM TIRF to detect antibody-target binding (figure 4a). We merged and processed the Cy3 and Cy5 channels (figure 4b,c), and correlation analysis showed significant levels of specific antibody binding (figure 4d and see the electronic supplementary material for details).
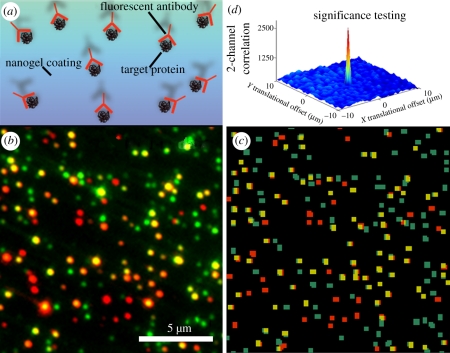
Figure 4.
Nanogel coatings can be utilized for sensitive and specific, digital antibody binding. (a) Cy3-target proteins were immobilized to a nanogel-coated surface and then bound by Cy5-antibody. (b) Merged raw image of red and green TIRF channels (15 µm × 15 µm). (c) Merged red and green TIRF channels after image processing. TIRF imaging was able to detect a high number of binding events (yellow) in which target proteins (green) were accessible to binding by antibodies (red). (d) The correlation between detection antibodies and individual analyte proteins tethered to nanogel-coated surfaces indicates high target protein accessibility and compatibility with TIRF detection.
By analysing single molecules in the two channels, we observed that the antibodies bound to their targets with high specificity: 68 ± 1% of antibodies were bound specifically to tethered target proteins. We also found that the antibodies were able to bind to target molecules sensitively: a substantial fraction of targets was bound by antibody (50.3 ± 5.9%) and this fractional binding is high compared with values we obtained previously using the covalently coupled BSA surface (31 ± 6%) [4]. Notably, this is not the maximum fractional binding since more will bind with higher antibody concentrations. Also, this level of fractional binding is a lower bound estimate, since some target molecules may be invalid (e.g. denatured proteins).
These results indicate that the nanogel coating allows targets to be immobilized and bound with high antibody occupancy and low background. Thus, nanogel coatings are highly suited for performing digital antibody binding experiments and should be generally applicable to SM in vitro binding studies.
3.5. Investigation into the role of albumin in nanogel capping and cross-linking
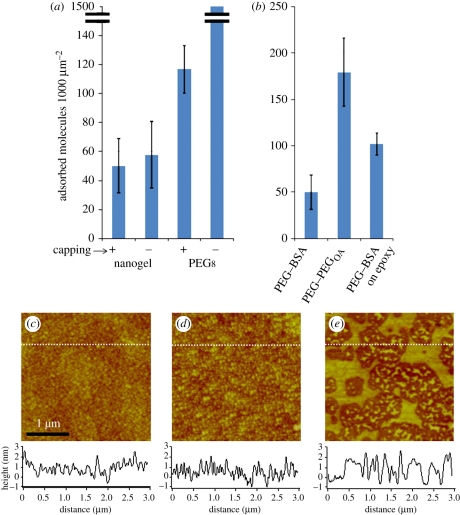
Since the final steps of both the nanogel and the multi-arm PEG coatings involve capping with BSA, we wanted to investigate to what extent the performance of the coatings depends on capping. We performed SM adsorption measurements on nanogel and multi-arm PEG coatings with and without capping steps. We found the nanogel coating behaved the same with and without the capping step, whereas the protein resistance of the multi-arm PEG coating was reliant on capping (figure 5a). This is presumably because the uncapped nanogel, which is formed from a partially polymerized solution containing a wide size-range of PEG–BSA complexes, creates a surface that is more densely packed than that of the uncapped multi-arm PEG.
Figure 5.
(a) Adsorption onto nanogel-coated and multi-arm PEG coated (PEG8) surfaces was measured with and without the use of a final BSA capping step. The nanogel-coated surfaces provided low antibody adsorption even without the capping step. (b) We measured adsorption onto the standard nanogel-coated surface (PEG–BSA). We also measured adsorption onto a surface coated with a nanogel solution formed with an alternative cross-linker to BSA (PEG–PEGOA). Finally, we measured adsorption onto a surface that was coated by coupling PEG–BSA nanogels to epoxysilanated glass (as opposed to the standard, mercaptosilanated glass). We conclude that nanogel cross-linkers and coupling chemistries can have a significant effect on adsorption performance. (c–e) We assessed the topology of (c) the standard, thiol-reacted nanogel surface, (d) the epoxy-coupled nanogel surface and (e) the multi-arm PEG-coated surface. Cross-sectional height analysis is depicted in charts below. The thiol-reacted nanogel coating created the most connected surfaces while multi-arm PEG monolayer coatings generated the least connected surfaces.
Next, since the BSA molecules in the nanogel act as a multi-functional amine cross-linker, we wanted to investigate whether a multi-arm amine-terminated PEG could be substituted for and provide the same performance as BSA. We synthesized PEG–PEGOA nanogels by substituting BSA with PEG-octoamine as has been described in previous work [10]. We applied this coating and the PEG–BSA nanogel coating to glass and found that adsorption using the alternative cross-linker detracted from the performance of the coating (figure 5b). This result points to the possibility that the PEG–BSA nanogel performance is contingent on a high molecular weight cross-linking molecule—in this case, BSA.
3.6. Investigation into alternative coupling chemistries
We next explored the use of an alternative method for coupling nanogel solutions to glass substrates with the hope of further reducing protein adsorption. In our previously described experiments, we prepared surfaces by reacting the nanogel-containing solutions with mercaptosilanated glass [8]. Coupling was achieved because the vinylsulfone groups in the PEG portion of the nanogels react with the mercaptosilanated surfaces. An alternative coupling strategy is to react the amine groups in the BSA portion of the nanogel with an epoxysilanted surface.
We prepared surfaces using this alternative coupling chemistry and measured protein adsorption. The epoxide-reacted nanogel-coating adsorbed antibodies at a density of 103 molecules per 1000 µm2 image (figure 5b), so this surface adsorbed approximately two times more protein than did the thiol-reacted nanogel surfaces (p = 5.6 × 10−4, t-test). We concluded that thiol-coupled nanogels are more resistant to protein adsorption than epoxide-coupled nanogels.
We were surprised to observe this difference in performance simply owing to coupling chemistry. To try to understand this, we used AFM to characterize their morphologies (see the electronic supplementary material). The standard, thiol-reacted nanogel surface displayed a homogeneous background (roughness value Rq = 0.480 nm), suggesting that the nanogels form a continuous matrix on the surface (figure 5c). The nanogels on the epoxide-reacted surface appeared to be generally smaller and less homogeneous (roughness value Rq = 0.673; figure 5d). By contrast, the multi-arm PEG-coated surface formed a less-connected and non-homogeneous surface (roughness value Rq = 0.823; figure 5e). (Force curve analysis performed on the thiol-reacted nanogel coating, the epoxy-reacted nanogel coating and the multi-arm PEG coating showed detachment forces of 19.087, 41.584 and 34.225 nN, respectively.) We believe the high connectivity of the thiol-reacted nanogels, reflected by roughness measurements, contributes to the lower protein adsorption when observed.
4. Discussion
Under the SM detection regime, nanogel coatings provided significantly lower protein adsorption than BSA-coated or multi-arm PEG monolayer-coated substrates. Moreover, there was a consistent gain in performance across three protein types and DNA, labelled by dyes and fluorescent proteins. The nanogel surface was resilient to surfactant and was compatible with a SM antibody binding experiment. We did not seek to optimize the fractional binding in these immunoassays (which depends on antibody concentration), but for the concentration of antibody used here, the fraction of ligands bound was at least as high as that obtained previously in digital antibody binding experiments using BSA-coated surfaces [4]. Since the nanogel surface outperformed the covalently coupled BSA and the multi-arm PEG SM surfaces in adsorption and resilience [4,7], we conclude that the nanogel surface should be a useful coating for an array of SM studies.
Surfactants can be useful in in vitro binding experiments, such as immunoassays, for performing stringent washes and for un-binding antibodies from their epitopes in repeated-binding experiments. A caveat is that surfactants can be detrimental to SM surface coatings. We studied the effect of SDS surfactant on the nanogel coating and the BSA coating and found that the nanogel-coated surfaces were unaffected by treatment with surfactant while the BSA-coated surfaces were substantially affected. This is surprising because the nanogels are partially composed of albumin. It can be inferred that the BSA-only coatings undergo some level of denaturation by the surfactant conditions. If this is the case, then the PEG macromolecules in the nanogels may stabilize the albumin molecules within them and enable them to withstand the denaturing conditions of the surfactant. This highlights another improved behaviour that the combination of albumin and PEG in a nanogel can confer over BSA-only surfaces. It is promising, since it may enable lower-background and repeated-binding digital antibody binding studies.
SM methods play a prominent role in the understanding of many biological systems [12–17]. Looking forward, SM antibody detection may provide strategies for parallel detection of proteins. The recent development of massively parallel SM DNA analysis technologies by Helicos Biosciences and Pacific Biosciences was facilitated by surfaces resistant to nucleic acid adsorption [18,19]. Promisingly, the nanogel coating characterized here shows similar levels of background adsorption to that of such DNA technologies and also benefits from resilience to wash steps. Therefore, PEG–BSA nanogels provide a thin, resilient coating technique that should benefit SM protein analysis, and we hope that this coating methodology will spark an increase in SM antibody binding studies.
Acknowledgements
We thank Jim Havranek and Ben Borgo for providing the mCherry-E. coli methionine aminopeptidase fusion protein. We thank David Mayhew, Francesco Vallania, Jessica Hoisington-Lopez, Maximiliaan Schillebeeckx, Michael Brooks, Scott Higdon, Todd Druley, Xuhua Chen and Yue Yun for helpful discussions. This work was supported by an Innovative Molecular Analysis Technologies grant from the National Cancer Institute (1R21CA151197-01) and a Technology Development in Epigenetics grant from the National Institutes of Health (1R01DA025744-01).
References
- 1.Mikhailenko S. V., Oguchi Y., Ishiwata S. 2011. Insights into the mechanisms of myosin and kinesin molecular motors from the single-molecule unbinding force measurements. J. R. Soc. Interface 7(Suppl. 3), S295–S306 10.1098/rsif.2010.0107.focus (doi:10.1098/rsif.2010.0107.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heyes C. D., Kobitski A. Y., Amirgoulova E. V., Nienhaus G. U. 2004. Biocompatible surfaces for specific tethering of individual protein molecules. J. Phys. Chem. B 108, 13 387–13 394 10.1021/jp049057o (doi:10.1021/jp049057o) [DOI] [Google Scholar]
- 3.Yanker D. M., Maurer J. A. 2008. Direct printing of trichlorosilanes on glass for selective protein adsorption and cell growth. Mol. Biosyst. 4, 502–504 10.1039/b801161c (doi:10.1039/b801161c) [DOI] [PubMed] [Google Scholar]
- 4.Tessler L. A., Reifenberger J. G., Mitra R. D. 2009. Protein quantification in complex mixtures by solid phase single-molecule counting. Anal. Chem. 81, 7141–7148 10.1021/ac901068x (doi:10.1021/ac901068x) [DOI] [PubMed] [Google Scholar]
- 5.Agrawal A., Deo R., Wang G. D., Wang M. D., Nie S. M. 2008. Nanometer-scale mapping and single-molecule detection with color-coded nanoparticle probes. Proc. Natl Acad. Sci. USA 105, 3298–3303 10.1073/pnas.0712351105 (doi:10.1073/pnas.0712351105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy R., Hohng S., Ha T. 2008. A practical guide to single-molecule FRET. Nat. Method. 5, 507–516 10.1038/nmeth.1208 (doi:10.1038/nmeth.1208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groll J., Amirgoulova E. V., Ameringer T., Heyes C. D., Rocker C., Nienhaus G. U., Moller M. 2004. Biofunctionalized, ultrathin coatings of cross-linked star-shaped poly(ethylene oxide) allow reversible folding of immobilized proteins. J. Am. Chem. Soc. 126, 4234–4239 10.1021/ja0318028 (doi:10.1021/ja0318028) [DOI] [PubMed] [Google Scholar]
- 8.Scott E. A., Nichols M. D., Cordova L. H., George B. J., Jun Y. S., Elbert D. L. 2008. Protein adsorption and cell adhesion on nanoscale bioactive coatings formed from poly(ethylene glycol) and albumin microgels. Biomaterials 29, 4481–4493 10.1016/j.biomaterials.2008.08.003 (doi:10.1016/j.biomaterials.2008.08.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbert D. L., Hubbell J. A. 2001. Conjugate addition reactions combined with free-radical cross-linking for the design of materials for tissue engineering. Biomacromolecules 2, 430–441 10.1021/bm0056299 (doi:10.1021/bm0056299) [DOI] [PubMed] [Google Scholar]
- 10.Wacker B. K., Scott E. A., Kaneda M. M., Alford S. K., Elbert D. L. 2006. Delivery of sphingosine 1-phosphate from poly(ethylene glycol) hydrogels. Biomacromolecules 7, 1335–1343 10.1021/bm050948r (doi:10.1021/bm050948r) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeung Y. G., Stanley E. R. 2009. A solution for stripping antibodies from polyvinylidene fluoride immunoblots for multiple reprobing. Anal. Biochem. 389, 89–91 10.1016/j.ab.2009.03.017 (doi:10.1016/j.ab.2009.03.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wayment J. R., Harris J. M. 2009. Biotin–avidin binding kinetics measured by single-molecule imaging. Anal. Chem. 81, 336–342 10.1021/ac801818t (doi:10.1021/ac801818t) [DOI] [PubMed] [Google Scholar]
- 13.Roy R., Kozlov A. G., Lohman T. M., Ha T. 2009. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461, 1092–1097 10.1038/nature08442 (doi:10.1038/nature08442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Yeh Y. S., Barbara P. F. 2009. HIV-1 nucleocapsid protein bends double-stranded nucleic acids. J. Am. Chem. Soc. 131, 15 534–15 543 10.1021/ja9070046 (doi:10.1021/ja9070046) [DOI] [PubMed] [Google Scholar]
- 15.Wu J. Y., Stone M. D., Zhuang X. 2009. A single-molecule assay for telomerase structure-function analysis. Nucleic Acids Res. 38, e16. 10.1093/nar/gkp1033 (doi:10.1093/nar/gkp1033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deniz A. A., Mukhopadhyay S., Lemke E. A. 2008. Single-molecule biophysics: at the interface of biology, physics and chemistry. J. R. Soc. Interface 5, 15–45 10.1098/rsif.2007.1021 (doi:10.1098/rsif.2007.1021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reifenberger J. G., Toprak E., Kim H., Safer D., Sweeney H. L., Selvin P. R. 2009. Myosin VI undergoes a 180° power stroke implying an uncoupling of the front lever arm. Proc. Natl Acad. Sci. USA 106, 18 255–18 260 10.1073/pnas.0900005106 (doi:10.1073/pnas.0900005106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapranov P., et al. 2010. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature 466, 642–646 10.1038/nature09190 (doi:10.1038/nature09190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flusberg B. A., Webster D. R., Lee J. H., Travers K. J., Olivares E. C., Clark T. A., Korlach J., Turner S. W. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Method. 7, 461–465 10.1038/nmeth.1459 (doi:10.1038/nmeth.1459) [DOI] [PMC free article] [PubMed] [Google Scholar]