Abstract
Differentiation of the embryonic termini in Drosophila depends on signaling by the Tor RTK, which induces terminal gene expression by inactivating at the embryonic poles a uniformly distributed repressor activity that involves the Gro corepressor. Here, we identify a new gene, cic, that acts as a repressor of terminal genes regulated by the Tor pathway. cic also mediates repression along the dorsoventral axis, a process that requires the Dorsal morphogen and Gro, and which is also inhibited by Tor signaling at the termini. cic encodes an HMG-box transcription factor that interacts with Gro in vitro. We present evidence that Tor signaling regulates terminal patterning by inactivating Cic at the embryo poles. cic has been evolutionarily conserved, suggesting that Cic-like proteins may act as repressors regulated by RTK signaling in other organisms.
Keywords: Drosophila, RTK signaling, repression, capicua, groucho, torso
Signaling by receptor tyrosine kinases (RTKs) is an important mechanism linking extracellular signals (e.g., growth factors) to changes in gene expression and cell behavior. RTK signaling involves a pathway of transducing proteins including Ras, Raf, and mitogen-associated protein kinase (MAPK), which ultimately affects expression of target genes by phosphorylation of specific transcription factors. This transduction pathway has been highly conserved during evolution and controls multiple developmental processes in all animals. One such process that has been studied in detail is the development of terminal structures (the anterior and posterior poles) of the Drosophila embryo (Duffy and Perrimon 1994). Terminal development requires a cascade of maternally acting genes including torso (tor), which encodes an RTK uniformly distributed along the plasma membrane of the early embryo. Tor activation occurs exclusively at the embryonic poles by a ligand produced locally through the action of the fs(1)Nasrat, fs(1) pole-hole, torso-like, and trunk genes (Duffy and Perrimon 1994).
Tor signaling proceeds via the Ras/Raf/MAPK pathway to regulate expression of the zygotic genes tailless (tll) and huckebein (hkb), which are specifically expressed at each pole of the embryo (Duffy and Perrimon 1994, and references therein). These genes encode transcription factors that initiate the developmental programs leading to differentiation of head and tail structures. Recently, it has been shown that Tor signaling does not activate terminal gene expression directly; rather, it functions by antagonizing at the poles a uniformly distributed repressor activity, allowing other maternal factors to activate transcription locally (Rusch and Levine 1994; Liaw et al. 1995; Paroush et al. 1997). Evidence for this view comes from the identification of regulatory elements in the tll promoter (called tor response elements, tor-REs) that confer terminal-specific expression and that, when mutated, cause severe derepression of tll transcription (Liaw et al. 1995).
Additional evidence for the regulation of tll and hkb by relief of repression derives from the role of the Groucho (Gro) corepressor in this process (Paroush et al. 1997). Gro is a nuclear WD-repeat protein that does not bind DNA but interacts with a variety of DNA-bound transcriptional repressors (Fisher and Caudy 1998; Parkhurst 1998). These associations recruit Gro to target promoters, bringing about transcriptional repression. Gro has been shown to participate in terminal development by restricting the expression of tll and hkb to the embryonic termini: Embryos deprived of maternal Gro function show derepression of tll and hkb toward the middle of the embryo (Paroush et al. 1997). Thus, it has been proposed that Gro forms part of a repressor complex specifically inactivated by Tor signaling at the embryo poles (Paroush et al. 1997). However, the specific target of Tor signal inhibition remains unknown.
Similarly, the identity of the factor(s) that recruit Gro to terminal promoters is uncertain. Several Gro partners have been identified to date, for example, Hairy, Runt, Engrailed, Dorsal, and dTCF/pangolin (Cavallo et al. 1998; Fisher and Caudy 1998; Parkhurst 1998; Roose et al. 1998), but none of these proteins seem to play a role in terminal patterning. Also, previous studies identified two putative repressors of tll, GAGA, and NTF-1/Grainyhead, but there is no evidence that they function via Gro (Liaw et al. 1995; Paroush et al. 1997). Thus, it is thought that Gro is recruited to the tll and hkb promoters by an as yet unknown repressor factor(s) (Paroush et al. 1997; Fisher and Caudy 1998; Parkhurst 1998).
In this work, we identify a novel gene, capicua (cic), that acts as a repressor of tll and hkb expression. In addition, cic mediates ventral repression of the dorsally expressed gene zerknüllt (zen), a process that also requires Gro and the rel factor Dorsal, and which is also inhibited by Tor signaling at the embryonic termini. cic encodes a putative transcription factor with a DNA-binding domain of the HMG box class. The Cic protein interacts with Gro in vitro, suggesting that both factors function in the same protein complex. Evidence is presented that Cic is the target inhibited by the Tor signal at the embryonic poles. Finally, Cic has been conserved during evolution, suggesting that Cic-like proteins may also function in repressor processes regulated by RTK signaling in other species.
Results and Discussion
Identification of cic, a novel maternal effect mutation affecting terminal development
We isolated cic in a P-element screen for female sterile mutations affecting the anteroposterior pattern of embryos. Females homozygous for the cic mutation (cic1) are fully viable and produce embryos that form head and tail structures but lack most of the segmented trunk (Fig. 1A,B; hence, the name capicua, the Catalan for head-and-tail). Hereafter, we refer to embryos lacking maternal cic function as cic mutant embryos. The phenotype of cic1 mutant embryos is rather uniform: Most embryos (>80%) retain only 1–3 partial abdominal denticle belts at 25°C, whereas the rest of the embryos do not show any signs of abdominal segmentation. This latter phenotype is shown by virtually all embryos from females carrying the cic1 allele in trans with a deficiency of the region, suggesting that cic1 is a strong hypomorph. The cic1 phenotype is similar to that of embryos from females carrying dominant gain-of-function mutations in tor (torgof; Klinger et al. 1988) and other components of the Tor RTK pathway (see Duffy and Perrimon 1994). These mutations cause constitutive Tor RTK signaling in all regions of the embryo, leading to ectopic tll and hkb expression, and the subsequent differentiation of the segmented trunk as terminal structures. However, the above results with deficiencies of the cic region indicate that cic1 is a recessive loss-of-function mutation (see legend to Fig. 1).
Figure 1.
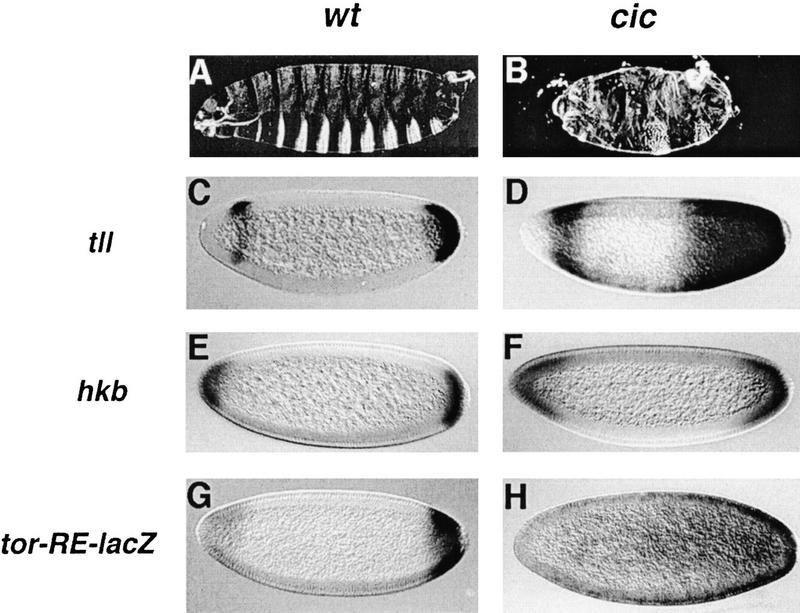
cic is required for repression of tll and hkb during terminal development. Cuticle phenotypes of embryos derived from wild-type (A) and homozygous cic1 (B) females. Note the strong suppression of trunk segmentation in the cic1 mutant embryo. Embryos from females carrying the cic1 allele in trans with a deficiency of the cic region show a slightly more severe phenotype (not shown). (C–H) RNA expression patterns of tll (C,D), hkb (E,F), and tor-RE-lacZ (G,H) in embryos derived from wild-type (C,E,G) and homozygous cic1 (D,F,H) females. Derepression is observed in all cases in cic1 mutant embryos. In this and following figures, anterior is to the left and dorsal is up.
Given the similarities between the torgof and cic1 phenotypes, we examined the expression patterns of tll and hkb in cic1 embryos. Expression of both genes expands toward the center of such embryos, predominantly in the posterior domain (Fig. 1C–F). The expanded expression of tll and hkb is very similar to that observed in torgof and gro mutant embryos (Weigel et al. 1990; Brunner et al. 1994; Paroush et al. 1997). We also analyzed the expression of a lacZ transgene under the control of a tor-RE from the tll promoter (see Introduction) that drives terminal-specific transcription (Liaw et al. 1995; construct G22) (Fig. 1G). In cic1 mutant embryos, expression of this construct is derepressed toward the middle of the embryo (Fig. 1H). Together, these results suggest that the cic gene is normally required to restrict tll and hkb expression to the embryonic poles.
cic could affect tll and hkb expression by restricting Tor signaling to the embryonic poles (e.g., by limiting the domain of Tor receptor activation, or the domain of Tor signal transduction inside of the embryo). Alternatively, cic could function, like gro, as a repressor of tll and hkb downstream of the Tor pathway. To help distinguish between these possibilities, we performed epistasis analyses using loss-of-function mutations in tor, Draf, and Dsor (encoding a Drosophila MEK, or MAPK kinase, homolog, Tsuda et al. 1993; see Fig. 2A), which normally cause a phenotype complementary to that of cic1, that is, absence of terminal structures. Embryos from females homozygous for cic1 and tor are identical to those from cic1 females alone (Fig. 2B). Likewise, cic1 females carrying loss-of-function clones of Draf or Dsor in the germ line produce embryos that display the cic phenotype (Fig. 2C). Thus, cic acts genetically downstream of Draf and Dsor. In addition, we examined directly the domain of Tor signal activity using a monoclonal antibody against the active, diphosphorylated form of Drosophila MAPK (known as Erk; Brunner et al. 1994; Gabay et al. 1997), and found a normal pattern of Erk activation in cic1 embryos (Fig. 2, cf. D and E). This shows that derepression of tll and hkb in cic1 mutant embryos is not due to an expanded domain of Tor signaling, suggesting that cic is part of the activity that represses tll and hkb in the central region of the embryo and is inhibited by Tor signaling at the embryonic poles.
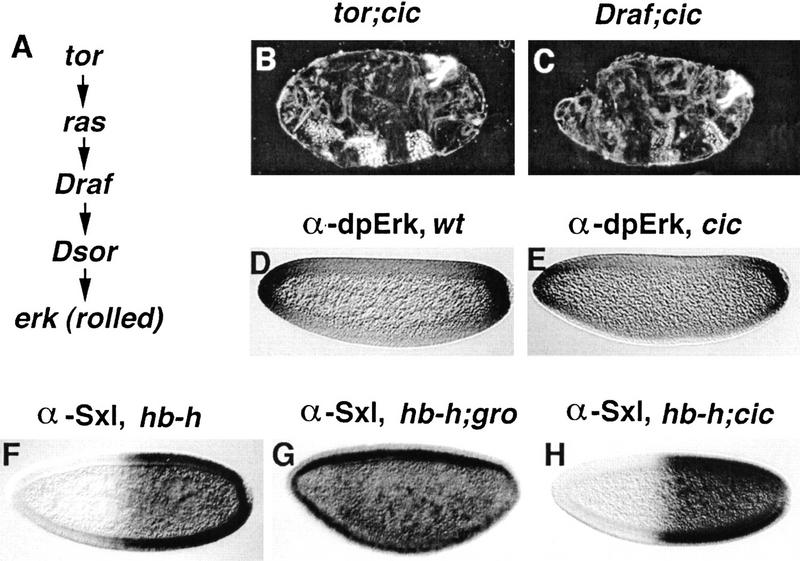
Figure 2.
cic acts downstream of the Tor pathway and is not required for Gro activity. (A) Diagram of the Tor cascade. (B,C) Cuticle phenotypes of embryos derived from doubly homozygous tor; cic females (B) and from cic1 females carrying germ-line clones of Draf (C); the embryos display a cic-like phenotype, suggesting that cic acts downstream of Draf. The roles of cic, tor, and Draf in terminal patterning are strictly maternal and the mutations do not show paternal rescue. (D,E) Antibody staining for activated Erk protein in wild-type (D) and cic1 (E) embryos; the similar staining indicates that the domain of Tor signaling is unaffected in cic1 mutant embryos. (F–H) Effects on Sex-lethal (Sxl) expression of ectopic hairy (h) activity driven by the hunchback (hb) promoter in otherwise wild-type (F), groE48 (G), and cic1 (H) embryos. Embryos were stained with an antibody against the active Sxl protein. Repression of Sxl by Hairy requires gro, however the Hairy/Gro complex does not require cic function.
The similar effects of cic and gro on terminal patterning raise the possibility that cic is necessary for Gro corepressor activity in general. However, two lines of evidence argue against this idea. First, Gro participates in many developmental processes, whereas the role of cic appears restricted to terminal and dorsoventral patterning (see below). Second, we have confirmed that Gro-dependent repression by Hairy in a sex determination assay (Paroush et al. 1994; Jiménez et al. 1997) does not require cic function (Fig. 2F–H). These results indicate that cic does not generally affect Gro activity.
cic also affects repression along the dorsoventral axis of the embryo
Tor signaling at the embryo poles also regulates repressor processes that operate during dorsoventral patterning (Casanova 1991; Rusch and Levine 1994). Such patterning depends on the Dorsal morphogen, a rel domain factor that accumulates in ventral nuclei of early embryos and acts as both an activator and repressor of transcription: It activates ventral-specific genes, for example, twist (twi), and represses dorsal-specific genes such as zen (see Rusch and Levine 1996; and references therein). Repression by Dorsal requires its association with Gro and other postulated corepressors that bind next to Dorsal in the zen promoter (Jiang et al. 1993; Kirov et al. 1993; Dubnicoff et al. 1997; Valentine et al. 1998; see below). This repressor complex is under negative regulation by Tor signaling at the embryonic termini, allowing zen expression at each pole of the embryo (Casanova 1991; Rusch and Levine 1994).
The mechanism of repression by Dorsal is not fully understood. Valentine et al. (1998) recently showed that Dead-Ringer (Dri) and Cut function as corepressors that assist Dorsal (and Gro) in its function as a repressor. However, the effects of removing either of these two factors appears weaker than those caused by the loss of Dorsal or Gro function, suggesting that other factors may also contribute to Dorsal repression. Because cic is involved in a Gro-mediated process that is inactivated by Tor signaling, we wondered if cic could also be involved in Dorsal repression. Consistent with this idea, zen expression is expanded ventrally in cic1 mutant embryos (Fig. 3A,B). Although this expansion is not as strong as in dorsal or gro mutants, ectopic zen transcripts are clearly detected in lateral and ventral regions of the embryo, especially in its posterior half. In contrast, activation of twi by Dorsal is normal in cic1 embryos (Fig. 3C,D), suggesting that cic only participates in repression, not activation, by Dorsal.
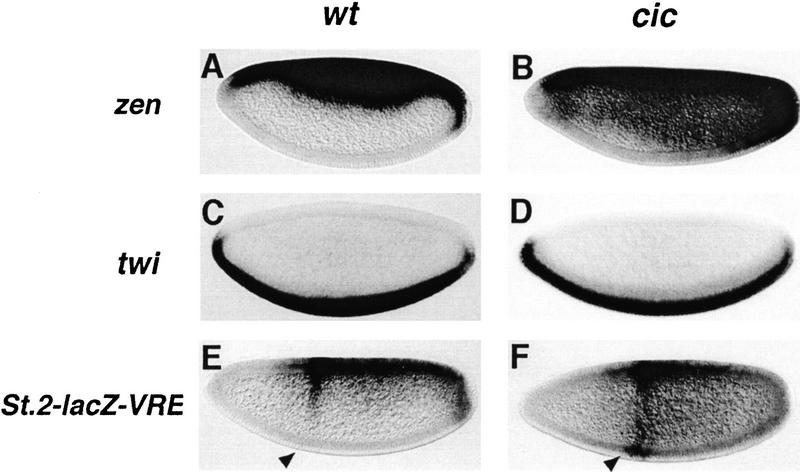
Figure 3.
cic is required for repression of zen and acts through the VRE. RNA expression patterns of zen (A,B), twi (C,D), and St.2-lacZ-VRE (E,F) in embryos derived from wild-type (A,C,E) and homozygous cic1 (B,D,F) females. Derepression of zen and lacZ expression driven by the eve stripe 2 enhancer (arrowhead) is observed in cic1 embryos. In contrast, the similar pattern of twi expression in wild-type and cic1 embryos indicates that activation of twi by Dorsal is independent of cic.
To test further the role of cic in ventral repression of zen, we analyzed a lacZ transgene carrying an even-skipped (eve) stripe 2 enhancer coupled to a silencer from the zen promoter, the zen Ventral Repression Element (VRE), which includes binding sites for Dorsal and adjacent regulatory sites (Cai et al. 1996; see Materials and Methods). In wild-type embryos, lacZ expression directed by the eve stripe 2 enhancer is repressed ventrally by the VRE (Fig. 3E). This repression is clearly attenuated in cic1 mutant embryos (Fig. 3F), permitting stripe 2 activation in the ventral-most side of the embryo. In addition, we observe significant ectopic lacZ expression in ventral and lateral regions of the embryo, as expected if repression by Dorsal bound to the VRE is switched in favor of activation (Jiang et al. 1993; Kirov et al. 1993; Dubnicoff et al. 1997; Valentine et al. 1998). These results suggest that cic encodes one of the cofactors required for VRE activity and the conversion of Dorsal from an activator to a repressor of transcription. Because Dri and Cut also function as Dorsal corepressors, it appears that this role is shared by several factors with overlapping activities.
cic encodes an evolutionarily conserved HMG box protein
The cic1 mutation was recovered in a P-element screen, but it is not caused by the insertion of a functional P element. We therefore isolated the cic gene by positional cloning. We mapped cic to chromosomal position 92D1-2 using standard recombination and deficiency tests. We then searched for DNA polymorphisms in this region specific to the cic1 chromosome and found that the cic1 allele is associated with the insertion of a 1.5-kb hobo transposon. This transposon maps ∼300 bp away from a previously described P element, P(PZ) bwk8482, which causes the female sterile mutation bullwinkle (bwk, Rittenhouse and Berg 1995). P(PZ) bwk8482 produces a phenotype different from cic1 and complements cic1, indicating that the two mutations affect different genetic functions (see below).
The cic1-specific hobo element is inserted in the 5′ untranslated region of a novel gene, which corresponds to EST clone LD05430 from the Berkeley Genome Project (Fig. 4A). Several lines of evidence confirm that this gene is cic. First, molecular analyses show that the hobo insertion disrupts the cic transcript (data not shown). Second, the cic1 mutation is not complemented by small deficiencies (<500 bp) that span the hobo insertion site (Fig. 4A). Finally, we rescued the cic phenotype by P-element transformation with a genomic fragment containing the cic gene (Fig. 4A). This fragment does not rescue the bwk phenotype, again showing that bwk and cic represent separate gene functions. Elucidation of the relationship between cic and bwk at the molecular level will require the cloning of bwk.
Figure 4.
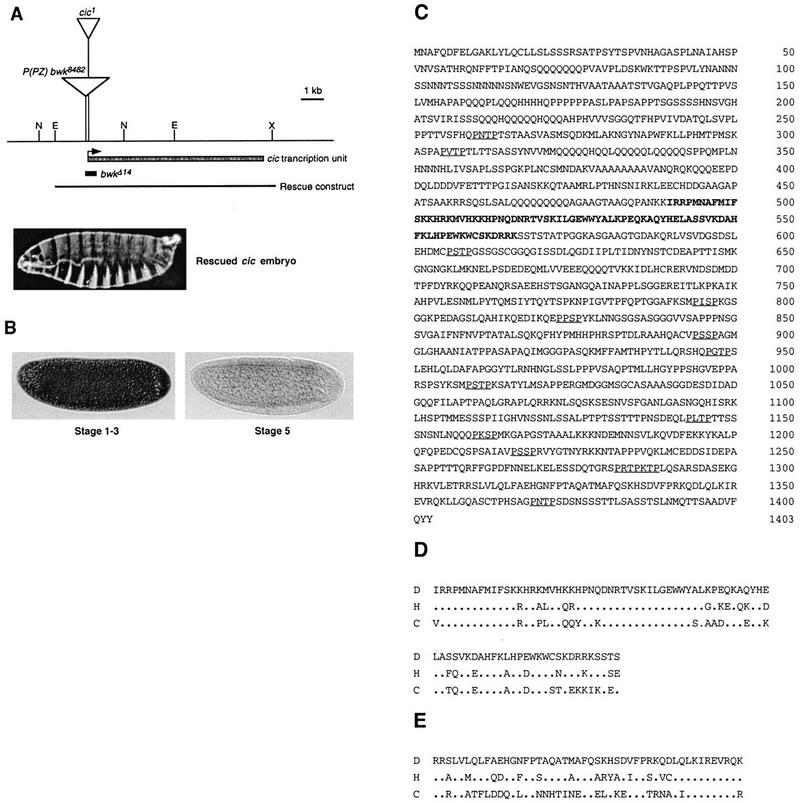
cic encodes an evolutionarily conserved HMG box transcription factor. (A) Diagram of the cic genomic region showing the insertion sites of the hobo element causing the cic1 mutation and the nearby P(PZ) bwk8482 P element (transposons are not represented at scale). The cic transcription unit spans ∼8 kb and includes at least two introns. We have mapped in detail only one intron in the 5′ region of the gene (not shown). The diagram shows the region deleted in a small deficiency caused by imprecise excision of P(PZ) bwk8482, bwkΔ14 (see Materials and Methods), which does not complement cic1. The genomic construct used to rescue the cic phenotype and a rescued embryo are also shown. (E) EcoRI; (N) NotI; (X) XbaI. (B) Maternal expression of cic. cic transcripts accumulate in early blastoderm embryos (stages 1–3) but become undetectable after the onset of gastrulation (stage 5). In situ hybridizations were carried out with an anti-sense cic RNA probe. (C) Amino acid sequence of the deduced Cic protein. The HMG box domain is shown in bold. Possible MAPK phosphorylation sites (P-X-S/T-P) are underlined. Note the presence of multiple homopolymeric stretches in the protein, particularly in the amino-terminal domain. (D) Sequence alignment of HMG box domains from Drosophila Cic (D) and two related proteins from humans (H; GenBank accession no. AB002304) and C. elegans (C; GenBank accession no. Z50797). Periods indicate identical residues. (E) Sequence alignment of the Cic carboxy-terminal domain (amino acids 1308–1355) with the corresponding region of the human and C. elegans proteins; the strong conservation indicates that these proteins are true orthologs. We have not detected significant similarities outside of the HMG box and carboxy-terminal domains.
We analyzed cic expression by in situ hybridization and found that cic mRNA is present at high levels in early blastoderm embryos (stage 1–3; Fig. 4B), consistent with a maternal expression of the gene. The cic transcripts decay rapidly so that they are barely detected by the onset of gastrulation (late stage 5; Fig. 4B) and at later stages of embryogenesis (not shown). These results, together with the strictly maternal effect character of the cic1 mutation, argue that cic function is restricted to terminal and dorsoventral patterning of the early embryo.
Sequencing of cic cDNA clones shows that it encodes a putative transcription factor with a DNA-binding domain of the HMG box class (Fig. 4C). HMG box proteins are thought to function as architectural factors that induce bending of the target DNA and facilitate the assembly of multiprotein regulatory complexes at promoters (Grosschedl et al. 1994). The Cic HMG box domain contains several amino acid residues shared by a group of HMG box factors that bind sequence specifically to DNA, such as the LEF-1/TCF-1 factors, SRY, and the SRY-related Sox proteins (Grosschedl et al. 1994). Similarity searches against protein databases reveal that Cic defines a new subfamily within this group, which includes two related HMG box proteins from humans and Caenorhabditis elegans (Fig. 4D). The similarity between Cic and these proteins is particularly strong in the HMG box domain (75% and 67% identity with the human and C. elegans sequences, respectively), but also extends to other regions of the proteins (Fig. 4E), suggesting that they represent true orthologs.
Cic interacts with Gro in vitro
Our results indicate that Cic functions in two Gro-dependent repressor processes inactivated by Torso signaling. We therefore examined whether Cic interacts with Gro in vitro. We expressed in bacteria three different fragments of Cic (amino-terminal, central, and carboxy-terminal, see Fig. 5) as GST fusions and assayed their ability to bind radiolabeled Gro protein. The carboxy-terminal portion of Cic interacts with Gro, whereas the amino-terminal and central regions of the protein show little or no binding (Fig. 5). The binding of Cic to Gro is weaker than that of Hairy, but stronger than the Dorsal/Gro interaction in the same assay (Fig. 5). We also find that the interaction of Cic with Gro does not depend on the conserved carboxy-terminal domain of Cic (Fig. 4E; data not shown), indicating that this domain mediates another aspect of Cic function. Taken together, the results support the idea that Cic and Gro form a repressor complex inactivated by Torso signaling during terminal and dorsoventral patterning (see below).
Figure 5.
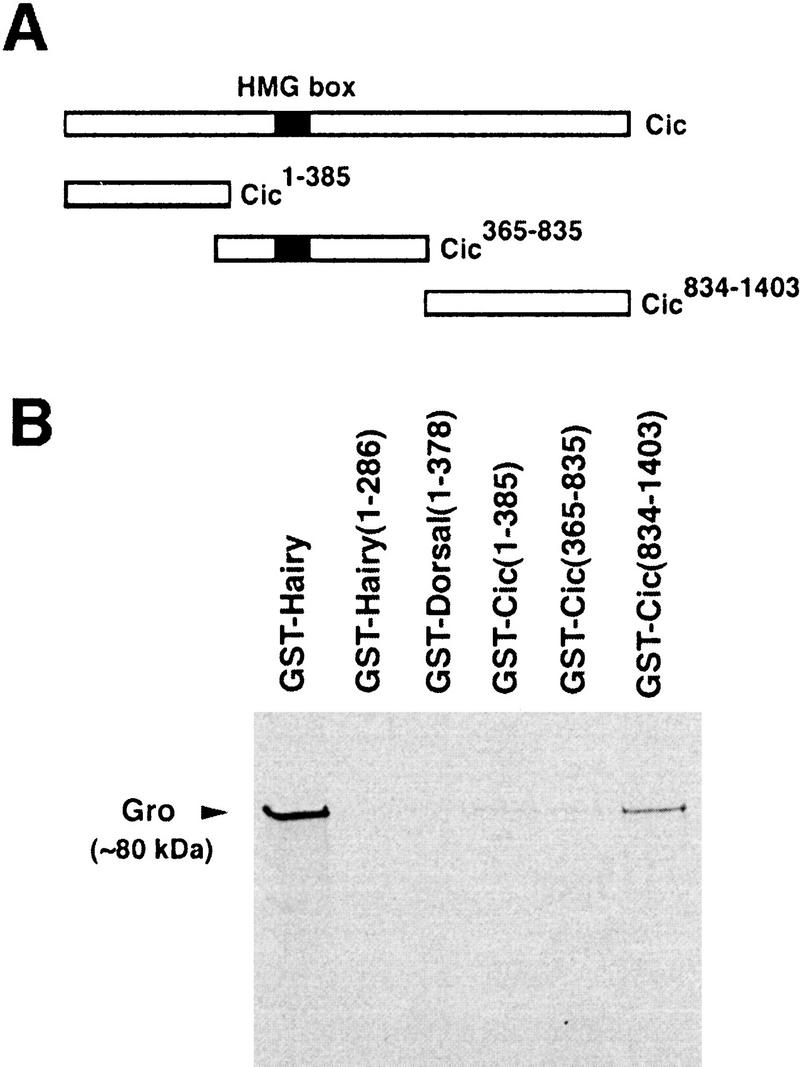
Cic binds to Gro in vitro. (A) Diagram of the Cic protein and three fragments tested for interaction with Gro. (B) 35S-labeled Gro was incubated with similar amounts of the indicated GST fusions bound to glutathione-Sepharose beads. After washing the beads, the retained Gro protein was detected by autoradiography. The Cic834–1403 domain binds to Gro, whereas the other two domains show little or no binding. GST fusions of Hairy and Dorsal1–378 were used as positive controls (Paroush et al. 1994; Dubnicoff et al. 1997), whereas GST–Hairy1–286 (a mutant lacking the carboxy-terminal Gro-binding motif) was the negative control (Jiménez et al. 1997). Binding of Gro to Dorsal1–378 appears very weak in our assay. We have not observed significant similarities between the Cic834–1403 domain and other Gro-binding motifs.
Cic is under negative post-transcriptional regulation by the Torso pathway
What is the actual target of Tor signal inactivation at the embryonic poles? Previous work has shown that the Drosophila Yan Ets-like repressor factor is degraded in response to RTK signaling during eye development (Rebay and Rubin 1995). Thus, it is possible that the target of Tor signaling is similarly inactivated at the embryo poles. The Gro protein is uniformly distributed in the blastoderm embryo and does not show down-regulation at the termini (Delidakis et al. 1991). Also, Gro corepressor activity during sex determination is not inhibited by Tor signaling, arguing that Gro is not the target of the Tor signal (Paroush et al. 1997). To monitor the pattern of Cic distribution in embryos, we raised a polyclonal antibody against an HMG box-containing fragment of the protein (see Materials and Methods). This antibody revealed a distinctive nuclear signal in wild-type but not cic1 blastoderm embryos (Fig. 6A,B), confirming both that Cic is a nuclear protein and that the cic1 allele is a strong loss-of-function mutation. Remarkably, Cic is distributed asymmetrically in blastoderm embryos, being present in nuclei from the presumptive trunk but absent at each pole of the embryo (Fig. 6A). Because cic mRNA is uniformly distributed in the embryo (Fig. 4B), the exclusion of the protein from the poles argues that Cic is under negative post-transcriptional regulation by the Tor signal transduction pathway.
Figure 6.
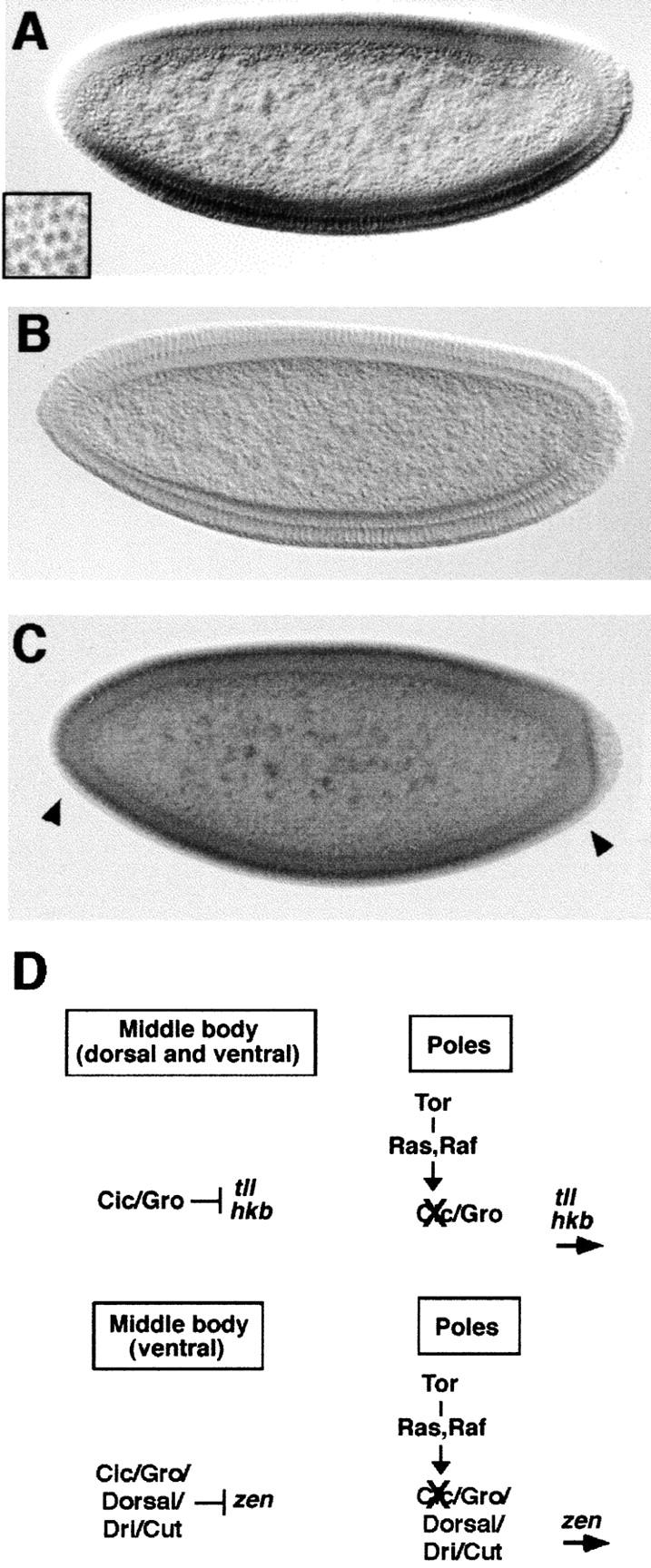
Cic is negatively regulated by the Tor pathway. (A) Pattern of Cic protein distribution in wild-type embryos. The protein localizes predominantly in the nucleus (inset; surface view of blastoderm embryo) and is present in medial but not terminal regions of the embryo. (B,C) Pattern of Cic protein in cic1 (B) and tor mutant embryos (C). Note the lack of staining in the cic1 background, and the accumulation of Cic protein at the poles of tor mutant embryos (arrowheads). (D) Model for Cic function in terminal and dorsoventral patterning. Cic associates with Gro to form a protein complex that represses tll and hkb expression in the central region of the embryo. The Tor RTK pathway inactivates Cic at the embryo poles, thus allowing activation of tll and hkb by other maternal factors. A similar mechanism operates in the regulation of zen, except that repression in middle regions of the embryo is only ventral and also requires Dorsal and other corepressors such as Dri and Cut. In both cases, repression could involve additional factors.
To test this idea, we examined the distribution of Cic in tor mutant embryos. In such embryos, the Cic protein is detected not only in medial regions of the embryo but also at the termini (Fig. 6C), implying that Tor signaling inhibits accumulation of Cic protein at the embryo poles. These results suggest that Cic is the target inactivated by the Tor signal, possibly via MAPK phosphorylation and subsequent degradation of the protein, as in the case of Yan (Rebay and Rubin 1995). Consistent with this idea, the Cic protein sequence includes 14 consensus MAPK phosphorylation sites (Fig. 4C). Future studies should define the mechanism by which Tor signaling regulates Cic accumulation and the functional domains of the protein involved in this control.
Possible mechanism of Cic action
How does Cic mediate repression of terminal and dorsoventral genes? Because we have shown that reporter constructs carrying the tll tor-RE and zen VRE are derepressed in cic mutant embryos, the simplest model is that Cic binds to these regulatory elements and recruits Gro for repression of these genes. Consistent with this idea, there are striking similarities between the consensus DNA-binding site for HMG box proteins (Grosschedl et al. 1994) and sequence elements within the tor-RE and VRE known to mediate transcriptional repression (Jiang et al. 1993; Kirov et al. 1993; Liaw et al. 1995). However, although these elements are bound by control HMG box proteins (our unpublished observations), we have not detected specific binding of Cic to them. Perhaps Cic has a very low affinity for DNA and/or requires the presence of accessory factors for efficient DNA binding. Several HMG box proteins rely on interactions with partner proteins to increase their affinity for DNA (Kjaerulff et al. 1997; Kamachi et al. 1999). Clearly, identification of the precise molecular mechanism of Cic function will require further analyses of its ability to interact with target sequences in terminal and dorsal-specific genes.
That Cic functions in association with other factors is consistent with previous studies that have implicated several proteins in Gro-dependent repression of terminal and dorsal-specific genes (Liaw et al. 1995; Valentine et al. 1998). For example, it has been shown that Dri and Cut are two of the cofactors required for repression by Dorsal through the zen VRE (Valentine et al. 1998), and our results indicate that Cic also contributes to switching Dorsal from an activator to a repressor of transcription. Similarly, the dramatic effects of Cic on terminal patterning indicate that both Cic and Gro are essential components in the repression of terminal genes. We still do not understand how the activity of all these factors is coordinated in vivo. Nevertheless, our results showing that Cic is under negative post-transcriptional control by the Tor RTK pathway, suggest that it functions as the regulatory element that links Tor signaling to the mechanism of repression (see Fig. 6D).
Finally, the Cic protein has been conserved widely during evolution, being present in organisms as different as C. elegans and humans. Thus, Cic-like proteins may also mediate the formation of multiprotein repressor complexes in other animals, and function as targets of RTK signaling to regulate cellular responses by relief of repression, rather than by direct activation of gene expression.
Materials and methods
Fly stocks
The cic1 mutation was isolated in a P-element screen for female-sterile mutations affecting anteroposterior polarity of embryos; the mutation is homozygous viable and 100% female sterile. Flies carrying the tor-RE-lacZ transgene (construct G22 in Liaw et al. 1995) were kindly provided by J. Lengyel (UCLA). The St.2-lacZ-VRE stock (construct St.2-lacZ-VRE 600; forward orientation) was a gift of H. Cai (University of Georgia, Athens). Germ-line clones of Draf and Dsor were generated with the DrafLE78 and Dsor1 alleles in combination with the ovoD-FLP-FRT system (Chou et al. 1993). tor mutant embryos were obtained from homozygous torXR1 mothers. Imprecise excisions of the bwk8482 P-element insertion were recovered as described (Rittenhouse and Berg 1995), and characterized by PCR. For rescue of the cic1 mutation, a genomic EcoRI–XbaI fragment was assembled into pCaSpeR4, and the resulting construct was used to transform y w flies.
Embryo analyses
Patterns of gene expression were determined by whole-mount in situ hybridization with digoxygenin-labeled DNA (tll) or RNA (hkb, zen, lacZ, cic) probes. Immunohistochemical detection of Erk and Sxl was performed with monoclonal antibodies against the diphosphorylated form of Erk (Sigma) and the active Sxl protein. For detection of Cic protein, a polyclonal antibody was generated in rabbits by injecting an HMG-box containing fragment of Cic (Cic365–582) fused to GST. Signals were detected with appropriate secondary antibodies coupled to alkaline phosphatase (in situ hybridizations) or HRP peroxidase (immunostainings).
Cloning of cic
cic was mapped to chromosomal position 92D1-2. Several genomic fragments from this region were isolated by plasmid rescue with different P-element insertions in this location. One such fragment recovered from the P(PZ) bwk8482 P-element insertion (Rittenhouse and Berg 1995) was found to span the insertion site of a hobo element specific to the cic1 chromosome. Sequencing of this site showed that the hobo element is inserted in the 5′ untranslated region of the LD05430 transcript. A second cic cDNA almost identical to the LD05430 clone was isolated by screening an embryonic cDNA library (gift of N. Brown, University of Cambridge, UK).
In vitro binding experiments
cDNA fragments encoding Cic1–385, Cic365–835, and Cic834–1403 were cloned into pGEX2T (Cic834–1403) or pZEX (Cic1–385 and Cic365–835; see Jiménez et al. 1997). Expression of GST fusions in Escherichia coli strain SRP84 and binding assays to radiolabeled Gro protein were carried out as described (Paroush et al. 1994; Jiménez et al. 1997).
Acknowledgments
We thank N. Martín for technical assistance, S. González-Crespo and J. Bernués for discussions, M. Erdélyi and P. Zavorszky for their help in the genetic screen, C. Berg, M. Levine, H. Cai, J. Lengyel, and Y. Kamachi for fly stocks and DNA clones and S. González-Crespo, A. González-Reyes, E. Sánchez-Herrero, and G. Morata for comments on the manuscript. G.J. is supported by a contract from the Spanish Ministerio de Educación y Cultura (MEC). A.G. was supported by fellowships from the Ministère de la Recherche et du Travail and the EMBL. This work was funded by the Spanish MEC and the Generalitat de Catalunya (C.R. Biotecnologia).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
The EMBL accession no. for the cic cDNA sequence is AJ252268.
Footnotes
E-MAIL jcrbmc@cid.csic.es; FAX 34 93 2045904.
References
- Brunner D, Oellers N, Szabad J, Biggs WH, Zipursky SL, Hafen E. A gain of function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signalling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- Cai HN, Arnosti D, Levine M. Long-range repression in the Drosophila embryo. Proc Natl Acad Sci. 1996;93:9309–9314. doi: 10.1073/pnas.93.18.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J. Interaction between torso and dorsal, two elements of different transduction pathways in the Drosophila embryo. Mech Dev. 1991;36:41–45. doi: 10.1016/0925-4773(91)90070-m. [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Delidakis C, Preiss A, Hartley DA, Artavanis-Tsakonas S. Two genetically and molecularly distinct functions involved in early neurogenesis reside within the Enhancer-of-split locus of Drosophila melanogaster. Genetics. 1991;129:803–823. doi: 10.1093/genetics/129.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ. Conversion of Dorsal from an activator to a repressor by the global corepressor Groucho. Genes & Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB, Perrimon N. The Torso pathway in Drosophila: Lessons on receptor tyrosine kinase signaling and pattern formation. Dev Biol. 1994;166:380–395. doi: 10.1006/dbio.1994.1324. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes & Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo B-Z. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development. 1997;124:3535–3541. doi: 10.1242/dev.124.18.3535. [DOI] [PubMed] [Google Scholar]
- Grosschedl R, Giese K, Pagel J. HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Jiang J, Cai H, Zhou Q, Levine M. Conversion of a dorsal-dependent silencer into an enhancer: Evidence for dorsal corepressors. EMBO J. 1993;12:3201–3209. doi: 10.1002/j.1460-2075.1993.tb05989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes & Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Cheah KSE, Kondoh H. Mechanism of regulatory target selection by the Sox High-Mobility-Group domain proteins as revealed by comparison of Sox1/2/3 and Sox9. Mol Cell Biol. 1999;19:107–120. doi: 10.1128/mcb.19.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov N, Zhelnin L, Shah J, Rushlow C. Conversion of a silencer into an enhancer: Evidence for a co-repressor in dorsal-mediated repression in Drosophila. EMBO J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff S, Dooijes D, Clevers H, Nielsen O. Cell differentiation by interaction of two HMG-box proteins: Mat1-Mc activates M cell-specific genes in S. pombe by recruiting the ubiquitous transcription factor Ste11 to weak binding sites. EMBO J. 1997;16:4021–4033. doi: 10.1093/emboj/16.13.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger M, Erdélyi M, Szabad J, Nüsslein-Volhard C. Function of torso in determining the terminal anlagen of the Drosophila embryo. Nature. 1998;335:275–277. doi: 10.1038/335275a0. [DOI] [PubMed] [Google Scholar]
- Liaw G, Rudolph KM, Huang J-D, Dubnicoff T, Courey AJ, Lengyel JA. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes & Dev. 1995;9:3163–3176. doi: 10.1101/gad.9.24.3163. [DOI] [PubMed] [Google Scholar]
- Parkhurst SM. Groucho: Making its Marx as a transcriptional co-repressor. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RLJ, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation and sex-determination, and interacts directly with Hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Wainwright SM, Ish-Horowicz D. Torso signalling regulates terminal patterning in Drosophila by antagonizing Groucho-mediated repression. Development. 1997;124:3827–3834. doi: 10.1242/dev.124.19.3827. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Rittenhouse KR, Berg CA. Mutations in the Drosophila gene bullwinkle cause the formation of abnormal eggshell structures and bicaudal embryos. Development. 1995;121:3023–3033. doi: 10.1242/dev.121.9.3023. [DOI] [PubMed] [Google Scholar]
- Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destrée O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Rusch J, Levine M. Regulation of the dorsal morphogen by the Toll and torso signaling pathways: A receptor tyrosine kinase selectively masks transcriptional repression. Genes & Dev. 1994;8:1247–1257. doi: 10.1101/gad.8.11.1247. [DOI] [PubMed] [Google Scholar]
- ————— Threshold responses to the dorsal regulatory gradient and the subdivision of primary tissue territories in the Drosophila embryo. Curr Opin Gen Dev. 1996;6:416–423. doi: 10.1016/s0959-437x(96)80062-1. [DOI] [PubMed] [Google Scholar]
- Tsuda L, Inoue YH, Yoo M-A, Mizuno M, Hata M, Lim Y-M, Adachi-Yamada T, Ryo H, Masamune Y, Nishida Y. A protein kinase similar to MAP kinase activator acts downstream of the Raf kinase in Drosophila. Cell. 1993;72:407–414. doi: 10.1016/0092-8674(93)90117-9. [DOI] [PubMed] [Google Scholar]
- Valentine SA, Chen G, Shandala T, Fernández J, Mische S, Saint R, Courey AJ. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Jürgens G, Klinger M, Jäckle H. Two gap genes mediate maternal terminal pattern formation in Drosophila. Science. 1990;248:495–498. doi: 10.1126/science.2158673. [DOI] [PubMed] [Google Scholar]