Abstract
Many indicators of protein evolutionary rate have been proposed, but some of them are interrelated. The purpose of this study is to disentangle their correlations. We assess the strength of each indicator by controlling for the other indicators under study. We find that the number of microRNA (miRNA) types that regulate a gene is the strongest rate indicator (a negative correlation), followed by disorder content (the percentage of disordered regions in a protein, a positive correlation); the strength of disorder content as a rate indicator is substantially increased after controlling for the number of miRNA types. By dividing proteins into lowly and highly intrinsically disordered proteins (L-IDPs and H-IDPs), we find that proteins interacting with more H-IDPs tend to evolve more slowly, which largely explains the previous observation of a negative correlation between the number of protein–protein interactions and evolutionary rate. Moreover, all of the indicators examined here, except for the number of miRNA types, have different strengths in L-IDPs and in H-IDPs. Finally, the number of phosphorylation sites is weakly correlated with the number of miRNA types, and its strength as a rate indicator is substantially reduced when other indicators are considered. Our study reveals the relative strength of each rate indicator and increases our understanding of protein evolution.
Keywords: protein evolution, disordered proteins, microRNA regulation, protein–protein interaction, phosphorylation
Introduction
Many indicators of protein evolutionary rate have been identified, including protein connectivity, gene expression level, gene expression breadth, gene compactness, the number of microRNA (miRNA) types that regulate a gene, extracellularity and protein folding pattern (Fraser et al. 2002; Drummond et al. 2005; Chen and Chuang 2006; Liao et al. 2006, 2010; Makino and Gojobori 2006; Pal et al. 2006; Drummond and Wilke 2008; Cheng et al. 2009; Chen et al. 2010; Yang, Zhuang, et al. 2010). However, some of these indicators are interrelated (Liao et al. 2006, 2010; Liang and Li 2007). How the correlations of a factor with other factors affect the strength of the factor as an indicator of protein evolutionary rate should be investigated.
One factor that seems to be of particular interest is miRNAs, which are noncoding RNAs that target mRNAs, leading to mRNA destabilization and translational repression (see Guo et al. 2010). Genes targeted by more types of miRNAs may be subject to more functional constraints and therefore evolve more slowly (Cheng et al. 2009). However, the number of miRNA types of a gene (NmiR) has been shown to be positively correlated with protein connectivity, gene expression breadth, and the length of 3′ untranslated regions (3′UTRs) (Liang and Li 2007; Cheng et al. 2009). How much do these correlations contribute to the strength of NmiR as an indicator of protein evolutionary rate?
Another interesting observation is that genes encoding proteins with more intrinsically disordered regions (IDRs) tend to be targeted by more types of miRNAs, compared with genes encoding fewer IDRs (Liang and Li 2007; Chen et al. 2008; Edwards et al. 2009). IDRs are protein regions that fail to form compact 3D structures in native states. IDRs have been suggested to be associated with a wide variety of cellular functions (Uversky et al. 2005, 2008; Haynes and Iakoucheva 2006; Liu et al. 2006). First, IDRs have been demonstrated to correlate strongly with posttranslational modifications (Iakoucheva et al. 2004). Second, IDRs often contain short and degenerate linear motifs that promiscuously bind to certain protein domains with low affinities (see Vavouri et al. 2009). The low-affinity and promiscuous binding of IDRs may be the reason why proteins with more interacting partners (hubs) tend to contain more IDRs (Haynes et al. 2006). Because hubs tend to be more disordered and genes encoding more disordered proteins tend to be regulated by a larger number of miRNA types (NmiR) and because both protein connectivity and NmiR are negatively correlated with evolutionary rate, one may intuitively speculate that proteins with more IDRs, on average, have a lower evolutionary rate than those with fewer or no IDRs. However, IDRs tend to evolve faster than ordered regions, and proteins with more IDRs tend to evolve faster than proteins with fewer or no IDRs (Brown et al. 2002, 2010; Kim et al. 2008; Chen et al. 2010). The effect of these complicated relationships on the rate of protein evolution should be clarified.
In this study, we tried to assess the relative strengths of the two indicators, NmiR and disorder content (the percentage of IDRs in a protein) because they are correlated but have opposite effects on the protein evolution. In addition, we have also studied the following rate indicators: protein–protein interactions (PPIs), gene expression level, gene expression breadth, and the number of phosphorylation sites in a protein. Other factors have less abundant data and thus have not been considered here. For each indicator studied, we computed the partial correlation between the indicator and the rate of protein evolution by controlling for the other indicators under study. This approach may largely disentangle the effects of these interrelated indicators.
Materials and Methods
Data Used
Human protein sequences, human–mouse orthologs, and the human–mouse evolutionary rates, including the number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) were downloaded from the Ensembl Genome Browser at http://www.ensembl.org/ (version 56). For human genes with more than one isoform, only the longest isoform is chosen. To avoid the confounding factor of gene duplication, only human–mouse 1:1 orthologues were considered.
Only genes that had mRNA expression data in both of the two human mRNA expression data sets used here were included for analysis. The first data set was derived from six human tissues (heart, kidney, liver, muscle, spleen, and testis) using high-density exon arrays (Xing et al. 2007). The second data set was generated by Su et al. (2004) from 73 nonpathogenic human tissues (Gene Atlas V2, http://symatlas.gnf.org/). We aligned each probe set against the Ensembl coding sequences (CDSs) and only considered the probe sets that were 100% identical to the CDSs. We further removed probe sets that matched to more than one gene to avoid ambiguity (Chen et al. 2010). The expression level of a gene was defined as the average signal intensity across 6 and 73 examined tissues for the first and the second expression data sets, respectively. A gene is said to be expressed in a tissue if its average signal intensity in this tissue is greater than 200, as suggested by Su et al. (2004). The expression breadth of a gene was measured by the number of tissues that the gene is expressed in data of Su et al.
Human PPIs were downloaded from Bossi and Lehner (2009). They collected PPIs from 21 databases and only considered the interactions supported by at least one direct experimental validation. Their data set contained 10,229 proteins and 80,922 interactions. According to their definition, a PPI may occur in a tissue only if both of the interacting proteins are expressed in the same tissue (i.e., mRNA intensities of two interacting genes are both greater than 200). Because a PPI in our study could be present in at most 73 tissues, we defined narrowly expressed PPIs (“EB_Narrow”) as PPIs that are present in less than 15 tissues (∼20% of the total number of tissues considered). For the PPI analysis, we only considered proteins that at least one of their interacting partners is also present in our data set (5,124 proteins remain). To increase the reliability of our result, we used two definitions of hub proteins: 1) top 20% (PPIs across all tissues >18) and 2) top 35% (PPIs across all tissues >10) highly connected proteins (supplementary table S1, Supplementary Material online).
Experimentally validated phosphorylation sites of human proteins were obtained from Chen et al. (2010). This data set contained 15,914 phosphorylation sites on 4,398 proteins, collected from UniprotKB (http://www.uniprot.org/), PhosphoELM (Diella et al. 2008), and HPRD (Keshava Prasad et al. 2009).
miRNA Target Prediction
Human miRNA target predictions were downloaded from TargetScanHuman at http://www.targetscan.org/ (release 5.1), one of the most widely used miRNA target prediction tools. To obtain more reliable prediction results, we only considered miRNAs whose target sites are conserved across most mammals (defined by TargetScan). The complexity (level) of miRNA regulation of a gene was measured by the total number of distinct miRNA types by which the 3′UTR of this gene was targeted (NmiR).
Prediction of Disordered Residues and Protein Disorder Content
We predicted disorder potential for each residue of human proteins using DISOPRED2 (version 2.4) with default parameters (5% for false positive threshold) (Ward et al. 2004). DISPPRED2 is one of the top-ranking disorder prediction tools and has a lower level of false positive rate (Moult et al. 2007). The disorder content (i.e., percentage of IDRs, denoted “DisCont”) of each protein was estimated by dividing the number of disordered residues by the protein length. Proteins with <100 amino acids in length were not considered in the study because the estimate of disorder content in a short protein is subject to a large standard error. We classified proteins into three groups of similar size, according to their DisCont: lowly intrinsically disordered (DisCont < 18%; 3,258 proteins), moderately intrinsically disordered (18% ≤ DisCont < 43%; 3,372 proteins), and highly intrinsically disordered (DisCont ≥ 43%; 3,164 proteins), as described in Gsponer et al. (2008). To examine how disorder content influences the evolution of proteins and how it is related to miRNA regulation, we grouped lowly and moderately intrinsically disordered proteins together as L-IDPs and compared the evolution of L-IDPs with that of highly intrinsically disordered proteins (H-IDPs). L-IDPs are lumped together because they evolve at similar rates (P value = 0.67, Wilcoxon rank-sum test with continuity correction; supplementary fig. S1, Supplementary Material online), whereas H-IDPs evolve faster.
Results
The Number of miRNA Types Is a Stronger Indicator of Evolutionary Rate Than Disorder Content
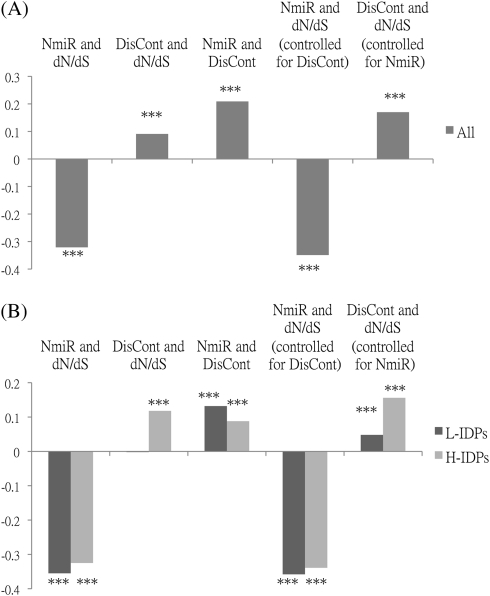
Previous reports have suggested that IDRs and miRNA regulation have opposite effects on protein evolutionary rate (dN/dS) (Kim et al. 2008; Cheng et al. 2009). But which factor is a better indicator of dN/dS? To answer this question, we compile a list of 9,794 human–mouse 1:1orthologous gene pairs to study the relationships between disorder content of the protein (DisCont) and dN/dS and between the number of miRNA types that regulate the gene (NmiR) and dN/dS. In agreement with previous results, dN/dS is negatively correlated with NmiR (Cheng et al. 2009) but positively correlated with DisCont (Kim et al. 2008) (fig. 1A and supplementary table S2, Supplementary Material online). Interestingly, the absolute value of the former correlation is nearly three times that of the latter. Because NmiR and DisCont are also correlated (Edwards et al. 2009), we compute the partial correlation between NmiR and dN/dS and that between DisCont and dN/dS by controlling for DisCont and NmiR, respectively. Interestingly, the partial correlation of DisCont almost doubles, whereas that of NmiR increases only slightly (fig. 1A and supplementary table S2, Supplementary Material online). However, the latter is still twice larger in absolute value than the former. Thus, NmiR is a far stronger indicator of evolutionary rate than DisCont. Also, the strength of DisCont as a rate indicator is reduced due to the correlation between DisCont and NmiR.
FIG. 1.
Spearman's rank correlations between evolutionary rate (dN/dS) and disorder content (DisCont) or the number of miRNA types (NmiR). (A) All: all proteins under study (9,794 proteins). (B) L-IDPs: lowly and moderately intrinsically disordered proteins (DisCont < 43%; 6,630 proteins); H-IDPs: highly intrinsically disordered proteins (DisCont ≥ 43%; 3,164 proteins). DisCont: the number of disordered residues divided by the protein length; NmiR: the number of miRNA types that regulate the gene under study. Significance:*P value ≤ 0.05, **P value ≤ 0.001,***P value ≤ 0.0001.
dN/dS Is Not Always Positively Correlated With Disorder Content
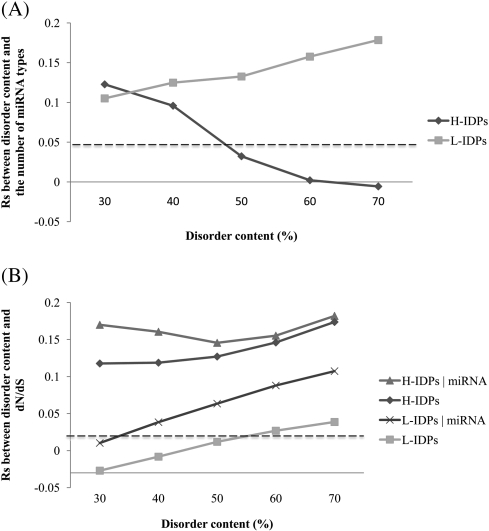
Previous findings that proteins with more IDRs tend to evolve faster than those with fewer or no IDRs (Kim et al. 2008) and that transcripts encoding proteins with more IDRs have a higher level of miRNA regulation (Liang and Li 2007) need clarification because genes with a higher level of miRNA regulation were reported to evolve more slowly (Cheng et al. 2009). To disentangle the related effects of DisCont and NmiR, we divide proteins into two groups, “H-IDPs” and “L-IDPs,” according to whether the DisCont is higher or lower than 43% (see Materials and Methods). Our correlation analyses reveal that dN/dS and NmiR are correlated in both groups, whereas dN/dS and DisCont are not correlated in L-IDPs (fig. 1B and supplementary table S2, Supplementary Material online). We next examine whether this lack of correlation is because NmiR has an opposite and stronger effect on dN/dS than does DisCont and because the correlation between NmiR and DisCont is much stronger in L-IDPs than in H-IDPs (fig. 1B and supplementary table S2, Supplementary Material online). We compute the partial correlations between dN/dS and DisCont for both groups by controlling for NmiR. Indeed, the partial correlation between dN/dS and DisCont in L-IDPs increases and becomes significant (fig. 1B and supplementary table S2, Supplementary Material online). We also try different disorder content thresholds for classifying the two groups and find similar results (fig. 2). Our results indicate that among less disordered proteins, those with higher disorder content do not have an elevated evolutionary rate if NmiR is not controlled.
FIG. 2.
Spearman's rank correlations (Rs) between disorder content and the number of miRNA types and between disorder content and evolutionary rate (dN/dS). Proteins are divided into “H-IDPs” and “L-IDPs” groups according to whether the disorder content is higher or lower than the given threshold (the x axis). For example, proteins in the “H-IDPs” group and the “L-IDPs” group with the threshold of 50% are the groups with a disorder content higher and lower than 50%, respectively. Each point stands for the Rs value of the corresponding group. Five disorder thresholds are examined here to see how the correlations change under different thresholds. In (B), “H-IDPs|miRNA” and “L-IDPs|miRNA” represent the partial correlations (controlled for the number of miRNA types that regulate the gene) for the H-IDPs and L-IDPs groups, respectively. In both (A) and (B), points above the dotted line represent correlations significantly greater than zero by Spearman's rank test (P < 0.05).
The Number of Highly Disordered Interaction Partners of a Protein and dN/dS Are Negatively Correlated
In addition to NmiR and DisCont, other factors (PPIs, gene expression level, and gene expression breadth) have been reported to be indicators of dN/dS (Bloom and Adami 2004; Fraser and Hirsh 2004; Drummond and Wilke 2008; Park and Choi 2010). Moreover, proteins having phosphorylated resiudes may evolve more slowly because they may be more functionally important: phosphorylated and nonphosphorylated forms can perform different functions or be transported to different localizations (Cohen 2000). Interestingly, the above indicators are interrelated (Liao et al. 2006; Liang and Li 2007; Cheng et al. 2009). To disentangle their relationship and assess the relative strength of each factor, we compute the partial correlation of each factor with dN/dS by controlling for the remaining factors. Proteins without PPI data are excluded to measure and control the effects of PPIs (5,124 proteins remain; see Materials and Methods). In addition to the total number of PPIs (PPIAll), two other indices are examined: the number of H-IDPs with which a protein interacts (PPIH-IDP) and the number of L-IDPs with which a protein interacts (PPIL-IDP) because features of the interaction partners of a protein may affect its evolutionary rate (Makino and Gojobori 2006).
When the relationships among these interrelated factors are not considered, all the indicators are negatively correlated with dN/dS, except that DisCont is positively correlated with dN/dS. However, only NmiR (ρ=−0.32), DisCont (ρ=−0.21), and PPIH-IDP (ρ=−0.13) remain as strong rate indicators after controlling for the other factors (table 1 and supplementary table S3, Supplementary Material online). The strengths of gene expression breadth and expression level become weak and that of PPIL-IDP, PPIAll, and the number of phosphorylation (NPhospho) disappear. Moreover, all the indicators, except for NmiR, have different strengths in L-IDPs and in H-IDPs. For example, strengths of DisCont and PPIH-IDP in H-IDPs are twice the corresponding values in L-IDPs; the strengths of expression level and expression breadth are significant in L-IDPs but disappear in H-IDPs (table 1 and supplementary table S3, Supplementary Material online). Unexpectedly, PPIL-IDP and PPIAll become as positive rate indicators in H-IDPs after controlling for the other factors (table 1 and supplementary table S3, Supplementary Material online). This suggests that the negative correlation between PPIAll (and also PPIL-IDP) and dN/dS is mainly because PPIAll (and also PPIL-IDP) is positively correlated with PPIH-IDP (ρ=0.77 for PPIAll; ρ=0.49 for PPIL-IDP) and because PPIH-IDP is a strong negative indicator. Therefore, proteins that interact with more H-IDPs tend to evolve more slowly.
Table 1.
Spearman's Rank Correlation Between an Indicator and Evolutionary Rate After Controlling for the Other Indicators.
| Indicator | Before Control |
After Controlk |
||||
| Alla | L-IDPsa | H-IDPsa | Alla | L-IDPsa | H-IDPsa | |
| NmiRb | −0.298*** | −0.344*** | −0.293*** | −0.316*** | −0.329*** | −0.297*** |
| DisContc | 0.113*** | 0.026 | 0.134*** | 0.209*** | 0.081*** | 0.197*** |
| PPIH-IDPd | −0.196*** | −0.206*** | −0.241*** | −0.128*** | −0.10*** | −0.201*** |
| PPIL-IDPe | −0.124*** | −0.156*** | −0.056* | 0.002 | −0.028 | 0.074* |
| PPIAllf | −0.171*** | −0.188*** | −0.160*** | 0.001 | −0.022 | 0.074* |
| NPhosphog | −0.060*** | −0.116*** | −0.021 | 0.001 | −0.031 | 0.030 |
| Expbreadthh | −0.110*** | −0.153*** | −0.016 | −0.017 | −0.055* | 0.045 |
| Explevel_xingi | −0.150*** | −0.178*** | −0.078** | −0.058*** | −0.085*** | −0.017 |
| Explevel_Suj | −0.129*** | −0.168*** | −0.038 | −0.049** | −0.067** | −0.044 |
All: all proteins under study that have PPI data (5,124 proteins); L-IDPs (DisCont < 43%; 3,300 proteins); and H-IDPs (DisCont ≥ 43%; 1,824 proteins).
NmiR: The number of miRNA types that regulate the gene under study.
DisCont: Disorder content, the number of disordered residues divided by protein length.
PPIH-IDP: The number of H-IDPs with which a protein interacts.
PPIL-IDP: The number of L-IDPs with which a protein interacts.
PPIAll: The total number of PPIs.
NPhospho: The number of experimentally verified phosphorylation sites.
Expbreadth: The number of tissues that a gene is expressed in data of Su et al.
Explevel_xing: The average signal intensity of a gene in data of Xing et al.
Explevel_Su: The average signal intensity of a gene in data of Su et al.
Partial correlation between evolutionary rate and the indicator under study by controlling for NmiR, PPIH-IDP, DisCont, PPIAll, NPhospho, Expbreadth, and Explevel_xing, except for the indicator under study.
Significance: *P value < 0.05, **P value ≤ 0.001, and ***P value ≤ 0.0001.
The Number of Phosphorylation Sites and the Number of miRNA Types Are Weakly Correlated
Phosphorylation plays an important role in protein function, stability, and localization (Cohen 2000). Therefore, it may be interesting to see if proteins with more phosphorylation sites are under more complex miRNA regulation. On the basis of experimentally validated phosphorylation sites, we find that the number of phosphorylation sites (NPhospho) and NmiR are positively correlated in both H-IDPs and L-IDPs (table 2). Because two-third of the proteins under study have no identified phosphorylation sites, we remove proteins without any experimentally validated phosphorylation sites (2,885 proteins remain). Now a positive correlation between NPhospho and NmiR is observed only in L-IDPs. Moreover, the partial correlations between NPhospho and NmiR in all proteins and in L-IDPs become weak after controlling for other factors (table 2), suggesting a weak tendency for proteins with more phosphorylation sites to have more complex miRNA regulation.
Table 2.
Spearman's Rank Correlation Between the Number of miRNA Types (NmiR) and the Number of Phosphorylation Sites (NPhospho).
| Before Control |
After Controlb |
|||||
| Alla | L-IDPsa | H-IDPsa | Alla | L-IDPsa | H-IDPsa | |
| All proteins (9,794 proteins) | 0.172*** | 0.123*** | 0.169*** | 0.076*** | 0.057*** | 0.085*** |
| Proteins with verified phosphorylation sites (2,885 proteins) | 0.109*** | 0.113*** | 0.022 | 0.044* | 0.073* | 0.001 |
All: all proteins under study; L-IDPs (DisCont < 43%; 3,300 proteins); and H-IDPs (DisCont ≥ 43%; 1,824 proteins).
Partial correlations: the partial correlation between NmiR and NPhospho is controlled for disorder content, total number of PPIs, expression breadth, expression level, and evolutionary rate.
Significance: *P value ≤ 0.05, **P value ≤ 0.001, and ***P value ≤ 0.0001.
The Number of miRNA Types and the Number of Tissue-Specific PPIs Are Weakly Correlated
It has been reported that hub proteins are subject to more complex miRNA regulation than non-hub proteins (Liang and Li 2007). However, whether the expression patterns of their PPIs affect the complexity of their miRNA regulation remains unclear. Because housekeeping proteins tend to have more PPIs (Lin et al. 2009) and have been suggested to be reused for tissue-specific processes (Bossi and Lehner 2009), we hypothesize that hub proteins with more tissue-specific PPIs need tighter regulation (i.e., under more complex miRNA regulation). To test this hypothesis, we focus on hub proteins (total number of interactions across all tissues >18) and calculate the number of their interactions that are narrowly expressed (EB_Narrow) (see Materials and Methods). We find that L-hubs (DisCont < 43%) and H-hubs (DisCont ≥ 43%), on average, have similar number of EB_Narrow PPIs (P value > 0.05 by Wilcoxon rank-sum test). However, the positive correlation between NmiR and the number of EB_ Narrow PPIs is only weakly significant in L-hubs (table 3). The trend remains when we control for other factors or choose another connectivity threshold for the definition of hubs (total number of interactions across all tissues >10) (table 3). In addition, we find that expression breadth and disorder content are slightly negatively correlated (ρ = −0.06; P value < 1.42 × 10−5by Spearman's correlation test), suggesting a slight tendency for highly disordered proteins to have a higher level of tissue specificity. These results indicate that the expression breadth or disorder content of the interacting partners of less disordered hubs may slightly increase the complexity of their miRNA regulation.
Table 3.
Spearman's Rank Correlation Between the Number of miRNA Types (NmiR) and the Number of Narrowly Expressed PPIsa in Hubs.
| Hub Definition | Before Control |
After Controlc |
||||
| All Hubs | L-Hubsb | H-Hubsb | All Hubs | L-Hubsb | H-Hubsb | |
| PPIs > 18 | 0.100** | 0.166*** | −0.052 | 0.104** | 0.155*** | −0.014 |
| PPIs > 10 | 0.024 | 0.064* | −0.057 | 0.042 | 0.072* | −0.025 |
The number of PPIs that are expressed in less than 15 tissues in data of Su et al.
L-hubs: hubs with DisCont < 43%; H-hubs: hubs with DisCont ≥ 43%.
The partial correlation is controlled for disorder content, the total number of PPIs, the number of phosphorylation sites, expression breadth, expression level, and evolutionary rate.
Significance: *Pvalue ≤ 0.05, **P value ≤ 0.001, and ***P value ≤ 0.0001.
Discussion
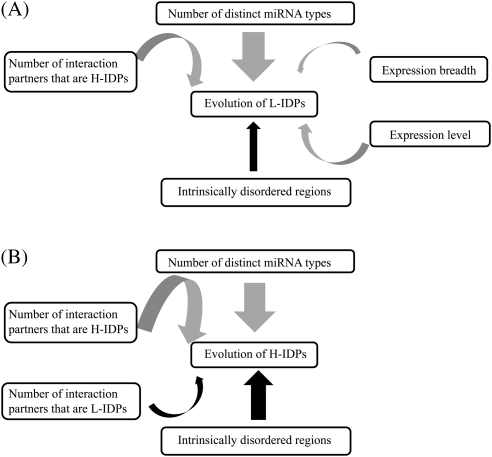
Prior studies have reported factors that can affect the evolutionary rate of proteins (e.g., the number of miRNA types, expression level, expression breadth, gene compactness, extracellularity and protein folding pattern) (Fraser et al. 2002; Drummond et al. 2005; Liao et al. 2006, 2010; Makino and Gojobori 2006; Pal et al. 2006; Drummond and Wilke 2008; Yang, Zhuang, et al. 2010). However, as some factors are correlated, the relative influence of a factor on protein evolution remains unclear. In this study, we clarify this issue by computing the partial correlation between a factor and protein evolutionary rate by controlling for the other factors. We summarize these correlations in fig. 3. We find that the number of miRNA types is the strongest indicator and its correlations with other indicators only mildly affect its strength. One possible scenario is that genes with higher number of miRNA types are more likely to be pleiotropic. Pleiotropic genes have been suggested to encode multifunctional products, which can be involved in different biological processes or have more interacting partners (He and Zhang 2006). Therefore, pleiotropic genes may need more precise regulation of gene expression, which is associated with a more complex miRNA regulation. Also, the multifunctional property of pleiotropic genes may impose stronger selective constraints on their sequence evolution, leading to the observation that genes with higher number of miRNA types evolve more slowly. Future studies on the relationships among miRNA regulation, pleiotropy, and protein evolution may help to examine this scenario.
FIG. 3.
Rate indicators of protein evolution in (A) L-IDPs and in (B) H-IDPs. Gray and black arrows, respectively, represent negative and positive correlations. The thickness of a line between two indicators indicates the strength of their correlation.
Because L-IDPs (lowly and moderately intrinsically disordered proteins) are more than 50% ordered, dN/dS may be dominated by the evolutionary pressures in the ordered regions in L-IDPs. Indeed, for lowly disordered proteins (DisCont < 18%; 3,258 proteins), disorder content and dN/dS are not correlated even after miRNA regulation is controlled (supplementary fig. S2, Supplementary Material online). The lack of correlation may suggest that dN/dS is dominated by constraints on ordered regions in a protein that is more than 72% ordered. For moderately disordered proteins (18% ≤ DisCont < 43%; 3,372 proteins), however, disorder content and dN/dS are weakly correlated before control, and the correlation almost doubles when miRNA regulation is controlled (supplementary fig. S2, Supplementary Material online). Hence, the result indicates that disorder content indeed affects dN/dS even when proteins are 57–82% ordered. The increase in correlation is mainly because disorder content and miRNA regulation are correlated and because miRNA regulation has an inversely stronger effect on evolutionary rate than disorder content.
For H-IDPs (disorder content ≥ 43%), however, disorder content is positively correlated with evolutionary rate even without controlling for miRNAs or other factors. This is mainly because the correlation between disorder content and the level of miRNA regulation is weaker and the strength of disorder content is stronger in H-IDPs, compared with that in L-IDPs. Thus, our results may explain the previous finding that proteins with a disorder content higher than 50% evolve faster than those with a disorder content lower than 50% (Kim et al. 2008). Furthermore, the less long-range interactions between residues in the protein in H-IDPs may lead to an elevated rate of sequence evolution. Long-range interactions in a protein are interactions between residues that are in contact with each other in the protein native structure but are distant to each other in the primary sequence (Gromiha and Selvaraj 2004). The long-range interactions between residues in a protein play a critical role in its folding, stability, and function, such as ligand binding, conformational changes, and catalysis (Gromiha and Selvaraj 1999, 2004; Yang, Welch, et al. 2010). Residue changes involving long-range interactions may be deleterious. Thus, the less long-range interactions in disordered proteins than in ordered proteins make changes in disordered proteins have a higher likelihood to survive in evolution.
In addition, we find that proteins interacting with more H-IDPs tend to evolve more slowly, shedding light on the functional role of the high disorder content of a protein's interacting partners. The lack of well-defined 3D structures of IDRs may create exposed hydrogen bonds for IDPs, which may increase the risk of protein aggregation (Fernandez and Chen 2009) or increase the ability to form protein complex with other H-IDPs. It has been shown that proteins that are involved in complex formation evolve more slowly than those that are not (Manna et al. 2009). Therefore, increasing disorder content is one way to increase interactions and the tendency of complex formation, which in turn constrains evolutionary rate. However, a high disorder content of its interacting partners could be another way for a protein to increase its “disorderness” without increasing its own disorder content. Through its interaction with a H-IDP, a protein could participate in complex formation or interact with other H-IDPs via the interactions between this H-IDP and other H-IDPs (the “hitchhiking effect” or “connections through connections”). The gain of function by interacting with H-IDPs may in turn impose selective constraints on the protein sequence evolution. Thus, the more H-IDPs that a protein interacts, the more likely it evolves slowly. This may explain the previous observation of a negative correlation between evolutionary rate and the total number of PPIs (Fraser et al. 2002) because the total number of PPIs and the number of highly disordered interaction partners of a protein are positively correlated (ρ = 0.77; P value < 2.2 × 10−16 by Spearman's correlation test). Unexpectedly, highly disordered proteins that interact with more L-IDPs tend to evolve faster. One possible scenario is that when the number of L-IDPs with which an H-IDP interacts increases, its chance to interact with other H-IDPs may be reduced.
We notice that the partial correlations between evolutionary rate and expression breadth and between evolutionary rate and expression level are not significant in H-IDPs. The trend remains similar when expression specificity (τ), defined in Yanai et al. (2005), is used, although for L-IDPs, the strength of expression specificity as a rate indicator is stronger than that of expression breath (supplementary tables S4 and S5, Supplementary Material online). There are two possible reasons. First, the expression levels of H-IDPs, on average, are lower than those of L-IDPs (supplementary fig. S3, Supplementary Material online). Their lower expression reduces the strength of expression breadth as an indicator of evolutionary rate because whether they are present in a tissue becomes difficult to determine and because they are more narrowly expressed (supplementary fig. S3, Supplementary Material online). Second, the resolution of microarray is not high enough to distinguish between expression levels among H-IDPs because of their lower expression and the smaller expression variance (P value < 1.65 × 10−6 by the Brown–Forsythe test) compared with those in L-IDPs. Future technologies with a higher resolution power may help to overcome this problem.
In our study, we did not remove orthologues with <50% sequence identity as commonly done in the literature. To see whether fast evolving proteins dominate our results, we also conducted the analyses only on those orthologues with <50% sequence identity. We found that only miRNA regulation complexity and protein disorder are the strong rate indicators in H-IDPs, whereas only expression breadth is a strong rate indicator in L-IDPs (supplementary tables S6 and S7, Supplementary Material online). However, because the number of fast evolving proteins is small compared with that of all proteins (319 vs. 9,794 for the study of the correlation of evolutionary rate with miRNA and with disorder content; 111 vs. 5,124 for the study of the relative strength of each rate indicator), our conclusions are not affected by fast evolving proteins.
We find that phosphorylated proteins tend to have a higher level of miRNA regulation of the gene and that the number of phosphorylation sites of a protein is correlated with the level of miRNA regulation in L-IDPs. There are two possible reasons. First, a protein with multiple phosphorylation sites may be recognized by different kinases, so it may be involved in various signaling pathways, may be transported to different cellular localizations, and may have distinct conformations (Cohen 2000; Gsponer et al. 2008). Second, phosphorylation could regulate protein half-life by resisting or promoting protein degradation (Lin et al. 2006). Thus, proteins with phosphorylation sites are dynamic in time and space in response to environmental conditions and developmental stages. miRNAs may serve as a mechanism to fine-tune the expression level of these functionally important proteins.
We also show that the number of tissue-specific PPIs is weakly but positively correlated with the level of miRNA regulation for hub proteins with disorder content <43%. Because miRNAs can be expressed in a tissue-specific manner, transcripts that need complex expression patterns to be reused as core modules of the PPI network may need a higher level of miRNA regulation to perform tissue-specific functions. In addition, because H-IDPs tend to be narrowly expressed, they may participate in network modularity and tissue-specific functions with hub proteins.
Supplementary Material
Supplementary tables S1–S7 and figures S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank the two reviewers and Dr. Ben-Yang Liao for valuable suggestions. This study was supported by National Science Council (NSC99-2628-B-001-009-MY3) and (NSC99-2628-B-001-008-MY3) and National Institute of Health grants (GM30998 and 5R01MH080425).
References
- Bloom JD, Adami C. Evolutionary rate depends on number of protein-protein interactions independently of gene expression level: response. BMC Evol Biol. 2004;4:14. doi: 10.1186/1471-2148-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Mol Syst Biol. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. 2010;27:609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55:104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- Chen FC, Chen CJ, Li WH, Chuang TJ. Gene family size conservation is a good indicator of evolutionary rates. Mol Biol Evol. 2010;27:1750–1758. doi: 10.1093/molbev/msq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FC, Chuang TJ. The effects of multiple features of alternatively spliced exons on the K(A)/K(S) ratio test. BMC Bioinformatics. 2006;7:259. doi: 10.1186/1471-2105-7-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liang H, Fernandez A. Protein structure protection commits gene expression patterns. Genome Biol. 2008;9:R107. doi: 10.1186/gb-2008-9-7-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Chen FC, Li WH. Phosphorylated and nonphosphorylated serine and threonine residues evolve at different rates in mammals. Mol Biol Evol. 2010;27:2548–2554. doi: 10.1093/molbev/msq142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Bhardwaj N, Gerstein M. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics. 2009;10:431. doi: 10.1186/1471-2164-10-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites—update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards YJ, Lobley AE, Pentony MM, Jones DT. Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Biol. 2009;10:R50. doi: 10.1186/gb-2009-10-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Chen J. Human capacitance to dosage imbalance: coping with inefficient selection. Genome Res. 2009;19:2185–2192. doi: 10.1101/gr.094441.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE. Evolutionary rate depends on number of protein-protein interactions independently of gene expression level. BMC Evol Biol. 2004;4:13. doi: 10.1186/1471-2148-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- Gromiha MM, Selvaraj S. Importance of long-range interactions in protein folding. Biophys Chem. 1999;77:49–68. doi: 10.1016/s0301-4622(99)00010-1. [DOI] [PubMed] [Google Scholar]
- Gromiha MM, Selvaraj S. Inter-residue interactions in protein folding and stability. Prog Biophys Mol Biol. 2004;86:235–277. doi: 10.1016/j.pbiomolbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34:305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Toward a molecular understanding of pleiotropy. Genetics. 2006;173:1885–1891. doi: 10.1534/genetics.106.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, et al. (30 co-authors) Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Sboner A, Xia Y, Gerstein M. The role of disorder in interaction networks: a structural analysis. Mol Syst Biol. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Li WH. MicroRNA regulation of human protein protein interaction network. RNA. 2007;13:1402–1408. doi: 10.1261/rna.634607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- Liao BY, Weng MP, Zhang J. Impact of extracellularity on the evolutionary rate of mammalian proteins. Genome Biol Evol. 2010;2:39–43. doi: 10.1093/gbe/evp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, Rustgi A, Fuchs SY, Diehl JA. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Liu WC, Hwang MJ. Topological and organizational properties of the products of house-keeping and tissue-specific genes in protein-protein interaction networks. BMC Syst Biol. 2009;3:32. doi: 10.1186/1752-0509-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, Gojobori T. The evolutionary rate of a protein is influenced by features of the interacting partners. Mol Biol Evol. 2006;23:784–789. doi: 10.1093/molbev/msj090. [DOI] [PubMed] [Google Scholar]
- Manna B, Bhattacharya T, Kahali B, Ghosh TC. Evolutionary constraints on hub and non-hub proteins in human protein interaction network: insight from protein connectivity and intrinsic disorder. Gene. 2009;434:50–55. doi: 10.1016/j.gene.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69(Suppl 8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ. An integrated view of protein evolution. Nat Rev Genet. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- Park SG, Choi SS. Expression breadth and expression abundance behave differently in correlations with evolutionary rates. BMC Evol Biol. 2010;10:241. doi: 10.1186/1471-2148-10-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, et al. (13 co-authors) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Xing Y, Ouyang Z, Kapur K, Scott MP, Wong WH. Assessing the conservation of mammalian gene expression using high-density exon arrays. Mol Biol Evol. 2007;24:1283–1285. doi: 10.1093/molbev/msm061. [DOI] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, et al. (12 co-authors) Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yang JR, Zhuang SM, Zhang J. Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Mol Syst Biol. 2010;6:421. doi: 10.1038/msb.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Welch JL, Arnold JJ, Boehr DD. Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry. 2010;49:9361–9371. doi: 10.1021/bi100833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.