Abstract
Mitochondrial genomes often contain large amounts of plastid DNA (ptDNA)-derived sequences (MTPTs). It has been suggested that the intercompartmental transfer of ptDNA is greatly reduced in species with only a single plastid per cell (monoplastidic) as compared with those with many plastids per cell (polyplastidic). This hypothesis has not been applied to the movement of DNA from plastids to mitochondria. By analyzing the organelle genomes from diverse mono- and polyplastidic taxa, I show that MTPTs are restricted to the mitochondrial genomes of species with many plastids per cell and are absent from those with one plastid per cell or with monoplastidic meristematic systems. Moreover, the most bloated mitochondrial genomes that were explored had the largest MTPT contents. These data, like previous results on ptDNA-derived sequences in nuclear genomes, support the hypothesis that plastid number and the forces governing the expansion and contraction of noncoding mitochondrial DNA (mtDNA) influence MTPT abundance. I also show that plastid genomes are depauperate in mtDNA-derived sequences (PTMTs), irrespective of the number of mitochondria per cell and plastid genome size, which may reflect the lack of a DNA uptake system in plastids.
Keywords: chloroplast, mitochondria, NUPT, NUMT, MTPT, PTMT
The cells of most plastid-bearing eukaryotes have three genetic compartments (mitochondrial, plastid, and nuclear), giving six possible directions for intercompartmental DNA transfer (Timmis et al. 2004; Kleine et al. 2009). The migration of mitochondrial, plastid, and nuclear DNA (mtDNA, ptDNA, and nucDNA) among these compartments can result in the functional relocation of genes, as exemplified in early mitochondrial and plastid evolution when there was an exodus of organelle genes to the nuclear genome (Gray et al. 1999; Archibald 2009). However, the majority of contemporary intracellular DNA transfer events give rise to noncoding sequences (Lopez et al. 1994; Richly and Leister 2004a, 2004b), which have accumulated to high levels in the organelle and nuclear genomes of some eukaryotic species but are conspicuously absent from those of others (Hazkani-Covo et al. 2010; Smith et al. 2011).
The limited transfer window hypothesis argues that species with a single plastid per cell (monoplastidic) experience less intercompartmental ptDNA transfer than those with many plastids per cell (polyplastidic) because they have fewer plastids to donate ptDNA, and lysis of the plastid would almost certainly result in death to the cell, unlike the case for polyplastidic taxa (Barbrook et al. 2006). Support for this hypothesis has come from the observation that the nuclear genomes of monoplastidic species contain few, if any, ptDNA-derived sequences (NUPTs), whereas those from polyplastidic taxa abound with NUPTs (Lister et al. 2003; Martin 2003; Richly and Leister 2004b; Lane 2011; Smith et al. 2011). The limited transfer window hypothesis has not yet been applied to the movement of ptDNA from plastids to mitochondria.
Mitochondrial genomes are often riddled with ptDNA-derived sequences, called mitochondrial plastid DNAs (MTPTs) (Ellis 1982; Knoop 2004; Wang et al. 2007). For example, ∼5% of both the rice and the corn mtDNAs are comprised of MTPTs, which are distributed among ∼30 different regions (Notsu et al. 2002; Clifton et al. 2004). Conversely, the mtDNAs from the liverwort Marchantia polymorpha and the plastid-bearing protists Chlamydomonas reinhardtii and Plasmodium falciparum contain no MTPTs (Gray and Boer 1988; Oda et al. 1992; Vaidya and Mather 2009). And although they can be active donors, plastid genomes from eukaryotes as a whole are typically depauperate in mtDNA-like sequences (PTMTs) (Kleine et al. 2009). The reasons why some mitochondrial genomes harbor ptDNA-like sequences, whereas others do not, remain unclear.
To accurately identify plastid-derived DNA within a mitochondrial genome, complete mtDNA and ptDNA sequences are required. In the past, the small number of species for which both of these data were available prevented broad-scale analyses of MTPTs. Here, I take advantage of a recent increase in the number of sequenced mitochondrial and plastid genomes from diverse mono- and polyplastidic taxa to explore the diversity in MTPT content among plastid-containing eukaryotes and the effect that plastid number has on inter-organelle ptDNA transfer.
As of 1 March 2011, there are 42 species in GenBank’s Entrez Organelle Genome Database with completely sequenced mitochondrial and plastid DNAs: 17 green algae, 15 land plants, 4 apicomplexans, 2 red algae, 1 glaucophyte, 1 cryptophyte, 1 haptophyte, and 1 stramenopile (table 1 and supplementary table S1, Supplementary Material online). Of these, 29 are monoplastidic or effectively so and 13 are polyplastidic. Twelve of the monoplastidic species are also monomitochondrial (the remaining 30 taxa are polymitochondrial). The number of organelles per cell for each of these species and the references used to determine these statistics are listed in table 1 and supplementary table S2 (Supplementary Material online), respectively. The haptophyte Emiliania huxleyi and the stramenopile Thalassiosira pseudonana can have up to two plastids per cell but for simplicity were treated here as monoplastidic. The lycophyte Selaginella moellendorffii, the moss Physcomitrella patens, and the liverwort M. polymorpha are polyplastidic, but for the purpose of this study, they were considered “effectively monoplastidic” because meiosis only occurs in cells that contain a single plastid (i.e., they have monoplastidic meristematic systems) (Brown and Lemmon 1990, 1997; Rudall and Bateman 2007).
Table 1.
Number and Total Amount (in Kilobases) of MTPTs and PTMTs in the Available Organelle Genome Sequences from Plastid-Harboring Eukaryotes
| Taxon | # Plastids per Cella | # Mitochondria per Cella | MTPTs |
PTMTs |
||||
| Number of Blast Hitsb | Accumulative Length (kb) | Average Length (kb) | Number of Blast Hitsb | Accumulative Length (kb) | Average Length (kb) | |||
| Land plants | ||||||||
| Arabidopsis thaliana | Multiple | Multiple | 12 | 2.2 | 0.18 | 0 | 0 | 0 |
| Brassica napus | Multiple | Multiple | 14 | 5.4 | 0.39 | 0 | 0 | 0 |
| Carica papaya | Multiple | Multiple | 17 | 15.1 | 0.89 | 0 | 0 | 0 |
| Cycas taitungensis | Multiple | Multiple | 25 | 7.4 | 0.3 | 0 | 0 | 0 |
| Marchantia polymorpha | Effectively monoplastidicc | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Nicotiana tabacum | Multiple | Multiple | 28 | 8.4 | 0.3 | 0 | 0 | 0 |
| Oryza sativa Indica | Multiple | Multiple | 27 | 12.8 | 0.47 | 0 | 0 | 0 |
| Oryza sativa Japonica | Multiple | Multiple | 27 | 12.9 | 0.48 | 0 | 0 | 0 |
| Physcomitrella patens | Effectively monoplastidicc | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Selaginella moellendorffii | Effectively monoplastidicc | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Sorghum bicolor | Multiple | Multiple | 33 | 15.0 | 0.45 | 0 | 0 | 0 |
| Triticum aestivum | Multiple | Multiple | 26 | 8.9 | 0.34 | 0 | 0 | 0 |
| Vigna radiata | Multiple | Multiple | 10 | 1.4 | 0.14 | 0 | 0 | 0 |
| Vitis vinifera | Multiple | Multiple | 31 | 15.8 | 0.51 | 0 | 0 | 0 |
| Zea mays | Multiple | Multiple | 31 | 10.3 | 0.33 | 0 | 0 | 0 |
| Green algae | ||||||||
| Chaetosphaeridium globosum | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Chara vulgaris | Multiple | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Chlamydomonas reinhardtii | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Chlorokybus atmophyticus | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Coccoymyxa sp. C-169d | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Dunaliella salina | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Helicosporidium sp. | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Mesostigma viride | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Micromonas sp. RCC299 | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Nephroselmis olivacea | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Oltmannsiellopsis viridis | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Ostreococcus tauri | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Pedinomonas minor | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Pseudendoclonium akinetumd | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Pycnococcus provasolii | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Scenedesmus obliquus | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Volvox carteri | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Red algae | ||||||||
| Cyanidioschyzon merolae | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Porphyra purpurea | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Glaucophyte | ||||||||
| Cyanophora paradoxa | Single | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Apicomplexans | ||||||||
| Babesia bovis | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Eimeria tenella | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Plasmodium falciparum | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Theileria parva | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Haptophyte | ||||||||
| Emiliania huxleyi | 1–2 | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Stramenopile | ||||||||
| Thalassiosira pseudonana | 1–2 | Multiple | 0 | 0 | 0 | 0 | 0 | 0 |
| Cryptophyte | ||||||||
| Rhodomonas salina | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
For references and notes on the number of organelles per cell, see supplementary table S2 (Supplementary Material online).
Blast parameters were as follows: BlastN (v2.2.23) with an expectation value of 0.0001, a word size of 11, match and mismatch scores of 2 and –3, respectively, and gap-cost values of 5 (existence) and 2 (extension). Multiple hits to the same organelle DNA region were counted only once. Regions of organelle DNA that contained tight clusters of MTPTs (i.e., sections of plastid-like DNA interrupted by sequence that did not show identity to ptDNA) were counted as separate hits.
M. polymorpha, P. patens, and S. moellendorffii contain cells that are polyplastidic, but for the purpose of this study, they were considered “effectively monoplastidic” because meiosis only occurs in cells that contain a single plastid (Brown and Lemmon 1990, 1997; Rudall and Bateman 2007).
Similar repeat elements were found in both the mitochondrial and the plastid genomes of these species, but because the origin of these elements are presently unknown, they were not considered in our MTPT/PTMT analyses.
Materials and Methods
Only species (and subspecies) for which complete mitochondrial and plastid genome sequences are available (as of 1 March 2011) (based on GenBank's Entrez Organelle Genome Database: http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome) were included in the MTPT/PTMT analyses. This is because organelle genomes often contain sequences (particularly intergenic and repetitive DNA) that are species specific. Moreover, organelle gene contents can vary significantly among certain lineages, especially green algae. Mitochondrial and plastid genomes were scanned for MTPTs and PTMTs with BlastN (v2.2.23) (Altschul et al. 1990) using an expectation value (E value) of 0.0001, a word size of 11, match and mismatch scores of 2 and –3, and gap-cost values of 5 (existence) and 2 (extension). Hits under 30 nt and showing less than 70% sequence identity to the query were ignored. Spurious hits, such as those where plastid genes matched to homologous genes in the mitochondrial genome (e.g., plastid-encoded ribosomal DNA [rDNA] matching to mitochondrial-encoded rDNA) were ignored. Regions of mtDNA that contained tight clusters of MTPTs (i.e., ptDNA-like sequences interrupted by mitochondrial sequence that did not show sequence identity to ptDNA) were counted as separate hits. Most of the ptDNA-derived sequences that were identified within the mitochondrial genomes represented noncoding DNA or pseudogenes; however, instances of functional relocation of ptDNA-encoded genes (e.g., transfer RNAs) to the mitochondrial genome were considered as MTPTs.
More Plastids, More MTPTs
Complete MTPT statistics for the various plastid-bearing species that were investigated are shown in table 1 and figure 1. None of the monoplastidic or effectively monoplastidic taxa had plastid-like DNA in their mitochondrial genomes, whereas all but one of the polyplastidic species contained MTPTs. The green alga Chara vulgaris was the only polyplastidic species that did not have MTPTs, a feature which may reflect the small number of plastids within its reproductive tissues (1–6) (Pickett-Heaps 1975; Renzaglia and Garbary 2001) and its increased ability to expunge excess mtDNA relative to other polyplastidic species (discussed further below). On average, the polyplastidic species had 9 kb of MTPTs, divided among 20 different loci. The largest numbers of MTPTs were found in the mitochondrial genomes of the common grape Vitis vinifera and the grass Sorghum bicolor, both of which had ∼15 kb of plastid-derived DNA, distributed among ∼30 different regions (table 1 and fig. 1). Unlike plastid number, being mono- or polymitochondrial appeared to have no effect on the number of observed MTPTs within the mitochondrial genomes (table 1). The results presented here on MTPTs are consistent with previous investigations on ptDNA-like sequences in nuclear genomes (NUPTs) (Smith et al. 2011), which showed that polyplastidic taxa had on average 80 times more NUPTs than species that were monoplastidic or effectively so. Together, the MTPT and NUPT data support the hypothesis that monoplastidy greatly reduces the potential for intercompartmental ptDNA migration.
FIG. 1.—
Bar graph showing the number and accumulative length of ptDNA-derived sequences (MTPTs) within the mitochondrial genomes of various plastid-harboring eukaryotes. For references and notes on the number of organelles per cell, see table 1 and supplementary table S2 (Supplementary Material online).
More Genome, More MTPTs
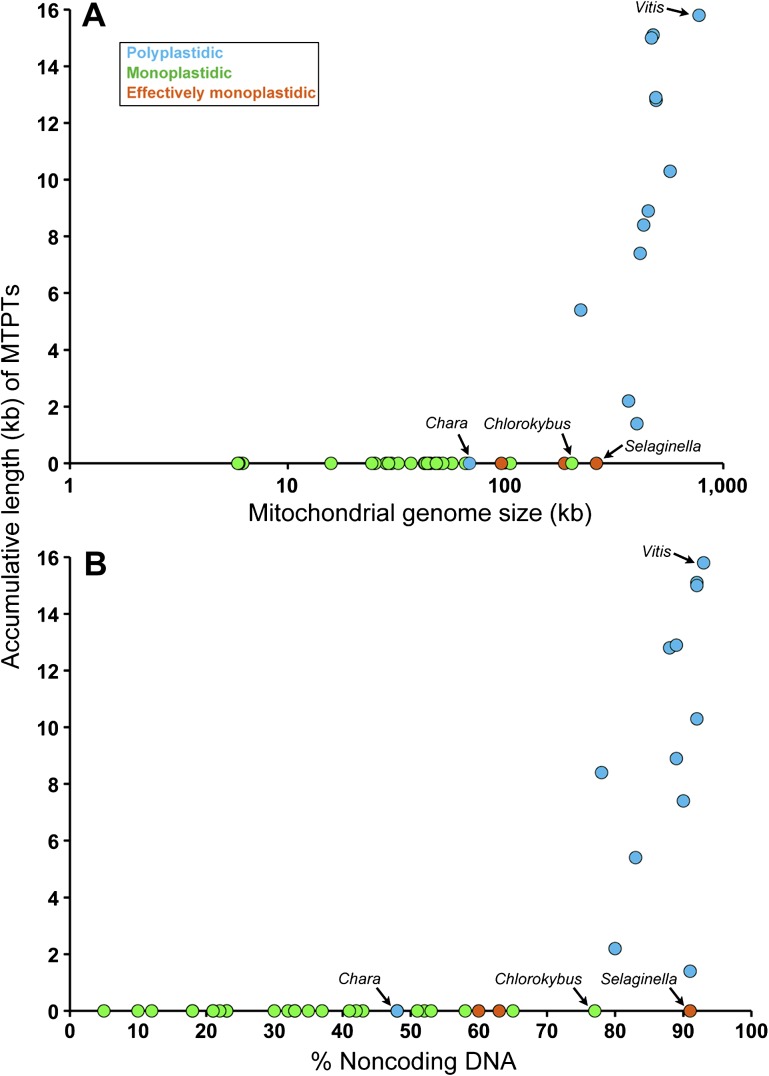
MTPT abundance scaled exponentially with both mitochondrial genome size and mtDNA noncoding content (fig. 2A and B, respectively). The most bloated mtDNAs that were explored—those belonging to the angiosperms V. vinifera, S. bicolor, Carica papaya, and Zea mays—had the largest MTPT contents. This relationship is not surprising given that other types of excess DNA, including introns, repetitive elements, and intergenic DNA, are known to mutually expand as the number of noncoding nucleotides in a mitochondrial genome increases (Lynch et al. 2006). It is thought that the burden of harboring surplus mtDNA is positively proportional to the mitochondrial mutation rate (Lynch et al. 2006); thus, the very low mtDNA mutation rates that are typically found in angiosperms (Lynch et al. 2006 and references therein) may promote the accumulation of MTPTs. In this context, it is noteworthy that angiosperm mtDNAs often contain nucDNA-derived sequences as well as genes horizontally acquired from other mitochondrial genomes (Richardson and Palmer 2007; Kleine et al. 2009), reinforcing the notion that they are sponges for all types of foreign DNA. Among the polyplastidic species that were investigated, the green alga C. vulgaris had the smallest and most compact mtDNA (supplementary table S1, Supplementary Material online), implying that it is more proficient than its land plant counterparts at eliminating (or preventing the accumulation of) excess nucleotides from its mitochondrial genome. If true, this may help explain why it was the only polyplastidic taxa without MTPTs. The relationship between MTPTs and genome size is in accordance with previous investigations on organelle-to-nucleus DNA transfers, which found that bloated nuclear genomes harbor more plastid- and mitochondrial-derived sequences than those that are compact (Hazkani-Covo et al. 2010; Smith et al. 2011). Ultimately, these data suggest that the probability of transferred organelle DNA persisting in a genome is largely governed by the forces controlling the expansion and contraction of bulk DNA.
FIG. 2.—
Plot of MTPT content versus mitochondrial genome size (log scale) (A) and mitochondrial genome noncoding DNA content (B). Species of particular interest are labeled on the plot. Data on mitochondrial genome size and noncoding DNA content are shown in supplementary table S1 (Supplementary Material online).
Plastids Can Dish It Out but Can't Take It
Not 1 of the 42 species employed here contained mtDNA-like sequences in its plastid genome, irrespective of the number of mitochondria per cell and plastid genome size (table 1 and supplementary table S1, Supplementary Material online). This strengthens the commonly held view that the exchange of DNA between plastids and mitochondria is a one-way street (Richardson and Palmer 2007; Kleine et al. 2009). It has been suggested that because plastid genomes are often more compact than their mitochondrial counterparts (particularly for land plants and certain lineages of green algae) they are more efficient than mtDNAs at purging excess nucleotides. However, this hypothesis seems unlikely when considering that no PTMTs were found within the ptDNA of the polymitochondrial green alga Volvox carteri, which boasts a plastid genome that is larger (>525 kb) and more expanded (∼80% noncoding) than many angiosperm mtDNAs (Smith and Lee 2010). As noted by previous investigators (Richardson and Palmer 2007), an intriguing observation concerning the presence of MTPTs in mtDNAs and the absence of PTMTs from ptDNAs is that mitochondria (at least those of land plants and mammals) have an active DNA import system (Koulintchenko et al. 2003, 2006), whereas no such system has been identified in plastids.
Concluding Remarks
As predicted by the limited transfer window hypothesis, MTPTs were abundant in polyplastidic species, particularly those with expanded mitochondrial genomes, and were absent from monoplastidic species as well as polyplastidic taxa with monoplastidic meristematic systems. Neither the polymitochondrial nor the monomitochondrial taxa contained mtDNA-derived sequences in their plastid genomes, which may reflect the absence of an efficient DNA import system within plastids. These results, like those on plastid-like sequences in nuclear genomes, support the idea that monoplastidy limits the potential for ptDNA to be donated to other organelles. The probability that ptDNA sequences will be taken up by those organelles and persist as MTPTs or NUPTs is defined by an organelle's ability (or lack thereof) to both import exogenous DNA and retain it within its genome.
Supplementary Material
Supplementary tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
I thank Robert W. Lee and Kate Crosby for helpful feedback throughout the data analysis and writing of the manuscript. Thanks also to Yoshihisa Hirakawa, Jan Janouškovec, and the rest of the Keeling lab for insightful discussions. D.R.S. is supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE. Monoplastidic cell division in lower land plants. Am J Bot. 1990;77:559–571. doi: 10.1002/j.1537-2197.1990.tb13588.x. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE. The quadripolar microtubule system in lower land plants. J Plant Res. 1997;110:93–106. doi: 10.1007/BF02506848. [DOI] [PubMed] [Google Scholar]
- Clifton SW, et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004;136:3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. Promiscuous DNA-chloroplast genes inside plant mitochondria. Nature. 1982;299:678–679. doi: 10.1038/299678a0. [DOI] [PubMed] [Google Scholar]
- Gray MW, Boer PH. Organization and expression of algal (Chlamydomonas reinhardtii) mitochondrial DNA. Philos Trans R Soc Lond B Biol Sci. 1988;319:135–147. doi: 10.1098/rstb.1988.0038. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLos Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via permeability transition pore complex. EMBO J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum Mol Genet. 2006;15:143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- Lane N. Plastids, genomes, and the probability of gene transfer. Genome Biol Evol. 2011;3:372–374. doi: 10.1093/gbe/evr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister DL, Bateman JM, Purton S, Howe CJ. DNA transfer from chloroplast to nucleus is much rarer in Chlamydomonas than in tobacco. Gene. 2003;316:33–38. doi: 10.1016/s0378-1119(03)00754-6. [DOI] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W, O'Brien SJ. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 1994;39:174–190. doi: 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- Martin W. Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci U S A. 2003;100:8612–8614. doi: 10.1073/pnas.1633606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notsu Y, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- Oda K, et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD. Green algae structure, reproduction and evolution in selected genera. Sunderland (MA): Sinauer; 1975. [Google Scholar]
- Renzaglia KS, Garbary DJ. Motile gametes of land plants: diversity, development, and evolution. CRC Crit Rev Plant Sci. 2001;20:107–213. [Google Scholar]
- Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58:1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol. 2004a;21:1081–1084. doi: 10.1093/molbev/msh110. [DOI] [PubMed] [Google Scholar]
- Richly E, Leister D. NUPTs in sequenced eukaryotes and their genomic organization in relation to NUMTs. Mol Biol Evol. 2004b;21:1972–1980. doi: 10.1093/molbev/msh210. [DOI] [PubMed] [Google Scholar]
- Rudall PJ, Bateman RM. Developmental bases for key innovations in the seed-plant microgametophyte. Trends Plant Sci. 2007;12:317–326. doi: 10.1016/j.tplants.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Smith DR, Crosby K, Lee RW. Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol Evol. 2011;3:365–371. doi: 10.1093/gbe/evr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW. Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol. 2010;27:2244–2256. doi: 10.1093/molbev/msq110. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Vaidya AB, Mather MW. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 2009;63:249–267. doi: 10.1146/annurev.micro.091208.073424. [DOI] [PubMed] [Google Scholar]
- Wang D, Wu Y, Shih AC, Wu C, Wang Y, Chaw S. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol Biol Evol. 2007;24:2040–2048. doi: 10.1093/molbev/msm133. [DOI] [PubMed] [Google Scholar]