Abstract
The genome sequence of the Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, is reported. With 1,191,693 nt in length and 1,023 predicted protein-coding genes, the Mamavirus has the largest genome among the known viruses. The genomes of the Mamavirus and the previously described Mimivirus are highly similar in both the protein-coding genes and the intergenic regions. However, the Mamavirus contains an extra 5′-terminal segment that encompasses primarily disrupted duplicates of genes present elsewhere in the genome. The Mamavirus also has several unique genes including a small regulatory polyA polymerase subunit that is shared with poxviruses. Detailed analysis of the protein sequences of the two Mimiviruses led to a substantial amendment of the functional annotation of the viral genomes.
Keywords: Mimivirus, viral genome, nucleocytoplasmic large DNA viruses
Acanthamoeba polyphaga mimivirus (APMV) has the largest viral genome sequenced so far (GenBank accession no. NC_006450) (Raoult et al. 2004). The analysis of the 1,181,404-bp linear double-stranded (ds) DNA of APMV revealed the conservation of several signature genes that are diagnostic of the nucleocytoplasmic large DNA viruses (NCLDVs), an expansive, apparently monophyletic group of viruses infecting eukaryotes that also include the Poxviridae, Phycodnaviridae, Iridoviridae, and Asfarviridae families (Iyer et al. 2001, 2006; Yutin et al. 2009; Koonin and Yutin 2010). However, in addition to genes that are shared with other NCLDV, APMV has been shown to possess a variety of genes that have not been previously detected in any viruses, in particular genes for components of the translation system such as aminoacyl-tRNA synthetases (Raoult et al. 2004; Colson and Raoult 2010). In phylogenetic trees of conserved NCLDV proteins, the APMV comprised a distinct branch, which together with the presence of numerous unique genes, suggests that it should be classified as the founding member of a new NCLDV family, the Mimiviridae (Koonin and Yutin 2010).
Until 2008, the APMV remained the only member of the Mimiviridae although numerous sequences homologous to portions of the Mimivirus genome have been identified in marine metagenomic samples (Monier et al. 2008). In 2008, a novel virus-like agent denoted the virophage has been isolated from amoebae infected with a giant virus that appeared to be a distinct strain of APMV and has been named the Mamavirus (La Scola et al. 2008). More recently, a group of closely related giant viruses have been isolated from diverse environmental samples, and preliminary sequence characterization has shown that these viruses were distinct members of Mimiviridae (La Scola et al. 2010). In addition, the genome sequence of a virus isolated from the marine microflagellate Cafeteria roenbergensis has been reported; this virus is more distantly related to the Mimiviruses and potentially represents a new genus of Mimiviridae or a sister family within the NCLDV (Fischer et al. 2010). Here, we briefly describe the complete genome sequence of the Mamavirus, its comparison with the APMV genome, and a reannotation of the Mimivirus gene complement. While this work was in progress, complete resequencing and reannotation of the APMV genome have been reported (Legendre et al. 2011). Therefore, here, we report most of the comparative genomic results for both the original and the new APMV sequences.
The Mamavirus was originally isolated from A. polyphaga after the amoebae were inoculated with water from a cooling tower located in Paris, France (La Scola et al. 2008). All subsequent work with the virus was performed on Acanthamoeba castellanii, so the virus was denoted A. castellanii mamavirus. The morphological features and cultural properties of the Mamavirus closely resembled those described of APMV and did not allow one to differentiate between the two viruses. The Mamavirus DNA was extracted by following the same procedure than was previously used for APMV (La Scola et al. 2008), and the genome was sequenced using the 454-Roche GS20 device as described previously (Raoult et al. 2004; Margulies et al. 2005).
The Mamavirus genome is 1,191,693 nt length which is 10,289 nt longer than the original APMV genome and 10,144 nt longer than the new version of the APMV genome (the Mamavirus genome sequence was deposited in GenBank with the accession number JF801956). As a result of the Mamavirus genome annotation (see supplementary methods and file 1, Supplementary Material online), 1,023 open reading frames (ORFs) were identified as putative protein-coding genes, with the average predicted protein size of 343 amino acids (aa). These genes are evenly distributed on both DNA strands, with 497 on the “direct” strand and 526 on the “reverse” strand. The mean size of intergenic regions is 133 ± 138 nt, with the predicted protein-coding density of 0.86 genes/kb (compared with 0.77 genes/kb for the “old” Mimivirus or 0.83 genes/kb for the “new” Mimivirus genome sequence). The ORFs were annotated with respect to the evolutionary conservation, protein domain content, and predicted functions by using PSI-BLAST search (Altschul et al. 1997) of the Refseq database at the NCBI, domain identification using RPS-BLAST search of the Conserved Domain Database (CDD) (Marchler-Bauer and Bryant 2004), and assignment of proteins to clusters of orthologous NCLDV genes (NCVOGs) (Yutin et al. 2009).
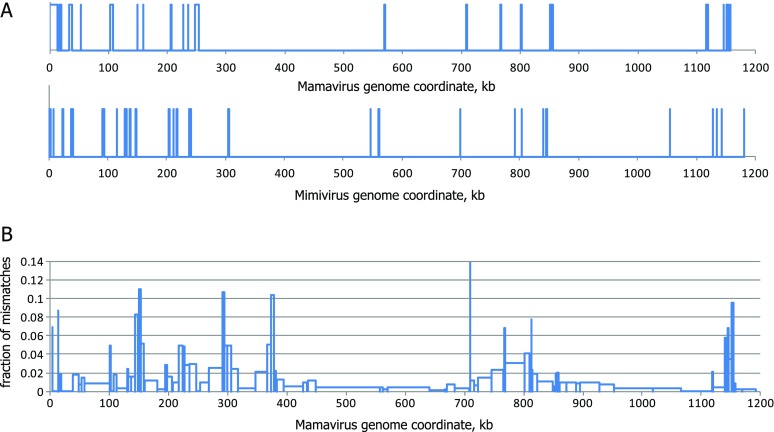
The alignment of the full-length genomes sequences of the Mamavirus and APMV that was constructed using the OWEN program (Ogurtsov et al. 2002) shows that the viral genomes are highly similar and collinear (fig. 1). Overall, after masking regions that were deemed unalignable (i.e., sequences longer than 200 nt containing gaps longer than 20 nt), the alignment contained approximately 99% identical nucleotides. Despite the overall high sequence conservation between the genomes of the Mamavirus and APMV, there were several unalignable regions that mostly concentrated in the terminal regions of the genomes, particularly, the 5′-region (fig. 1A). The Mamavirus genome contained a 5′-terminal segment of approximately 13 kb, for which there was no counterpart in the APMV genome, whereas the APMV genome contained an unalignable ∼900-nt-long 3′-terminal segment. The nucleotide mismatch fractions in aligned regions were nonuniformly distributed along the genome alignment, showing a pattern resembling the distribution of unaligned regions, with the highest level of divergence observed near the 5′-end (fig. 1B). This pattern of terminal divergence resembles the relationships between viral genomes in other groups of NCLDV, in particular, poxviruses (Senkevich et al. 1997).
FIG. 1.—
Schematic representation of the genome alignment of the Mamavirus and APMV. (A) The distributions of unaligned regions (longer than 200 nt, >20 nt gaps) in the Mamavirus and Mimivirus genomes. (B) The mean fraction of mismatches in aligned regions.
The 1,023 predicted proteins of the Mamavirus were compared with the predicted protein sequences of the APMV using an all-against-all BLASTP search which yielded 833 bidirectional best hits (BBHs) for which the lengths of the aligned protein sequences differed by less than 20% and which accordingly were classified as bona fide orthologous genes (supplementary file 1, Supplementary Material online). The Mamavirus and Mimivirus BBHs showed a mean amino acid identity of 98.3% (range from 64.5% to 100%) and a mean nucleotide identity of 98.8% (range from 82.3% to 100%), and the majority of the pairs had identity levels greater than 99% (supplementary file 1, Supplementary Material online). Given the overall high similarity of the genomes of the two viruses, the number of fully matching orthologs (BBH) was unexpectedly low. Most of the remaining ORFs failed to pass the similar length threshold due to frameshifts or unmatched stop codons that could reflect either the actual disruption of the respective genes or sequencing artifacts.
The new version of the APMV genome (Legendre et al. 2011) encompasses 1,018 genes of which 979 encode (predicted) proteins, 6 encode tRNAs, and the remaining 33 appear to encode other noncoding (nc) RNAs. We repeated the comparative analysis of the Mamavirus and Mimivirus genomes using this new version of APMV. The comparison of the nucleotide sequences of the complete genomes yielded minimal differences from the above results obtained with the original APMV sequence (data not shown). The comparison of the encoded proteins produced more substantial changes. In particular, with the new version of the APMV genome, the number of protein-coding genes that satisfied our criteria for bona fide orthology (see above) increased from 833 to 879. This noticeable increase in the extent of detectable orthology reflects the new, improved, and more complete annotation of the APMV genome, in particular, the elimination of most of the frameshifts that were present in the original APMV genome sequence. Among the orthologous protein-coding genes, seven have changed their positions, presumably due to limited genome rearrangements that occurred after the radiation of APMV and the Mamavirus from their common ancestor (supplementary file 1, Supplementary Material online). The comparison of the Mamavirus genome with the new version of the APMV genome revealed 29 APMV ORFs and 46 Mamavirus ORFs that were partially or completely absent in the counterpart genome (i.e., did not have hits covering more than 20% of their lengths; supplementary file 1, Supplementary Material online).
Almost all unusual features detected in the APMV genome are also present in the Mamavirus genome including highly conserved genes for protein components of the translation system and six tRNAs (supplementary file 1, Supplementary Material online). The intein detected in the APMV DNA polymerase (Raoult et al. 2004) is present in the Mamavirus ortholog as well. The gene for the largest subunit of the DNA-directed RNA polymerase has an intron in the same position in both viruses; however, Mamavirus misses one of the three introns that are present in the gene for the second largest RNA polymerase subunit of APMV. One of the four paralogous capsid protein genes of APMV, MIMI_L425, contains two introns (Azza et al. 2009). The orthologous Mamavirus gene lacks these introns but carries its own unique intron (supplementary file 2, Supplementary Material online).
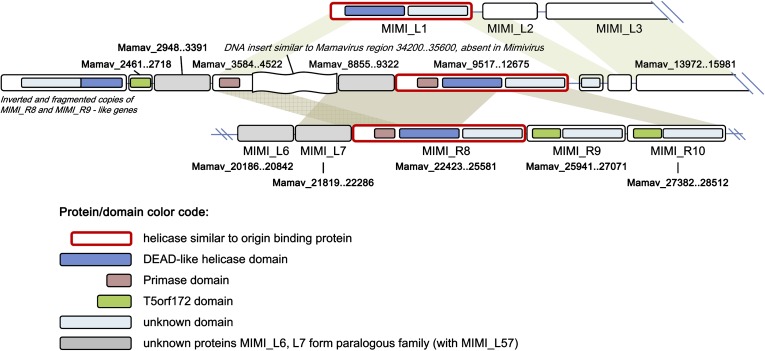
Most of the 46 “Mamavirus-only” predicted proteins are fragments, repeat rearrangements, or divergent paralogs of other proteins encoded elsewhere in both Mamavirus and Mimivirus genomes (supplementary file 1, Supplementary Material online). This trend was particularly obvious in the unique 5′-terminal 13-kb segment of the Mamavirus genome that harbors mostly short ORFs that appear to be truncated and diverged copies of other genes that are conserved between the two viruses (fig. 2). For example, between positions 9517 and 12675, a divergent protein similar to the origin-binding helicase is encoded (full-length match with MIMI_R8). Thus, the unique sequence segment in the Mamavirus genome mostly originated from duplications of other parts of the Mimi/Mamavirus genome, with some short regions apparently deleted in their original locations. However, a fragment between 4.5 and 9 kb might have been acquired by the Mamavirus from a source other than the common ancestor of the two Mimiviruses or else might have been lost in APMV: this sequence shows no similarity to any APMV sequences but is partially similar to another region of the Mamavirus genome (34–35.8 kb) which encodes uncharacterized predicted proteins.
FIG. 2.—
The unique 5′-terminal fragment of the Mamavirus genome: genome rearrangements and duplications. The figure shows a comparison of the 5′-end of the Mamavirus genome (middle) with the 5′-end of the APMV genome (top) and a downstream region that is conserved in both genomes (bottom; Mimivirus genomic positions 9500–18000). The genomic coordinates for all Mimivirus genes are shown. Shading shows homology between Mamavirus and APMV genes or domains.
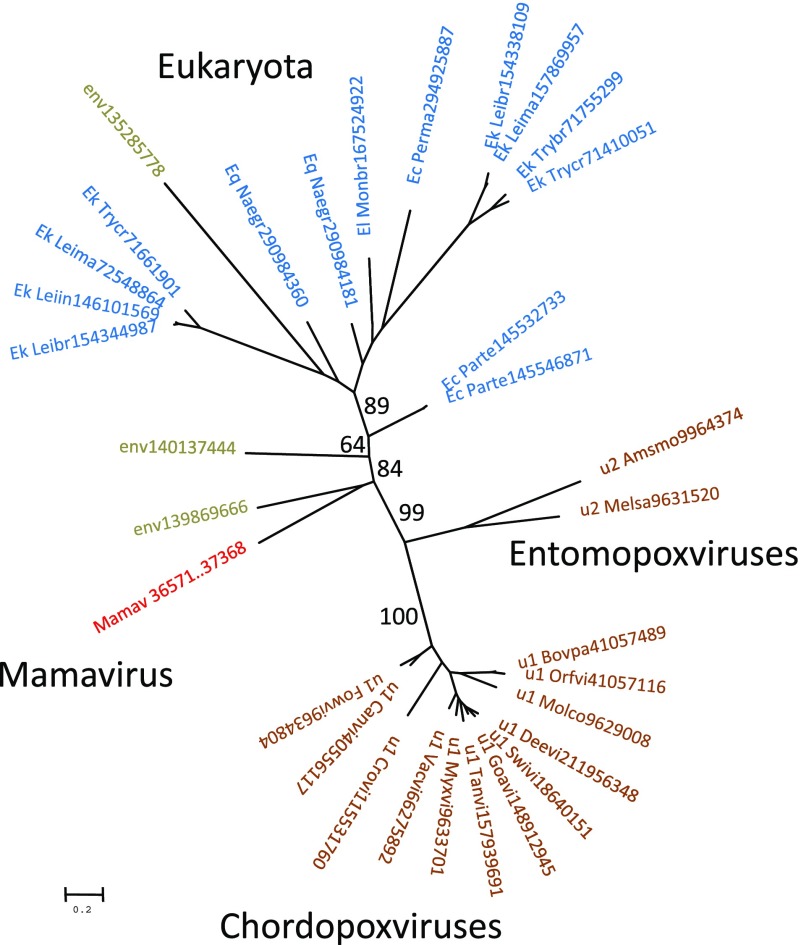
A predicted small regulatory subunit of polyA polymerase (PAPS) is encoded in the Mamavirus genome but is absent in APMV (in contrast, the large catalytic subunits are conserved). Among the other NCLDV, homologs of this protein are present only in poxviruses; in addition, homologs were detected in several unicellular eukaryotes including kinetoplastids, some ciliates (Paramecium but not Tetrahymena), the free-living excavate Naegleria gruberi, and the choanoflagellate Monosiga brevicolis (two paralogs). Phylogenetic analysis showed that the Mamavirus PAPS is distant from both poxviruses and Eukaryotes (fig. 3; see also supplementary file 3, Supplementary Material online). The distribution of the PAPS gene among viruses and eukaryotes in principle could be compatible with two alternative evolutionary scenarios: 1) independent acquisition from different eukaryotes and 2) presence in the ancestral NCLDV and subsequent loss in several virus lineages including APMV. The phylogenetic tree topology is compatible with the monophyly of all NCLDV PAPS and conversely does not suggest their origin from any specific lineage of eukaryotes (fig. 3), making the second scenario more likely. This scenario is compatible with the broader distribution of the catalytic subunit among the NCLDV (Iyer et al. 2001, 2006; Yutin and Koonin 2009; Yutin et al. 2009; Koonin and Yutin 2010). It seems most probable that the ancestral NCLDV encoded both subunits of polyA polymerase, and subsequently, most viruses have lost the gene for the regulatory subunit and some have lost both genes. This inferred evolutionary scenario resembles that for the NAD-dependent DNA ligase of the NCLDV (Yutin and Koonin 2009).
FIG. 3.—
Phylogenetic tree of the small regulatory subunit of polyA polymerase. The maximum-likelihood tree was constructed using TreeFinder (WAG matrix,G[Optimum]:4, 1,000 replicates, Search Depth 2; Jobb et al. 2004). The bootstrap support (expected-likelihood Weights) is shown for selected branches (percent). For each sequence, the species name abbreviation and the gene identification numbers are indicated; env stands for “marine metagenome.” Species abbreviations: Ec_Parte, Paramecium tetraurelia strain d4-2; Ec_Perma, Perkinsus marinus ATCC 50983; Ek_Leibr, Leishmania braziliensis MHOM/BR/75/M2904; Ek_Leiin, Leishmania infantum; Ek_Leima, Leishmania major strain Friedlin; Ek_Trybr, Trypanosoma brucei TREU927; Ek_Trycr, Trypanosoma cruzi strain CL Brener; El_Monbr, Monosiga brevicollis MX1; Eq_Naegr, Naegleria gruberi; u1_Bovpa, Bovine papular stomatitis virus; u1_Canvi, Canarypox virus; u1_Crovi, Crocodilepox virus; u1_Deevi, Deerpox virus W-1170-84; u1_Fowvi, Fowlpox virus; u1_Goavi, Goatpox virus Pellor; u1_Molco, Molluscum contagiosum virus subtype 1; u1_Myxvi, Myxoma virus; u1_Orfvi, Orf virus; u1_Swivi, Swinepox virus; u1_Tanvi, Tanapox virus; u1_Vacvi, Vaccinia virus; u2_Amsmo, Amsacta moorei entomopoxvirus “L”; u2_Melsa, Melanoplus sanguinipes entomopoxvirus.
Based on the Mamavirus–APMV protein comparisons and detailed examination of the homologs of all previously uncharacterized proteins, amendments to the annotations for 186 proteins were proposed (∼20% of the originally defined Mimivirus gene content; for the new version of the APMV genome (Legendre et al. 2011), the number of reannotated genes dropped to 159 or ∼16% of the complement of protein-coding genes) (supplementary file 1, Supplementary Material online) including functional predictions for many “hypothetical proteins.” These amended protein annotations include, among others, 16 helicases and 2 primases, 2 kinases, 7 endo- or exonucleases, 3 methyltransferases, and 5 ATP/GTPases; thus, the new annotations further increase the diversity of the functional repertoire of the Mimivirus. No functional annotation could be derived for any of the 75 new Mimivirus ORFs that have been recently identified by transcriptome analysis and predicted to encode proteins (Legendre et al. 2010, 2011).
Of the 33 ncRNAs annotated on the APMV genome (Legendre et al. 2011), 27 were represented by orthologs in the Mamavirus genome, with the nucleotide identity varying between 87% and 100%. For three APMV ncRNAs, there were no counterparts in the Mamavirus genome, and conversely, three ncRNAs were duplicated in the Mamavirus (supplementary file 1, Supplementary Material online). Finally, three putative ncRNA of APMV aligned with predicted protein-coding genes of the Mamavirus (supplementary file 1, Supplementary Material online). These are likely to be conserved protein-coding genes that have been misannotated as ncRNAs in APMV (Legendre et al. 2011).
Analysis of the RNA secondary structures using the RNAz and Afold programs (Ogurtsov et al. 2006; Gruber et al. 2010) showed that many of them fold in highly stable predicted structures and do not form alternative suboptimal structures in the range of 5% suboptimality (when folding within 5% of the minimum free energy is computed). These secondary structures are likely to be under strong selective pressure and might be crucial for the ncRNA functionality, similarly to other highly structured RNAs (Shabalina and Koonin 2008). In addition, we found that palindromic sequences present in the vicinity of the polyadenylation sites of APMV (Byrne et al. 2009) are perfectly conserved in the Mamavirus and so could be subject to selective constraint on the RNA structure.
Conclusions
The genomes of the two Mimivirus strains, the Mamavirus and APMV, are highly similar but show characteristic divergence in the terminal regions. The Mamavirus genome is the largest available virus genome, in part due to the presence of a 13-kb unique 5′-terminal region that apparently evolved by duplication of internal genomic sequences, possibly combined with the acquisition of a DNA fragment from an unknown source. These differences, however small, reveal pathways of Mimivirus genome evolution. A comprehensive comparative sequence analysis of the Mamavirus and APMV proteins led to a substantial amendment of the functional annotation of the Mimivirus genome and revealed several unique predicted proteins in the Mamavirus.
Supplementary Material
Supplementary methods and files 1–3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Lina Barrassi for technical assistance in the isolation of the Mamavirus. N.Y., S.A.S., and E.V.K. are supported by intramural funds of the US Department of Health and Human Services (National Library of Medicine).
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azza S, Cambillau C, Raoult D, Suzan-Monti M. Revised Mimivirus major capsid protein sequence reveals intron-containing gene structure and extra domain. BMC Mol Biol. 2009;10:39. doi: 10.1186/1471-2199-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne D, et al. The polyadenylation site of Mimivirus transcripts obeys a stringent ‘hairpin rule’. Genome Res. 2009;19:1233–1242. doi: 10.1101/gr.091561.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, Raoult D. Gene repertoire of amoeba-associated giant viruses. Intervirology. 2010;53:330–343. doi: 10.1159/000312918. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Findeiß S, Washietl S, Hofacker IL, Stadler PF. RNAZ 2.0: Improved noncoding RNA detection. Pac Symp Biocomput. 2010;15:69–79. [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci. U S A. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, et al. Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology. 2010;53:344–353. doi: 10.1159/000312919. [DOI] [PubMed] [Google Scholar]
- La Scola B, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- Legendre M, et al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010;20:664–674. doi: 10.1101/gr.102582.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Santini S, Rico A, Abergel C, Claverie JM. Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol J. 2011;8:99. doi: 10.1186/1743-422X-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Claverie JM, Ogata H. Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 2008;9:R106. doi: 10.1186/gb-2008-9-7-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogurtsov AY, Roytberg MA, Shabalina SA, Kondrashov AS. OWEN: aligning long collinear regions of genomes. Bioinformatics. 2002;18:1703–1704. doi: 10.1093/bioinformatics/18.12.1703. [DOI] [PubMed] [Google Scholar]
- Ogurtsov AY, Shabalina SA, Kondrashov AS, Roytberg MA. Analysis of internal loops within the RNA secondary structure in almost quadratic time. Bioinformatics. 2006;22:1317–1324. doi: 10.1093/bioinformatics/btl083. [DOI] [PubMed] [Google Scholar]
- Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Senkevich TG, Koonin EV, Bugert JJ, Darai G, Moss B. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: a case of hidden complexity. Biol Direct. 2009;4:51. doi: 10.1186/1745-6150-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]