Abstract
Activation of the Drosophila epidermal growth factor receptor (DER) by the transmembrane ligand, Spitz (Spi), requires two additional transmembrane proteins, Rhomboid and Star. Genetic evidence suggests that Rhomboid and Star facilitate DER signaling by processing membrane-bound Spi (mSpi) to an active, soluble form. To test this model, we use an assay based on Xenopus animal cap explants in which Spi activation of DER is Rhomboid and Star dependent. We show that Spi is on the cell surface but is kept in an inactive state by its cytoplasmic and transmembrane domains; Rhomboid and Star relieve this inhibition, allowing Spi to signal. We show further that Spi is likely to be cleaved within its transmembrane domain. However, a mutant form of mSpi that is not cleaved still signals to DER in a Rhomboid and Star-dependent manner. These results suggest strongly that Rhomboid and Star act primarily to present an active form of Spi to DER, leading secondarily to the processing of Spi into a secreted form.
Keywords: spitz, rhomboid, Star, EGFR, Drosophila, Xenopus animal caps
Cell fate decisions in embryonic development often depend on receptor tyrosine kinases that signal with precise temporal and spatial control. One striking example of this regulation is seen in the embryonic and adult development of Drosophila, in which multiple cell fate decisions require DER, the Drosophila epidermal growth factor receptor (EGFR) (Schweitzer and Shilo 1997). A number of genes have been identifed with phenotypes similar to DER, and were thus found to encode factors that regulate DER signaling. This group includes Spitz (Spi), a transmembrane ligand for DER that is similar to transforming growth factor-α (TGF-α), Rhomboid, a putative seven transmembrane domain protein, and Star, a single-pass transmembrane protein (Bier et al. 1990; Rutledge et al. 1992; Kolodkin et al. 1994) (Fig. 1A). As essential cofactors, Rhomboid and Star are thought to determine the pattern of DER activation during embryonic development by controlling the activity of the ligand and receptor that are ubiquitously expressed. The temporal and spatial expression of rhomboid is highly regulated, suggesting that it controls when and where DER is activated (Bier et al. 1990).
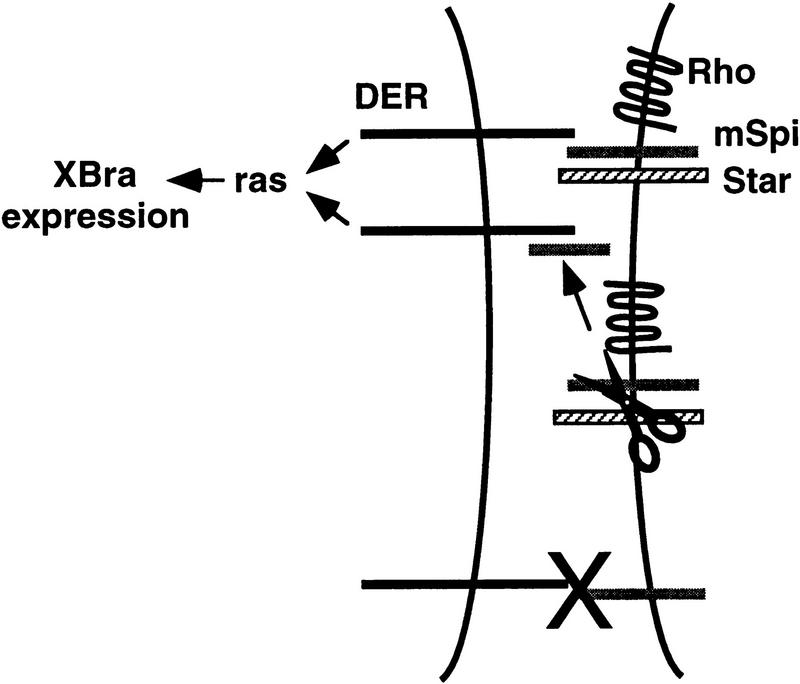
Figure 1.
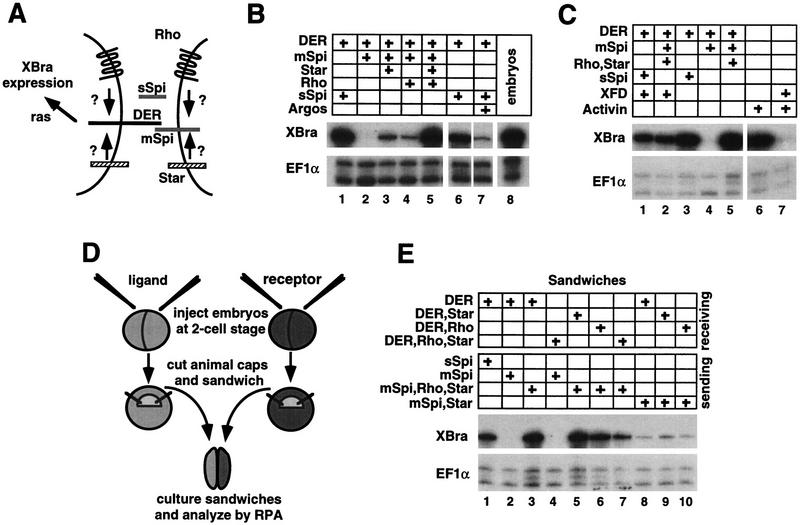
Rhomboid- (Rho) and Star-dependent mSpi activation of DER in Xenopus animal caps. (A) Models for Rhomboid and Star action. (B) RPA showing that Rhomboid and Star each weakly promote mSpi activation of DER, however, together they are synergistic (lanes 2–5). sSpi activates DER in the absence of Rhomboid and Star (lane 1). Argos represses sSpi activation of DER (lane 7). (C) Induction of XBra expression by DER is reduced in the presence of the dominant-negative FGF receptor, XFD (compare lane 1 with 3, and lane 2 with 5). In a positive control for the potency of XFD, lanes 6 and 7 show that XFD abolishes induction of XBra by activin. (D) Sandwich assay experimental design. (E) RPA of animal cap sandwiches showing that Rhomboid and Star must be coexpressed with the ligand, and not the receptor, for activation to occur (lanes 2–4). When Rhomboid and Star are present in both the signaling and receiving cells, the level of DER activation is attenuated (lanes 5–7).
Current evidence, primarily from genetic analyses, most strongly supports a model proposed by Shilo and colleagues in which Rhomboid and Star regulate processing of membrane-bound Spi (mSpi) to an active, soluble form (Golembo et al. 1996; Schweitzer et al. 1995b). rhomboid and Star can promote DER activation non cell autonomously (Golembo et al. 1996). Moreover, the requirement for rhomboid and Star for DER activation can be overcome by expressing just the soluble, extracellular portion of Spi (sSpi) (Schweitzer et al. 1995b). Alternative models, however, suggest that Rhomboid and Star regulate receptor function, or that they are involved in forming a complex at the cell surface that brings together receptor and ligand (Sturtevant et al. 1993, 1996; Stemerdink and Jacobs 1997; Guichard et al. 1999). To test these models, we have developed an assay based on Xenopus animal caps, in which Spi activates DER in a Rhomboid and Star-dependent manner. This assay not only allows the requirement for Rhomboid and Star to be analyzed in depth, but is amenable to a structure-function analysis of DER signaling. On the basis of the results from this assay, we propose that Rhomboid and Star control the pattern of DER activation by a novel mechanism involving ligand presentation and proteolysis.
Results
Xenopus animal cap assay for DER signaling
Rhomboid and Star-mediated DER signaling was analyzed by an assay in which Xenopus animal cap explants were isolated from embryos injected with in vitro synthesized DER, spi, rhomboid, and Star RNA (Fig. 1). This assay is based on the fact that DER activates the Ras pathway, which should lead to an up-regulation in the expression of the Ras target gene Xenopus Brachyury (XBra) in animal caps (Whitman and Melton 1992). Animal caps from injected embryos were allowed to develop until sibling embryos were late gastrulae (stage 11.5), when they were analyzed for XBra expression by RNAse protection assay (RPA). We found that expression of XBra could be induced in animal caps by DER but only under the same conditions that are required for the activation of DER in Drosophila (Fig. 1B). Thus, expression of XBra is not induced in animal caps that express DER alone, DER along with mSpi, or DER along with just Rhomboid and Star (Fig. 1B, lane 2; data not shown). In contrast, a high level of XBra expression is induced when animal caps express DER along with mSpi, Rhomboid, and Star (Fig. 1B, lane 5). The requirement for Rhomboid and Star for DER activation can be overcome in the animal cap assay, as in Drosophila, by expressing sSpi, an engineered form of Spi that contains just the extracellular domain (Fig. 1B, lane 1). In addition, DER activation can be blocked, as in Drosophila, by introducing the DER inhibitor, Argos (Schweitzer et al. 1995a; Fig. 1B, lane 7).
One possible contributing factor to the high levels of XBra induction that are achieved in the animal cap assay is that XBra expression could be amplified by an FGF-dependent positive regulatory loop. It has been shown that XBra, which is a T-Box-containing transcription factor, activates the fibroblast growth factor (FGF) gene, which in turn up-regulates XBra expression through activation of the FGF receptor (Tada et al. 1997). To test whether the level of XBra expression that is induced by DER is amplified through an FGF-dependent mechanism, RNAs encoding DER, Spi, Rhomboid, Star, and a dominant-negative form of the FGF-receptor, XFD, were coinjected (Amaya et al. 1991). In the presence of XFD, the level of XBra expression is reduced by ∼2.5-fold. This result shows that a positive regulatory loop between XBra and FGF amplifies the level of XBra expression that is induced by activation of DER, thus increasing the sensitivity of the animal cap assay (Fig. 1C). Together, these results demonstrate that regulation of Drosophila EGFR-signaling can be recapitulated in Xenopus animal caps, thus providing a faithful and highly sensitive assay with which to investigate the mechanism of action by Rhomboid and Star.
To test whether Rhomboid and Star are obligate cofactors in the animal cap assay, their abilities to promote DER activation were analyzed separately. We found that by themselves, Rhomboid and Star each weakly promote mSpi activation of DER, however, together they are strongly synergistic (Fig. 1B, lanes 3–5). Thus, both Rhomboid and Star may be required to achieve maximal levels of DER activation, but for lower levels of signaling, either one alone may be sufficient. It is possible that Rhomboid and Star are obligate cofactors but that there are homologous proteins present in the animal cap that fulfill the role of the missing component, albeit weakly. Alternatively, this result may reflect a way in which various levels of receptor activation may be achieved. In some settings, such as the Drosophila wing veins, rhomboid and Star are codependent, whereas in the eye, Star is sufficient and rhomboid function appears to be dispensible (Freeman et al. 1992a; Guichard et al. 1999).
Rhomboid and Star act in the signaling cells to promote DER activity
Next, we determined whether Rhomboid and Star are required for DER activity by acting in the signaling cell, the receiving cell, or in both cells. To do this, activation of XBra was measured in sandwiches that were made by combining an animal cap expressing DER with another animal cap expressing mSpi, in the presence or absence of Rhomboid and Star (Fig. 1D). When Rhomboid and Star are present in the receptor-expressing cells, mSpi fails to activate DER (Fig. 1E, lane 4). However, when Rhomboid and Star are present in the ligand-expressing cells, mSpi strongly activates DER (Fig. 1E, lane 3). It has been suggested that Rhomboid and Star may act as cell adhesion molecules to bring together the receptor and ligand into a cell surface complex (Stemerdink and Jacobs 1997; Sturtevant et al. 1993, 1996). To test this idea, sandwiches were made in which rhomboid and Star were expressed in both the sending and receiving cells. Interestingly, this configuration attenuated the level of DER signaling, with the strongest repression occuring when both Rhomboid and Star are present on both sides of the sandwich (Fig. 1E, lanes 5–7). It is an intriguing possibility that an interaction between Rhomboid and/or Star in trans may dampen the level of signal received by DER, providing another possible mechanism by which the level of DER activation could be finely tuned. Together, these results argue against models in which Rhomboid and Star regulate receptor function or act as cell adhesion molecules and support a model in which Rhomboid and Star potentiate DER activation by acting in the signaling cell.
mSpi is inactivated by sequences in its transmembrane and cytoplasmic domains
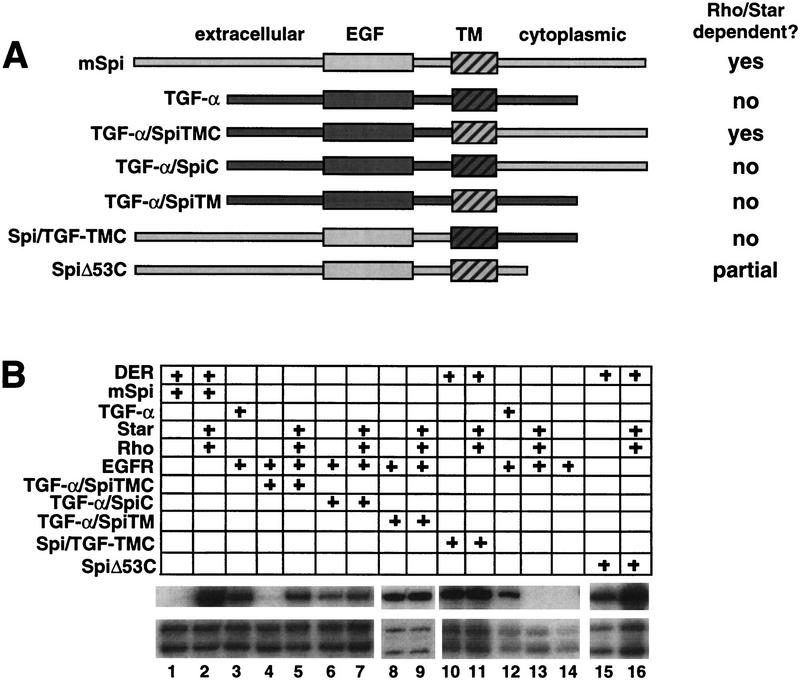
We next asked whether Rhomboid and Star potentiate DER signaling by changing the activity of its ligand, as suggested by the observation that sSpi does not require Rhomboid and Star to activate DER, whereas mSpi does. To address this question, we made a series of chimeras by replacing portions of human TGF-α, a vertebrate homolog of Spi, with the corresponding regions from mSpi (Fig. 2A). Human TGF-α alone strongly activates the human EGFR in the animal cap assay (Fig. 2B, lane 3). Strikingly, when the cytoplasmic (C) and transmembrane (TM) domains of TGF-α are replaced with those of mSpi (TGF-α/SpiTMC), the chimeric molecule activates the human EGFR only when Rhomboid and Star are present (Fig. 2B, lanes 4,5). In contrast, chimeric molecules in which the TGF-α C or TM domains are replaced separately with those of mSpi (TGF-α/SpiC and TGF-α/SpiTM, respectively) are constitutively active (Fig. 2B, lanes 6–9). Thus, together the mSpi TM and C domains are sufficient to confer Rhomboid and Star dependence on TGF-α. This result suggests that the C and TM domains maintain Spi in an inactive state, and that their ability to do so is transferrable to another EGFR ligand. As predicted by this interpretation, a membrane-bound form of Spi that activates DER signaling in the absence of Rhomboid and Star can be generated by replacing the mSpi TM and C domains with those of TGF-α (Spi/TGF-αTMC) (Fig. 2B, lanes 10,11). In addition, SpiΔ53C, a Spi mutant in which 53 carboxy-terminal residues are deleted and 17 cytoplasmic residues remain, exhibits some Rhomboid and Star-independent activity, providing further evidence that the C domain plays an inhibitory role (Fig. 2B, lanes 15,16). Together these results argue strongly that the C and TM domains of mSpi act to maintain an inactive state, with ligand activation occuring upon interaction with Rhomboid and Star.
Figure 2.
Analysis of TGF-α/Spi chimeras. (A) Schematic of TGF-α/Spi chimeric molecules. (B) RPA showing that the human EGFR is activated by human TGF-α (lane 3). Together the mSpi C and TM domains (TGF-α/SpiTMC) confer Rhomboid and Star dependence on TGF-α (lanes 4,5). Replacement of the TGF-α C or TM domains with those of mSpi results in constitutive activity (TGF-α/SpiC and TGF-α/SpiTM, lanes 6–9). The observation that TGF-α/SpiC exhibits reduced activity compared with TGF-α could reflect the loss of the TGF-α carboxy-terminal valines, which are normally required for targeting TGF-α to the cell surface (Briley et al. 1997). Spi/TGF-αTMC, in which the mSpi C and TM domains are replaced with those of TGF-α, is Rhomboid and Star independent (lanes 10,11). SpiΔ53C exhibits Rhomboid and Star-independent activity (lanes 15,16).
mSpi is on the cell surface in the presence and absence of Rhomboid and Star
One possible manner in which Rhomboid and Star could activate mSpi would be to target it to the cell surface by regulating its transit through the secretory pathway. For instance, transport of TGF-α to the cell surface requires TACIP18 (proTGF-α cytoplasmic domain interacting protein), a PDZ domain protein that interacts with the carboxy-terminal valine residues of proTGF-α (Fernandez-Larrea et al. 1999). To investigate this possibility, we biotinylated cell surface proteins on animal caps injected with RNA encoding tagged mSpi (mSpimyc) or with RNA encoding mSpimyc, Rhomboid, and Star. Protein lysates were incubated with streptavidin–agarose and then the bound fraction was eluted and analyzed by Western blotting with an anti-c-myc antibody. Biotinylated mSpi protein was recovered in each case, suggesting that it is unlikely that Rhomboid and Star are required to target mSpi to the cell surface (Fig. 3A). Furthermore, this observation implies that mSpi is at the cell surface but is inactive.
Figure 3.
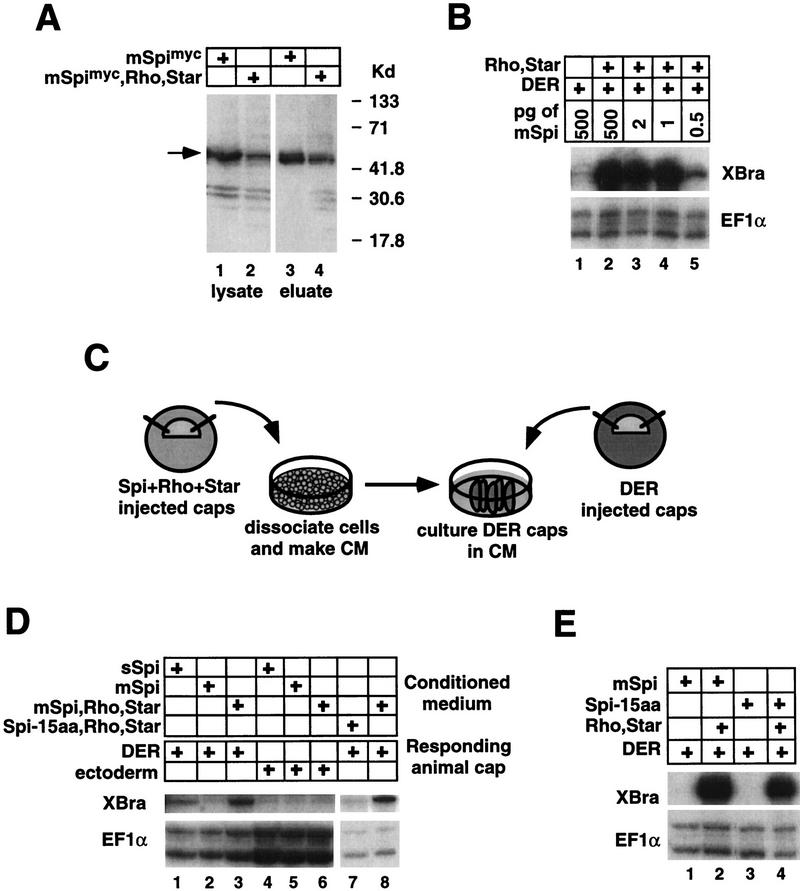
(A) Western analysis of biotinylated mSpimyc. mSpimyc can be biotinylated in the absence of Rhomboid and Star (cf. lanes 3 and 4). mSpimyc is detected by the anti-human c-myc antibody 9E10 and is indicated by an arrow. (Lanes 1,2) mSpimyc present in lysates prior to incubation with streptavidin-agarose (10% of the total lysate was loaded). (Lanes 3,4) mSpimyc eluted from streptavidin–agarose (the entire eluted fraction was loaded). (B) mSpi titration. Similar levels of DER activation are obtained over a range of 500 pg to 1 pg of injected mSpi RNA. (C) Experimental design for Rhomboid and Star-dependent production of Spi conditioned medium (CM). (D) RPA analysis of CM activities. sSpi CM activates DER independently of Rhomboid and Star (lane 1). mSpi CM induces XBra expression in DER-injected animal caps in a Rhomboid and Star-dependent manner (lanes 2,3), but not in uninjected animal caps (ectoderm, lanes 4–6). CM from animal caps expressing Spi-15aa, Rhomboid, and Star does not activate DER, whereas the positive control CM from animal caps expressing mSpi, Rhomboid, and Star efficiently activates DER (cf. lanes 7 and 8). (E) Spi-15aa strongly activates DER in a Rhomboid and Star-dependent manner, similar to mSpi.
Rhomboid and Star promote processing of mSpi to a soluble form
Another way in which Rhomboid and Star could lead to ligand activation is by promoting proteolytic processing, thus converting mSpi into a form similar to sSpi (Schweitzer et al. 1995b; Golembo et al. 1996). We first attempted to identify cleaved forms of Spi using versions with myc epitope tags at either terminus and by Western analysis. Comparison of protein lysates from embryos injected with mspimyc versus mspimyc, rhomboid, and Star did not reveal any Spi cleavage products (see Fig. 3A; data not shown). One possible explanation for this result is that mSpi proteolysis occurs at a low level and that only very small amounts of ligand are required to activate DER. A titration experiment supports this possibility as similar, high levels of DER activation are achieved over a range of 500 pg to only 1 pg of injected mspi RNA, thus below the limit of detection of Spi by Western analysis (Fig. 3B).
Given the possibility that only low levels of Spi are required to activate DER, we turned to a more sensitive assay to determine whether Rhomboid and Star promote proteolysis of Spi. Conditioned medium was prepared from dissociated animal cap cells from embryos injected with RNA encoding sSpi, or mSpi, or coinjected with RNAs encoding mSpi, Rhomboid, and Star. DER-injected animal caps were incubated in the conditioned medium and then analyzed for expression of XBra (Fig. 3C). The conditioned medium from animal caps expressing sSpi or mSpi/Rhomboid/Star contains an activity that activates DER, whereas that from animal caps expressing mSpi alone does not (Fig. 3D, lanes 1–3). In addition, the conditioned medium activity is DER dependent, as it is ineffective on uninjected animal caps (Fig. 3D, lanes 4–6). These results suggest that Rhomboid and Star activate mSpi by promoting its cleavage and secretion.
Spi does not need to be cleaved to activate DER
Next, we determined whether proteolytic processing is required for Rhomboid and Star activation of mSpi. To do this, we removed potential sites for processing of mSpi by deleting the sequences encoding the 15 amino acids (aa) between the Spi EGF and TM domains (Spi-15aa) (Fig. 3E). This region was selected because cleavage of TGF-α is known to take place within an analogous interval (Brachmann et al. 1989; Wong et al. 1989). When tested in the animal cap assay, Spi-15aa strongly activates DER in a Rhomboid and Star-dependent manner (Fig. 3E, lanes 3,4). In contrast, conditioned medium prepared from animal caps expressing Spi-15aa, Rhomboid, and Star does not contain any activity that activates DER, indicating that Spi-15aa is not cleaved (Fig. 3D, lane 7). Taken together, these results suggest that cleavage of mSpi depends on the sequence deleted in the Spi-15aa mutant; however, mSpi does not need to be cleaved to activate DER signaling. Thus, Rhomboid and Star may act to present mSpi to DER and subsequently facilitate or allow its cleavage.
Cleavage of mSpi may occur within its transmembrane domain
The results obtained with the Spi-15aa deletion mutant suggest that mSpi, like TGF-α, is processed to generate a soluble form. To examine the nature of this processing further, we next tested the possibility that it includes a cleavage within the transmembrane domain of mSpi. This possibility is suggested by the results obtained with the Spi/TGF-α chimeras, showing that the Spi transmembrane domain is important for Rhomboid and Star-dependent activation. Moreover, another multimembrane-spanning protein, Presenilin-1, mediates proteolyis of the β-amyloid precursor protein and Notch, both of which are cleaved within their transmembrane domains (De Strooper et al. 1999; Struhl and Greenwald 1999; Wolfe et al. 1999; Ye et al. 1999). If processing does lead to a cleavage in the membrane, we reasoned that this would release the intracellular domain of Spitz in a Rhomboid/Star-dependent manner. To detect this cleavage, we therefore generated a chimeric molecule in which the mSpi C domain is replaced with the myc-tagged, intracellular domain of the Xenopus Notch receptor (Spi/NICD) (Fig. 4A). The endogenous, γ-secretase-dependent Notch cleavage site (Schroeter et al. 1998) is not present in the Spi/NICD chimeric molecule (see Materials and Methods). If proteolytic processing of this molecule occurred within the Spi TM domain in a Rhomboid/Star-dependent manner, NICD may be released, translocate to the nucleus, and activate target genes (Fig. 4D; Lecourtois and Schweisguth 1998; Schroeter et al. 1998; Struhl and Adachi 1998). As a Notch target gene we used Xenopus Enhancer-of-split-related-1 (Esr-1) (Wettstein et al. 1997). Expression of Esr-1 was analyzed in animal caps that were coinjected with the neuralizing factor noggin, as Esr-1 is normally expressed in neural tissue and its induction by NICD is more robust in a noggin background (Wettstein et al. 1997).
Figure 4.
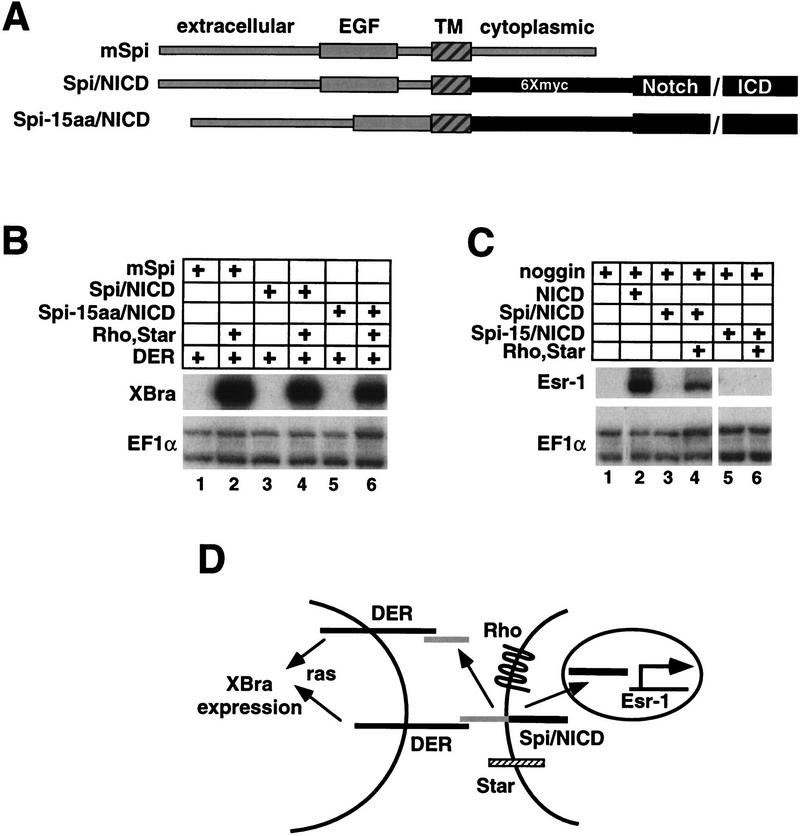
Analysis of Spi/NICD chimeras. (A) Spi/NICD chimeric molecules. (B) RPA showing that Spi/NICD and Spi-15aa/NICD activate DER in a Rhomboid and Star-dependent manner, similar to mSpi (lanes 1–6). (C) RPA showing that NICD induces Esr-1 expression (lane 2); however, the Spi/NICD chimeric molecule only activates Esr-1 when Rhomboid and Star are present (lanes 3,4). Spi-15aa/NICD does not activate Esr-1 (lanes 5,6). We note, induction of Esr-1 and XBra expression by Spi/NICD were analyzed as separate experimental samples because together XBra and noggin synergize to promote dorsal mesoderm formation, a background in which Esr-1 is poorly induced (Cunliffe and Smith 1994). (D) Model for Rhomboid and Star-dependent activation of Xbra and Esr-1 by the Spi/NICD chimera. Rhomboid and Star present Spi/NICD, and then cleavage releases both Spi and NICD to activate their respective targets. However, Spi does not need to be cleaved to be active.
When tested in the animal cap assay, Spi/NICD activates DER, but only in the presence of Rhomboid and Star, indicating that the Spi/NICD chimeric molecule still exhibits Rhomboid and Star-dependent Spi activity (Fig. 4B, lanes 1–4). We note, this result also indicates that the myc-tagged Xenopus NICD can effectively replace the Spi C domain, suggesting that the ability of the C domain to maintain Spi in an inactive state depends more on its structure than on its primary sequence. Significantly, Spi/NICD also activates the Notch target gene, Esr-1, in a Rhomboid and Star-dependent manner (Fig. 4C, lanes 1–4). This result suggests that Rhomboid and Star promote a proteolytic processing event within the Spi-TM domain that releases NICD (Fig. 4D). In addition, as Esr-1 induction is Rhomboid and Star dependent in the absence of DER, Rhomboid and Star can function independently of DER.
By analogy to the β-amyloid precursor protein and Notch, whose activites are regulated by multiple, interdependent cleavage events (Chan and Jan 1999), we decided to test the possibility that the 15 amino acids between the Spi EGF and TM domains that are required for production of soluble Spi are also required for the cleavage of the Spi/NICD chimeric molecule within its TM domain. Thus, we deleted the sequence encoding these 15 amino acids in the Spi/NICD chimera to produce Spi-15aa/NICD (Fig. 4A). This deletion mutant still strongly activates DER in a Rhomboid and Star-dependent manner (Fig. 4B, lanes 5,6), but no longer induces Esr-1, indicating that NICD is not released, and thus cleavage of this mutant does not occur (Fig. 4C, lanes 5,6). Thus, these results provide further independent evidence for our contention that Rhomboid and Star-dependent cleavage of mSpi requires the amino acids deleted in the Spi-15aa mutant, but mSpi need not be cleaved to activate DER signaling. Finally, these results suggest that there is a Rhomboid and Star-dependent cleavage event of mSpi within its TM domain. One possible explanation for these observations is that mSpi is cleaved both within the TM domain and within the 15 amino acids between the TM and EGF domains. Alternatively, a single cleavage of mSpi could occur within its TM domain that depends on the 15 amino acid interval.
Discussion
Our results suggest a novel regulatory mechanism of ligand presentation. mSpi is at the cell surface; however, its C and TM domains impose an inactive state in which cleavage and interaction with the receptor are prohibited. Subsequently, through an interaction with the Spi C and TM domains, Rhomboid and Star present an active form of mSpi, leading to, but not requiring, cleavage of its extracellular domain (Fig. 5).
Figure 5.
Model: mSpi is on the cell surface but it is inactive. Rhomboid and Star present an active form of mSpi, leading to, but not requiring, cleavage of its extracellular domain.
Several models could account for the Rhomboid and Star-dependent effects that we observed. One model is that Rhomboid and Star are required to direct mSpi to the proper compartment for signaling to occur. The results from the biotinylation experiments suggest strongly that Rhomboid and Star are not required for transport of mSpi to the cell surface, but it remains a possibility that Rhomboid and Star could play a role in localizing mSpi to specific cell surface microdomains such as lipid rafts (Brown and London 1998). An alternative class of models is that mSpi is at the cell surface and ready to signal, but that Rhomboid and Star are required for bringing mSpi into an active conformation. One version of this model is that Rhomboid and Star activate mSpi by promoting its oligomerization. However, this idea is difficult to reconcile with the observation that sSpi is active and either does not require oligomerization or oligomerizes independently of Rhomboid and Star. In addition, soluble EGF, which is similar to sSpi, binds as a monomer to the extracellular domain of the EGFR in a 1:1 ratio, suggesting that membrane-bound EGFR ligands may also bind the receptor as monomers (Lemmon et al. 1997). For these reasons, we favor an alternative model in which mSpi is present at the membrane in an inactive dimeric or oligomeric complex. Rhomboid and Star would be required to either prevent formation of this complex or to alter its conformation such that mSpi could be presented as an active form. This model is precedented by observations suggesting that a number of receptor tyrosine kinases exist as inactive dimers that are activated when specific inter-subunit conformational changes occur upon ligand binding (Jiang and Hunter 1999). Thus, formation of an inactive mSpi complex would be mediated by its C and TM domains and inhibited by an interaction between these domains and Rhomboid and Star. This model explains both why removal of these domains relieves the requirement for Rhomboid and Star, and transfer of these domains to TGF-α confers Rhomboid and Star dependence. Such a model also predicts that sSpi would be Rhomboid and Star independent.
Although the animal cap assay has allowed us to uncouple presentation from cleavage of Spi in vitro, in vivo cleavage may be necessary for normal development, as clonal analyses have revealed that Spi acts at a distance of several cell diameters (Freeman 1994; Tio et al. 1994). By analogy, in vitro TGF-α is active as both a membrane-bound and soluble form (Brachmann et al. 1989; Wong et al. 1989). However, examination of mutant mice lacking tumor necrosis factor converting enzyme (TACE), reveals that this enzyme plays a role in processing TGF-α, and that, despite the observation that membrane-anchored TGF-α is active in vitro, in vivo soluble TGF-α is essential for normal development (Peschon et al. 1998). It is certainly possible that in some developmental settings, secretion of Spi is crucial, whereas in other settings, it is sufficient that membrane-bound Spi activate DER only in neighboring cells. The discovery that expression of the DER-ligand vein is induced by Spi activation of DER suggests a mechanism by which membrane-bound Spi could effectively activate DER at a distance of several cell diameters (Golembo et al. 1999).
How do Rhomboid and Star promote cleavage of mSpi? Rhomboid and/or Star could play a passive role by making mSpi accessible to proteolysis upon presentation. Alternatively, Rhomboid and/or Star may actively facilitate Spi proteolysis either by activating or recruiting a protease or transporting Spi to the appropriate subcellular compartment. Similar roles have been proposed for the multimembrane-spanning proteins presenilin-1, which is required for proteolyis of β-amyloid precursor protein and Notch, and SREBP cleavage-activating protein (SCAP), which is required for the proteolyis of sterol regulatory element-binding protein (SREBP) (Sakai et al. 1998; De Strooper et al. 1999; Struhl and Greenwald 1999; Wolfe et al. 1999; Ye et al. 1999). It is also possible that Rhomboid and/or Star could themselves have proteolytic activity, as has been proposed for Presenilin-1 (Wolfe et al. 1999). A protease responsible for Spi cleavage is yet to be identified. Finally, although our study strongly suggests that presentation of Spi is inhibited by its C-domain, we have not addressed the question of whether proteolysis of Spi is also affected by the C-domain. For instance, proteolytic release of the extracellular domain of membrane bound neuregulin is dependent on its cytoplasmic domain (Liu et al. 1998). Future experiments will be aimed at determining whether Rhomboid and Star play a passive or an active role in the proteolysis of mSpi.
Another important question is, where exactly is the Spi cleavage site? Our study demonstrates that the 15 amino acid interval between the Spi EGF and TM domains is important for cleavage, although cleavage may also occur within its TM domain. The simplest model is that cleavage only occurs in the transmembrane domain, generating both a soluble extracellular and intracellular portion. In this model, the 15 amino acid interval between the Spi EGF and TM domains is required for recognition by a protease, perhaps in combination with Rhomboid and Star, which then cleaves in the transmembrane domain. Alternatively, processing of mSpi could be much more complicated, involving a cascade of cleavage events that are interdependent. In this model, for example, cleavage in the transmembrane domain may depend on a prior cleavage in the 15 amino acid interval. Cascades of proteolytic processing are evident in such examples as β-amyloid, Notch, and SREBP, as well as TGF-α, which undergoes two cleavages in its extracellular domain, the second of which is rate limiting (Massague 1990; Sakai et al. 1998; see references in De Strooper et al. 1999; Wolfe et al. 1999). It is interesting to speculate that the Spi C-domain that is released upon cleavage within the TM-domain may have a signaling function, as has been proposed for the cytoplasmic domain of TGF-α (Shum et al. 1994).
The EGFR belongs to a family of receptor tyrosine kinases that has been implicated in cellular proliferation, migration, and differentiation, as well as the generation and progression of tumors. Although there has been a great deal of progress toward understanding how signal transduction through these receptors is regulated, little is known about the mechanisms that control production of active forms of their ligands. The recent identification of both Caenorhabditis elegans and mammalian rhomboid homologs suggests that the function of Drosophila rhomboid may be conserved in evolution (Wilson et al. 1994; Pascall and Brown 1998). Studies of Drosophila rhomboid function will serve as a basis for understanding the Rhomboid family of transmembrane proteins and their roles in the regulation of ligand presentation and proteolysis.
Materials and methods
Construction of plasmids
The coding region of the human EGFR (Lin et al. 1984) was cloned into sp64 (Promega). Coding regions of spi (Rutledge et al. 1992), DER (Livneh et al. 1985), rho (Bier et al. 1990), Star (Kolodkin et al. 1994), argos (Freeman et al. 1992b), and human TGF-α (Jhappan et al. 1990), were cloned into the CS2+ expression vector (Turner and Weintraub 1994). mspi was truncated at the BglI site (nucleotide 621) to produce sspi. mspi was truncated at the PstI site (nucleotide 770) to produce spiΔ53C. mspimyc and sspimyc were made by cloning five consecutive myc tags into the BanI (nucleotide 328) site of mspi and sspi. Clones for chimeric molecules were constructed as follows, with the underlined amino acids inserted to produce restriction sites. Two weakly hydrophobic residues, Ala-138 and Ser-139, immediately amino-terminal to the predicted TM domain of Spi were included in the Spi TM domain. TGF-α/spiTMC, TGF-α amino acids 1–98/GluPhe/Spi amino acids 139–230. TGF-α/spiTM, TGF-α amino acids 1–98/GluPhe/Spi amino acids 139–164/GlySer/TGF-α amino acids 122–160. TGF-α/SpiC, TGF-α amino acids 1–121/GluPhe/Spi amino acids 161–230. Spi/TGF-αTMC, Spi amino acids 1–139/GluPhe/TGF-α amino acids 98–160. Spi-15 amino acids/NICD, Spi amino acids 1–122/GluPhe/Spi amino acids 139–160/GlySer/6Xmyc/Xenopus NICD amino acids 1751–2524. SpiDB/NICD, Spi amino acids 1–128/Spi amino acids 130–160/GlySer/6Xmyc/Xenopus NICD amino acids 1751–2524.
Isolation, treatment, and culturing of Xenopus animal caps
Two-cell stage Xenopus laevis embryos were injected in the animal region of each blastomere with 0.5 ng of capped synthetic RNAs encoding noggin (Lamb et al. 1993), XFD (Amaya et al. 1991), mSpi, Rhomboid, Star, Argos, Xenopus NICD (Wettstein et al. 1997), human TGF-α, human EGFR, and the chimeric molecules described herein. Animal caps were dissected at stage 9 and cultured until sibling controls reached stage 11.5 for XBra analysis and stage 13 for Esr-1 analysis. Some caps were treated with 5 ng/ml recombinant activin (provided by the Vale Laboratory, Salk Institute) in 0.5 × MMR, 0.1% BSA immediately after dissection.
RNase protection assay
RNA was isolated from eight animal caps per sample and analyzed by RNase protection assay, with 32P-labeled antisense RNA probes for XBra, EF-1α, and Esr-1 RNAs as described previously (Bhushan et al. 1994; Wettstein et al. 1997).
Biotinylation and Western analysis
A total of 40 animal caps each from embryos injected with sspimyc, mspimyc, or mspimyc, rhomboid, and Star were incubated in 1.5 mg/ml EZlink sulfo-NHS-SS-Biotin (Pierce) in 0.7× PBS, 0.5 mm MgSO4, 1 mm CaCl2 for 15 min, washed in 0.5× MMR, and homogenized in 200 μl of 1% Triton-X 100, 10 mm HEPES (pH 7.4), 150 mm NaCl, 100 mm CaCl2, 2 mm MgCl2, and protease inhibitors. Lysates were incubated with 20 μl of streptavidin–agarose (Pierce) overnight. Streptavidin–agarose bound proteins were washed in homogenization buffer, and eluted in reducing SDS sample buffer. Protein samples were resolved by SDS-PAGE on a 10%–20% gel, transferred to P-Immobilon (Millipore), and detected with the anti-human c-Myc antibody 9E10 and a peroxidase-conjugated secondary antibody with enhanced chemiluminescence (ECL, Amersham).
Preparation of conditioned medium
A total of 30 injected animal caps were dissociated in 200 μl of 1× Calcium-Magnesium Free Media (88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, and 7.5 mm HEPES) in 0.1% BSA. After 3 hr, the supernatant was diluted 1:1 with 1× MMR and passed over a 0.22 μm Millex-GV4 filter (Millipore).
Acknowledgments
We thank T. Hunter, G. Gill, E. Lamar, and G. Lemke for critical reading of this manuscript; E. Bier, A. Guichard, G. Merlino, M. Freeman, G. Gill, and E. Adamson for clones; R. Bradley and M. Perez for help with Westerns; A. Guichard, E. Bier, J.C. de la Torre, C. Ghiglione, and J. Posakony for helpful discussions. This work was supported by a grant from the NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kintner@salk.edu; FAX (858) 450-2172.
References
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Lin HY, Lodish HF, Kintner CR. The transforming growth factor beta type II receptor can replace the activin type II receptor in inducing mesoderm. Mol Cell Biol. 1994;14:4280–4285. doi: 10.1128/mcb.14.6.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Jan LY, Jan YN. rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes & Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- Brachmann R, Lindquist PB, Nagashima M, Kohr W, Lipari T, Napier M, Derynck R. Transmembrane TGF-alpha precursors activate EGF/TGF-alpha receptors. Cell. 1989;56:691–700. doi: 10.1016/0092-8674(89)90591-6. [DOI] [PubMed] [Google Scholar]
- Briley GP, Hissong MA, Chiu ML, Lee DC. The carboxyl-terminal valine residues of proTGF alpha are required for its efficient maturation and intracellular routing. Mol Biol Cell. 1997;8:1619–1631. doi: 10.1091/mbc.8.8.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Chan YM, Jan YN. Presenilins, processing of beta-amyloid precursor protein, and notch signaling. Neuron. 1999;23:201–204. doi: 10.1016/s0896-6273(00)80771-0. [DOI] [PubMed] [Google Scholar]
- Cunliffe V, Smith JC. Specification of mesodermal pattern in Xenopus laevis by interactions between Brachyury, noggin and Xwnt-8. EMBO J. 1994;13:349–359. doi: 10.1002/j.1460-2075.1994.tb06268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Fernandez-Larrea J, Merlos-Suarez A, Urena JM, Baselga J, Arribas J. A role for a PDZ protein in the early secretory pathway for the targeting of proTGF-alpha to the cell surface. Mol Cell. 1999;3:423–433. doi: 10.1016/s1097-2765(00)80470-0. [DOI] [PubMed] [Google Scholar]
- Freeman M. The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech Dev. 1994;48:25–33. doi: 10.1016/0925-4773(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Freeman M, Kimmel BE, Rubin GM. Identifying targets of the rough homeobox gene of Drosophila: Evidence that rhomboid functions in eye development. Development. 1992a;116:335–346. doi: 10.1242/dev.116.2.335. [DOI] [PubMed] [Google Scholar]
- Freeman M, Klambt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992b;69:963–975. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Golembo M, Raz E, Shilo BZ. The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development. 1996;122:3363–3370. doi: 10.1242/dev.122.11.3363. [DOI] [PubMed] [Google Scholar]
- Golembo M, Yarnitzky T, Volk T, Shilo BZ. Vein expression is induced by the EGF receptor pathway to provide a positive feedback loop in patterning the Drosophila embryonic ventral ectoderm. Genes & Dev. 1999;13:158–162. doi: 10.1101/gad.13.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard A, Biehs B, Sturtevant MA, Wickline L, Chacko J, Howard K, Bier E. rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development. 1999;126:2663–2676. doi: 10.1242/dev.126.12.2663. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Stahle C, Harkins RN, Fausto N, Smith GH, Merlino GT. TGF alpha overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell. 1990;61:1137–1146. doi: 10.1016/0092-8674(90)90076-q. [DOI] [PubMed] [Google Scholar]
- Jiang G, Hunter T. When dimerization is not enough. Curr Biol. 1999;12:R568–R571. doi: 10.1016/s0960-9822(99)80357-1. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Pickup AT, Lin DM, Goodman CS, Banerjee U. Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila. Development. 1994;120:1731–1745. doi: 10.1242/dev.120.7.1731. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Knecht AK, Smith WC, Stachel SE, Economides AN, Stahl N, Yancopolous GD, Harland RM. Neural induction by the secreted polypeptide noggin. Science. 1993;262:713–718. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Bu Z, Ladbury JE, Zhou M, Pinchasi D, Lax I, Engelman DM, Schlessinger J. Two EGF molecules contribute additively to stabilization of the EGFR dimer. EMBO J. 1997;16:281–294. doi: 10.1093/emboj/16.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CR, Chen WS, Kruiger W, Stolarsky LS, Weber W, Evans RM, Verma IM, Gill GN, Rosenfeld MG. Expression cloning of human EGF receptor complementary DNA: Gene amplification and three related messenger RNA products in A431 cells. Science. 1984;224:843–848. doi: 10.1126/science.6326261. [DOI] [PubMed] [Google Scholar]
- Liu X, Hwang H, Cao L, Wen D, Liu N, Graham RM, Zhou M. Release of the neuregulin functional polypeptide requires its cytoplasmic tail. J Cell Biol. 1998b;273:34335–34340. doi: 10.1074/jbc.273.51.34335. [DOI] [PubMed] [Google Scholar]
- Livneh E, Glazer L, Segal D, Schlessinger J, Shilo BZ. The Drosophila EGF receptor gene homolog: Conservation of both hormone binding and kinase domains. Cell. 1985;40:599–607. doi: 10.1016/0092-8674(85)90208-9. [DOI] [PubMed] [Google Scholar]
- Massague J. Transforming growth factor-alpha. A model for membrane-anchored growth factors. J Biol Chem. 1990;265:21393–21396. [PubMed] [Google Scholar]
- Pascall JC, Brown KD. Characterization of a mammalian cDNA encoding a protein with high sequence similarity to the Drosophila regulatory protein Rhomboid. FEBS Lett. 1998;429:337–340. doi: 10.1016/s0014-5793(98)00622-x. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;6:1503–1517. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Rutledge BJ, Zhang K, Bier E, Jan YN, Perrimon N. The Drosophila spitz gene encodes a putative EGF-like growth factor involved in dorsal-ventral axis formation and neurogenesis. Genes & Dev. 1992;6:1503–1517. doi: 10.1101/gad.6.8.1503. [DOI] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Goldstein JL, Brown MS. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site- 1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shilo BZ. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 1997;13:191–196. doi: 10.1016/s0168-9525(97)01091-3. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Howes R, Smith R, Shilo BZ, Freeman M. Inhibition of Drosophila EGF receptor activation by the secreted protein Argos. Nature. 1995a;376:699–702. doi: 10.1038/376699a0. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes & Dev. 1995b;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- Shum L, Reeves SA, Kuo AC, Fromer ES, Derynck R. Association of the transmembrane TGF-alpha precursor with a protein J. Cell Biol. 1994;125:903–916. doi: 10.1083/jcb.125.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemerdink S, Jacobs JR. Argos and Spitz group genes function to regulate midline glial cell number in Drosophila embryos. Development. 1997;124:3787–3796. doi: 10.1242/dev.124.19.3787. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, Bier E. The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes & Dev. 1993;7:961–973. doi: 10.1101/gad.7.6.961. [DOI] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, O'Neill JW, Biehs B, Colley N, Bier E. The Drosophila rhomboid protein is concentrated in patches at the apical cell surface. Dev Biol. 1996;174:298–309. doi: 10.1006/dbio.1996.0075. [DOI] [PubMed] [Google Scholar]
- Tada M, O'Reilly MA, Smith JC. Analysis of competence and of Brachyury autoinduction by use of hormone-inducible Xbra. Development. 1997;124:2225–2234. doi: 10.1242/dev.124.11.2225. [DOI] [PubMed] [Google Scholar]
- Tio M, Ma C, Moses K. spitz, a Drosophila homolog of transforming growth factor-alpha, is required in the founding photoreceptor cells of the compound eye facets. Mech Dev. 1994;48:13–23. doi: 10.1016/0925-4773(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes & Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Whitman M, Melton DA. Involvement of p21ras in Xenopus mesoderm induction. Nature. 1992;357:252–254. doi: 10.1038/357252a0. [DOI] [PubMed] [Google Scholar]
- Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- Wong ST, Winchell LF, McCune BK, Earp HS, Teixido J, Massague J, Herman B, Lee DC. The TGF-alpha precursor expressed on the cell surface binds to the EGF receptor on adjacent cells, leading to signal transduction. Cell. 1989;56:495–506. doi: 10.1016/0092-8674(89)90252-3. [DOI] [PubMed] [Google Scholar]
- Ye Y, Lukinova N, Fortini ME. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]