Abstract
The RecA protein of Escherichia coli, which has crucial roles in homologous recombination, DNA damage repair, induction of the SOS response, and SOS mutagenesis, was found to catalyze assimilation of complementary RNA into a homologous region of a DNA duplex (R-loop). The reaction strictly requires a region of mismatch in the duplex, which may serve as a nucleation site for RecA protein polymerization. The optimum conditions for the assimilation reaction resemble those for the previously studied RecA protein-catalyzed homologous pairing and strand exchange reaction between two DNA molecules. Our finding lends strong support to the proposal that RecA protein-catalyzed assimilation of a transcript into duplex DNA results in formation of an R-loop at certain regions of the chromosome and that, when stabilized, the R-loop can serve as an origin of chromosome replication.
Keywords: RecA protein, R-loop, cSDR, RNase H, DNA replication
The RecA protein of Escherichia coli is a multifunctional protein involved in many vital functions including homologous recombination, DNA damage repair, SOS induction upon DNA damage, and SOS mutagenesis (for review, see Kowalczykowski et al. 1994). The protein sequence is strongly conserved through evolution and analogs have been found in many species, from archaeabacteria to humans. RecA protein's ability to search for sequence homology (homologous pairing) and to promote strand exchange has a crucial role in homologous recombination. This homologous pairing and strand exchange function of RecA protein has been proposed to catalyze formation of an R-loop, a DNA duplex structure with RNA hybridizing to one strand displacing the opposite strand (for review, see Asai and Kogoma 1994).
Certain R-loops, when stabilized in the absence of ribonuclease H (RNase H, an endonuclease specific to RNA–DNA hybrids) in rnhA mutants, could serve as a site (oriK) for initiation of DNA replication (for review, see Kogoma 1997). Similar structures have been proposed to be involved in initiation of ColE1-type plasmid replication (Itoh and Tomizawa 1980) and bacteriophage T4 DNA replication (Kreuzer and Morrical 1994). An involvement of R-loops in initiation of chromosome replication at the origin, oriC, has also been suggested (Baker and Kornberg 1988). Furthermore, evidence has been presented that persistent R-loops are lethal if they are not removed (Itaya and Crouch 1991; Kogoma et al. 1993). How R-loops are formed and the factors involved in the process is not known. In this work we have directly examined whether RecA protein is capable of catalyzing invasion of an RNA transcript into duplex DNA in vitro.
Results
R-loop formation reaction
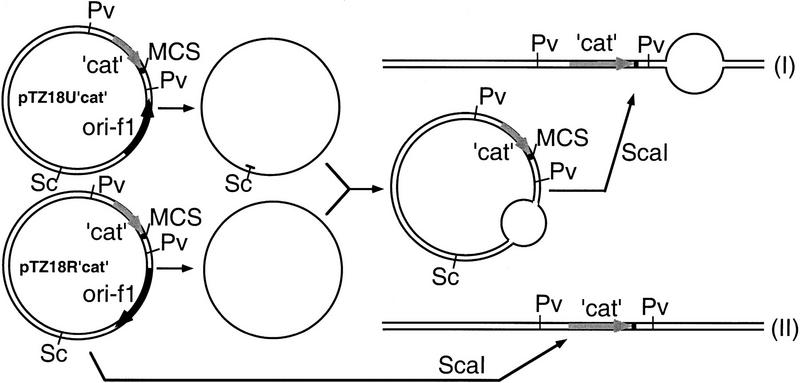
The DNA duplex used for R-loop formation was generated by the annealing of two complementary strands that were prepared from two plasmids, pTZ18Ucat and pTZ18Rcat, each carrying a truncated chloramphenicol transacetylase (cat) gene and the phage f1 origin of replication in the opposite orientations (Fig. 1). The single-stranded DNA (ss DNA) derived from pTZ18Ucat was linearized by cleaving it with ScaI after annealing a synthetic oligonucleotide at the ScaI restriction site. The two strands are complementary to each other except for the origin region (455 bp). Thus, annealing of the two strands resulted in a circular duplex with a region of mismatch 95 bp downstream of the cat sequence. The circular heteroduplex (a total length of 3.4 kb) was linearized by digestion with ScaI (Fig. 1, duplex I). As a control, an intact duplex (with no mismatch) was also employed (Fig. 1, duplex II). The RNA transcript (305 nucleotides) used was prepared by in vitro transcription of the cat sequence and was radiolabeled with [32P]UTP.
Figure 1.
Heteroduplex DNA substrate for RNA hybridization reactions. The scheme of construction for heteroduplex used in R-loop formation reactions (duplex I) and control reactions (duplex II) are shown (see Materials and Methods for details). (Sc) ScaI; (Pv) PvuII.
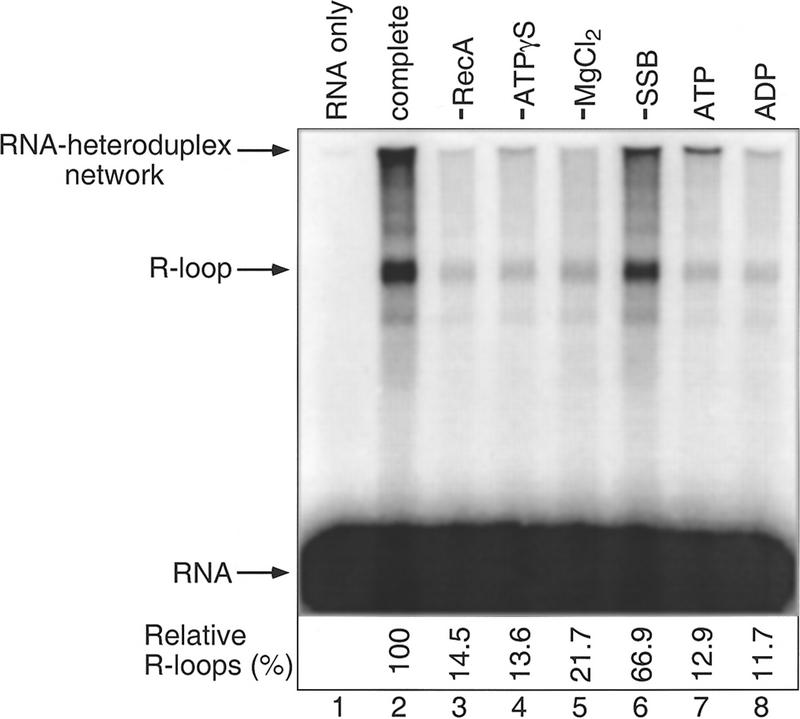
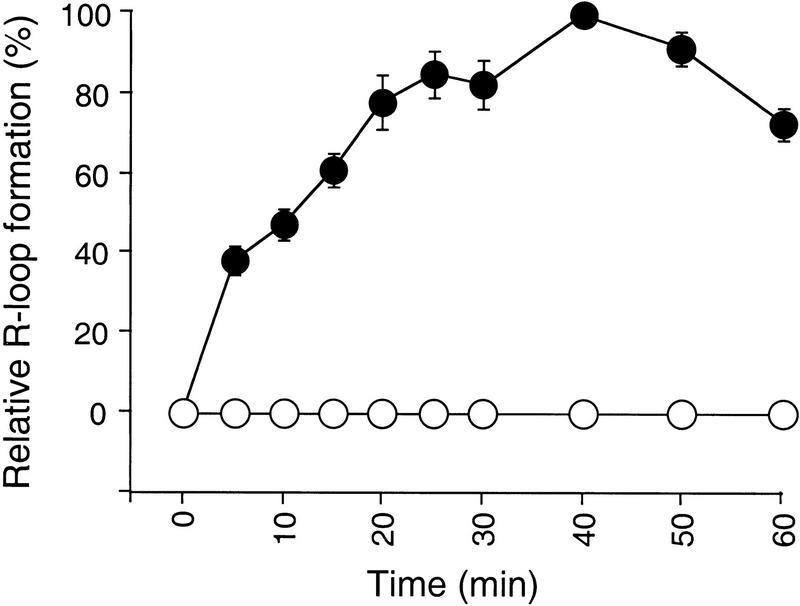
The heteroduplex (hd) DNA and transcript were paired under conditions similar to those developed for the RecA protein-catalyzed strand exchange reaction between circular ssDNA and linear duplex DNA (Cox and Lehman 1981). Thus, hd DNA was preincubated at 37°C in the presence of RecA protein and MgCl2, and the reaction was initiated by addition of 32P-labeled RNA transcript, ATPγS, and ssDNA-binding (SSB) protein. After 30 min, the reaction was terminated, and the reaction product was analyzed by agarose gel electrophoresis followed by autoradiography. In the complete reaction, the product contained radioactive material that migrated much more slowly than free labeled RNA (Fig. 2, lane 2). The position of this radioactive band (R-loop) corresponded with the mobility of hd DNA. Such slow migrating bands could not be detected in the absence of hd DNA (Fig. 2, lane 1) or with a hd DNA that lacked the cat sequence (data not shown). Radioactive material was also found above the hybrid band and in the well of the gel. Because this material was sensitive to RNase HI (see below), it was presumably labeled RNA hybridizing to other forms of DNA including multimers. For this reason, we presume that it represents a homology-dependent RNA-hd DNA network. The faint band just below the R-loop band corresponds to RNA binding to residual complimentary ss pTZ18Rcat DNA from incomplete heteroduplex DNA formation. All RNase HI-sensitive material combined, accounted for ∼38% of the total RNA. The R-loop reaction proceeded linearly with time, reaching a maximum at ∼40 min, after which time it slightly declined (Fig. 3). The control DNA duplex without mismatch (duplex II) was completely inactive for the reaction (Fig. 3).
Figure 2.
RecA protein-dependent R-loop formation. The R-loop formation reaction was carried out as described in Materials and Methods. Standard reaction conditions were used and, where indicated, a reaction component was omitted. In the reactions in lanes 7 and 8, ATPγS was replaced with ATP (3.0 mm) and ADP (3.0 mm), respectively. The relative R-loops (%) shown at the bottom of the gel were calculated by dividing the intensities of the R-loop bands by that in the complete reaction (lane 2).
Figure 3.
Time-course of R-loop formation. The R-loop formation reaction was carried out using either duplex I (●) or duplex II (○) as the DNA substrate (see Fig. 1). Samples were taken at the indicated times and analyzed for R-loop formation as described in Fig. 2. The data with duplex I are averages of two independent experiments ± variation.
R-loop formation is RecA protein-dependent
The R-loop reaction was dependent on RecA protein for optimum efficiency (Fig. 2, lane 3). In the absence of RecA protein, a faint band was seen consistently, its intensity amounting to 14%–15% of the complete reaction. The reaction also depended on ATPγS and MgCl2 (Fig. 2, lanes 4,5). The optimum concentration of MgCl2 was between 10 and 15 mm, which is in the optimal range for RecA protein-catalyzed strand exchange reactions with ssDNA (Cox and Lehman 1982). Concentrations below 3 mm were ineffective (data not shown). SSB protein was not essential (Fig. 2, lane 6) but was shown to stimulate the reaction. This is not surprising, given the suggested opposing roles of SSB as a stabilizer of ssDNA, as well as a direct competitor with RecA protein for ssDNA. As ATPγS, a nonhydrolyzable ATP analog, was effective, the reaction does not require the energy derived from NTP hydrolysis. Neither ATP (with no regenerating system) nor ADP was effective (Fig. 2, lanes 7,8). However, in the presence of a regenerating system, ATP could support the reaction (see below). These data suggest that ATP is required as a cofactor for RecA protein-catalyzed R-loop formation and is consistent with a model proposed by Kowalczykowski and Krupp (1995) in which ATP hydrolysis is required only for the redistribution of RecA molecules over large stretches of DNA.
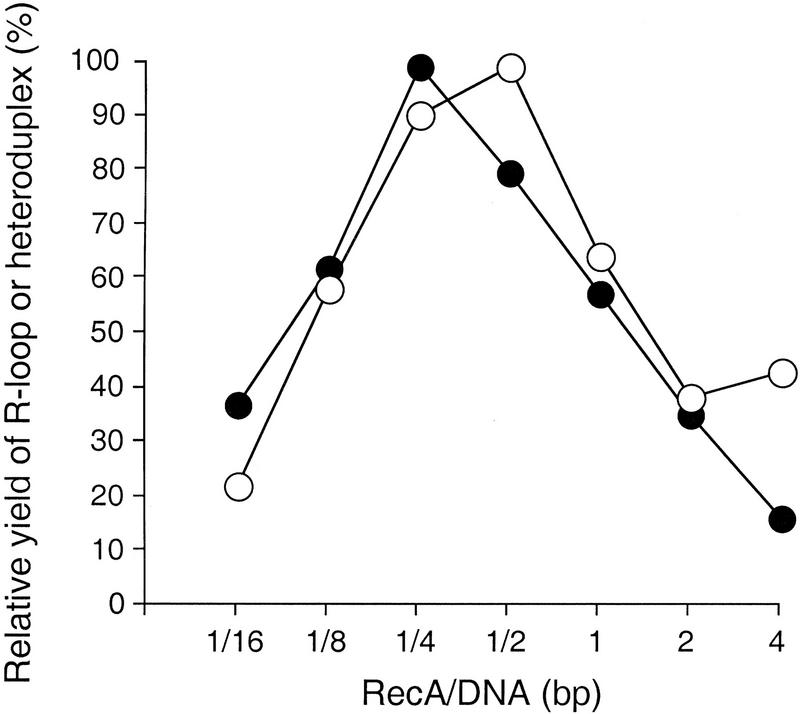
Strand exchange between circular ssDNA and linear double-stranded DNA (dsDNA) proceeds after RecA protein binds to ssDNA to form a nucleoprotein filament that contains one RecA monomer for every 3 nucleotides (Cox and Lehman 1981; Bryant et al. 1984; Tsang et al. 1985). We compared optimum RecA protein/DNA ratios for R-loop formation and for DNA strand exchange. Consistent with the previous reports, we found that an optimum ratio for the strand exchange reaction was between 2 and 4 nucleotides of ssDNA for every monomer of RecA protein (Fig. 4). For the R-loop reaction, the optimum ratio was 4 bp of dsDNA for each monomer of RecA protein (Fig. 4).
Figure 4.
Optimal molar ratios of RecA protein to DNA for R-loop and strand exchange reactions. The reaction for R-loop formation (●) was carried out as described in Materials and Methods except that the amount of RecA protein was varied as indicated. The relative amounts of R-loops generated were estimated by a PhosphorImager. The reaction for strand exchange (○) between circular ssDNA (from pTZ18Rcat) and linear dsDNA (pTZ18Rcat linearized at the ScaI site) was carried out as described previously (Cox and Lehman 1981). The products were separated by agarose electrophoresis and the relative amounts of heteroduplex products were quantified with a scanner. The abscissa is the molar ratio of RecA monomer/dsDNA (bp) for R-loop formation and RecA monomer/ssDNA (nucleotides) for DNA strand exchange.
The RNA associated with the hd DNA is an R-loop
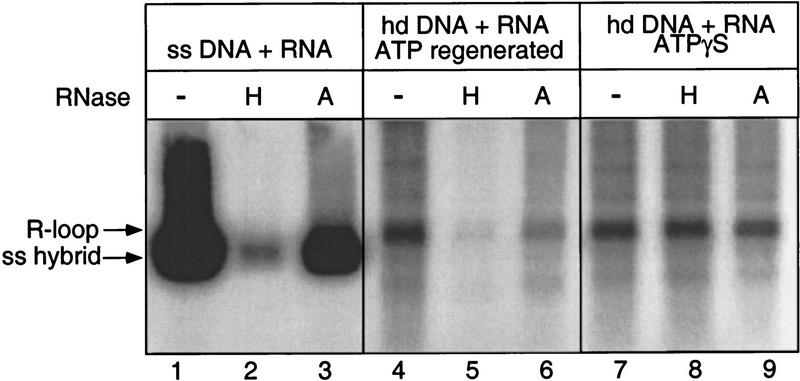
If the radioactive material comigrating with the hd DNA is an R-loop, then this material should be more sensitive to RNase HI (specific for hybridized RNA) than to RNase A (specific for free RNA). When R-loops were made in the presence of ATP and a regenerating system, the RNA associated with the hd DNA was nearly fully digested by RNase HI (Fig. 5, lane 5). This material showed partial resistance to RNase A, as did the hybrid that was made between circular ssDNA and the RNA transcript (Fig. 5, lanes 6,3). Considering the length of the RNA used, it is likely that portions of the RNA remain unpaired or are transiently displaced due to reinvasion by the displaced ssDNA produced during the strand exchange reaction. This would explain our findings that R-loops (and other RNA–dsDNA hybrid complexes) tend to be more sensitive to RNase A treatment than are RNA hybrids formed with ssDNA. Overall, these results strongly suggest that the RNA comigrating with the hd DNA is an R-loop. Thus, RecA protein is capable of assimilating RNA into duplex DNA under these conditions. Interestingly, when the energy source was replaced with ATPγS, the product was insensitive to RNase HI (Fig. 5, lane 8), suggesting that in the absence of ATP hydrolysis, RecA protein remains bound to the R-loop, thus inhibiting the action of RNase HI on the transcript. Supporting this reasoning, after deproteinization by treatment with SDS and proteinase K, the product became sensitive to RNase HI (data not shown).
Figure 5.
Sensitivity of R-loop formation to RNase HI. The products of the hybridization reactions were treated with no enzyme (−), RNase HI (H) (0.25 U/μl; U.S. Biochemical) or RNase A (A) (10 μg/μl). After the treatment, samples were deproteinized with SDS and proteinase K followed by phenol treatment. The R-loops used for samples shown in lanes 7–9 were prepared in a complete reaction as described in Materials and Methods. The R-loops in lanes 4–6 were made in a reaction in which ATPγS was replaced with ATP (3.0 mm) and a regenerating system (8 mm phosphocreatine plus 10 U/ml creatinephosphokinase). The single-stranded hybrid in lanes 1–3 was prepared by annealing of circular ssDNA derived from pTZ18Rcat and 32P-labeled RNA under the condition described previously (Kirkpatrick et al. 1992).
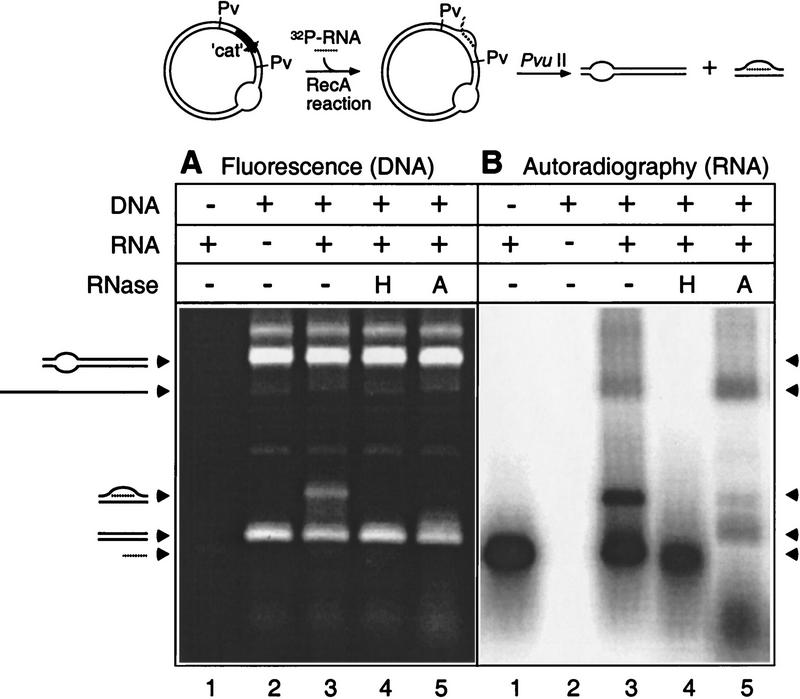
To more directly examine R-loop formation, the reaction was repeated using an identical heteroduplex DNA substrate (duplex I) except that it was not linearized by digestion with ScaI (see Fig. 1). After the R-loop formation reaction, the product was subsequently digested with PvuII, resulting in a 0.6-kb fragment containing the cat sequence (site of RNA hybridization) and a 2.8-kb fragment containing the rest of the pTZ18 vector sequence (Fig. 6, top). Any RNA hybridizing to the cat sequence would be visible as a shift in electrophoretic mobility of the 0.6-kb DNA fragment. Reaction products were separated by electrophoresis and were stained with the DNA-specific dye Vistra Green (Fig. 6A). To give the corresponding locations of RNA, the gel was blotted onto a membrane and exposed to a PhosphorImager screen to detect the positions of radiolabeled RNA (Fig. 6B). A significant portion of the 0.6-kb fragment was shifted in the gel in the presence of the RNA (Fig. 6A, cf. lanes 2 and 3) and was restored to the faster migrating position in the presence of RNase HI (Fig. 6A, lane 4). The shifted fragment was somewhat less sensitive to RNase A (Fig. 6A, lane 5), which further suggests that the RNA is in a hybrid form. Autoradiography of this gel (Fig. 6B) showed that the shifted 0.6-kb hybrid band (R-loop) in Figure 6A, lane 3, corresponds to the location of labeled RNA (Fig. 6B, lane 3). Note that formation of the R-loop was specific for the 0.6-kb PvuII fragment containing the cat gene; no labeled RNA was detected annealing to the large 2.8-kb heteroduplex PvuII fragment, which did not contain the cat sequence. The slowest migrating radioactive material in the reaction (Fig. 6B, lanes 3,5) corresponds to RNA annealing with complimentary ss pTZ18Rcat DNA that remained from incomplete hd DNA formation.
Figure 6.
R-loop gel shift assay. The R-loop formation reaction was carried out under standard reaction conditions except that circular heteroduplex (not ScaI digested) was used. To enhance product formation, RNA concentrations were increased, resulting in an equal molecular ratio of DNA to RNA in the reaction. The reaction was terminated and followed by phenol extraction and ethanol precipitation. The product was then digested with PvuII and subsequently treated with no enzyme (−), RNase HI (H) (1 U/μl), or RNase A (A) (1.0 μg/ml) (note that concentrations of RNase were optimized for this experiment). (A) Samples were electrophoresed, stained with Vistra Green (Amersham Life Sciences, Arlington Heights, IL), and DNA fluorescence was measured on a PhosphorImager. (B) The gel was then blotted onto a nylon membrane and subjected to autoradiography. All samples were treated identically with the following exceptions: (Lane 1) DNA omitted; (lane 2) RNA omitted; (lane 3) complete reaction; (lanes 4,5) RNase HI and RNase A treated, respectively. Arrowheads indicate positions of relevant species generated. From top to bottom, these are 2.8-kb PvuII vector fragment, leftover single-stranded pTZ18Rcat DNA from heteroduplex formation, R-loop, 0.6-kb PvuII cat fragment, free RNA transcript.
Discussion
Role of RecA protein in R-loop formation
Kirkpatrick et al. (1992) demonstrated that RecA protein promotes annealing (hybridization) between ssDNA and complementary RNA in the presence of ATPγS. RecA protein does so by first binding to ssDNA at a low MgCl2 concentration (1 mm) to form a nucleoprotein filament. Complementary RNA, in turn, interacts with the RecA protein–ssDNA complex and annealing ensues at a high MgCl2 concentration (12 mm). High concentrations of MgCl2 during the preincubation with RecA protein and ssDNA, which are unfavorable for nucleoprotein filament formation, severely inhibit the hybridization reaction (Kirkpatrick et al. 1992). In this work we determined that assimilation of complementary RNA into linear duplex DNA, by RecA protein to form an R-loop, has an optimum MgCl2 concentration in the range of 10–15 mm (data not shown). Low concentrations are ineffective. This novel reaction by RecA protein strictly requires a region of single-strandedness in the duplex DNA, which we generated by introducing a hd DNA bubble 95 bp downstream of the R-loop site. As RecA protein has been shown to not bind RNA well under the conditions used, that is, in the presence of high concentrations of MgCl2 and SSB protein (Kirkpatrick et al. 1992), RecA protein most likely first binds to the ssDNA region (mismatch) of the heteroduplex DNA substrate. Thus, in the presence of ATP, the mismatch region serves as a nucleation site for RecA polymerization, which then extends to the duplex region (Shaner and Radding 1987). The resulting nucleoprotein filament consisting of duplex DNA, RecA protein, and bound ATP facilitates a homology search reaction and catalyzes the eventual assimilation of the RNA transcript into the DNA duplex.
Recent findings by the Kowalczykowski laboratory support this interpretation, in which these works show that RecA protein polymerized along a dsDNA fragment can promote pairing and strand exchange with homologous ssRNA (Zaitsev and Kowalczykowski, 2000). Furthermore, we found that the optimum RecA protein/DNA ratio for our reaction, one monomer of RecA protein to 4 bp of DNA for R-loop formation, is in the correct range of stoichiometries (3–6 bp per RecA monomer) determined from direct binding assays or from dsDNA-binding assays (Kowalczykowski et al. 1987; Zaitsev and Kowalczykowski 1999).
Biological significance of R-loops
In the case of ColE1 plasmid replication, pairing between a stretch of 6 guanosine ribonucleotides of a nascent transcript and a stretch of 6 cytosine deoxyribonucleotides of the template DNA strand was proposed to prevent rewinding of the duplex behind the transcribing RNA polymerase, allowing formation of a persistent hybrid (R-loop) downstream (Masukata and Tomizawa, 1990). A similar mechanism (wedging) of R-loop formation at the origin of T4 DNA replication was also suggested (Mosig et al. 1995). Our in vitro demonstration that RecA protein can catalyze the assimilation of an RNA transcript into duplex DNA supports the proposal that RecA protein-catalyzed assimilation of a transcript into duplex DNA can result in formation of an R-loop at specific regions of the chromosome.
In E. coli rnhA mutants, we proposed that R-loops serve as origins of chromosome replication, leading to one form of recombination-dependent replication known as constitutive stable DNA replication (cSDR) (Kogoma et al. 1994; Hong et al. 1995). In this model (Fig. 7), the initial strand opening for initiation is achieved by RecA protein-facilitated hybridization of an RNA transcript to the template, displacing the other strand. The invasion event appears to be modulated by the supercoiling state of the region, which may explain why cSDR initiates at discrete sites on the chromosome (for review, see Kogoma 1997). The resulting structure, an R-loop, is stabilized owing to the absence of RNase H. DNA polymerase I then synthesizes DNA from the 3′ end of the hybridized RNA, enlarging the loop. PriA-catalyzed priming loads DnaB helicase, followed by DNA polymerase III replisome assembly. A second replication fork is established when DNA polymerase I extends the 3′ end of the newly synthesized lagging strand, whereas another removes the RNA using 5′ to 3′ exonuclease activity. This mechanism results in bidirectional replication initiated at an oriK site. In rnhA mutants, initiation of DNA replication at oriK strictly requires the homologous pairing and strand exchange function of RecA protein (Kogoma et al. 1985, 1994). Recently, RecG protein, a junction-specific helicase, was shown to resolve R-loops (Fukuoh et al. 1997), a result that is also consistent with the prior genetic finding that mutational inactivation of RecG activates oriK initiation (Hong et al. 1995). The in vivo conditions that stimulate RecA protein-catalyzed R-loop formation remain to be investigated; however, it is noteworthy that cSDR-like replication can be activated in rapidly growing wild-type E. coli cells at the time of entry to stationary phase (Hong et al. 1996).
Figure 7.
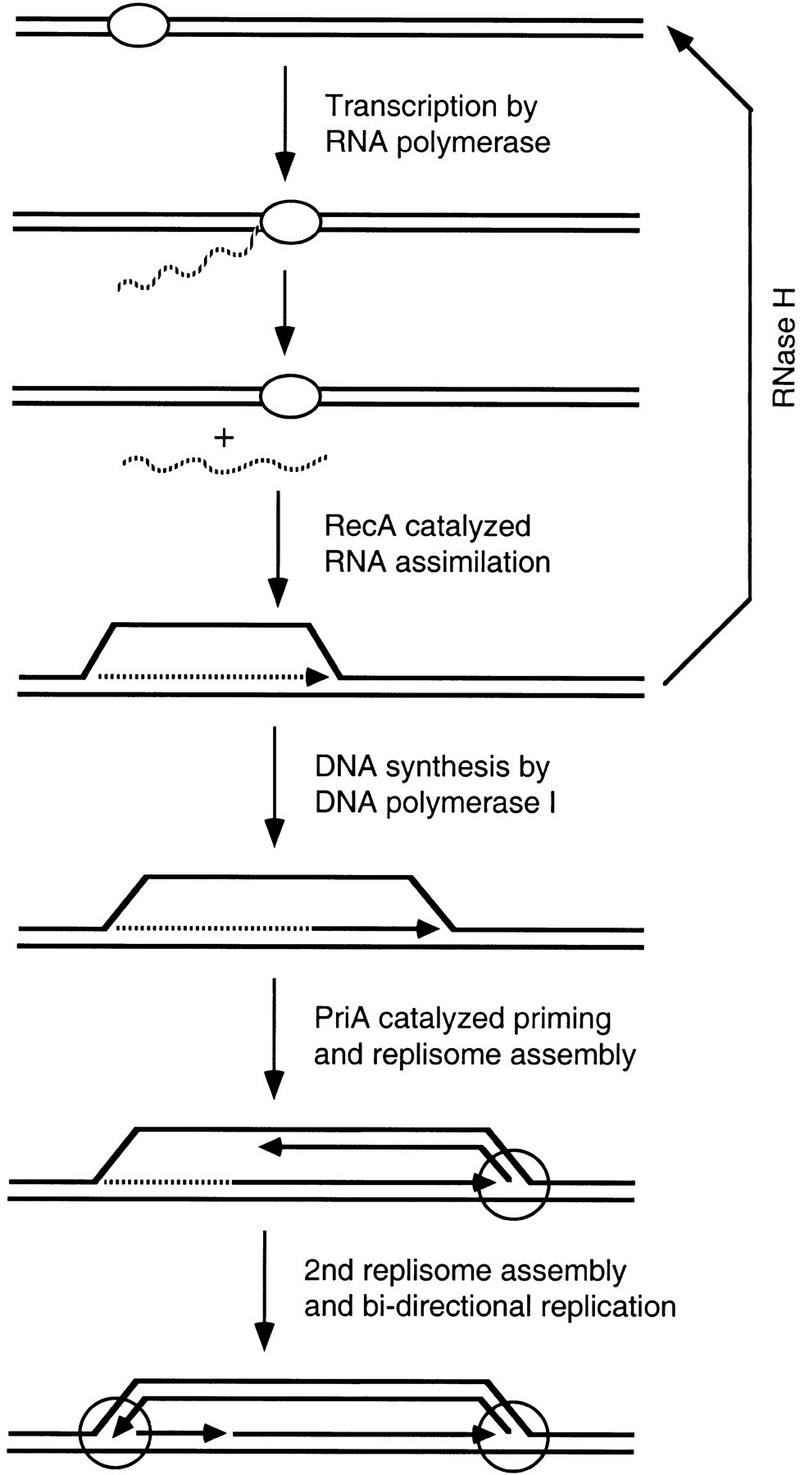
A model for initiation of recombination-dependent replication mediated by RecA. Solid and broken lines represent DNA and RNA strands, respectively. Small arrows indicate 3′ ends. Oval and large circles represent RNA polymerase and replication forks, respectively.
Materials and methods
DNA and RNA substrates
The construction of heteroduplex DNA substrates is illustrated in Figure 1. A 302-bp EcoRI–NcoI fragment (shaded arrow) of the cat gene from pNK2884 (Kleckner et al. 1991) was cloned between the EcoRI and BamHI sites within the multicloning sites (MCS) of pTZ18U and pTZ18R (U.S. Biochemical, Cleveland, OH). Circular ssDNA was prepared by the infection of two strains harboring either of the resultant plasmids with an M13 helper phage. Circular ssDNA from pTZ18Ucat was linearized by cleaving it with ScaI after annealing a complimentary 15-mer (5′-GTGAGTACTCAACCA-3′) at the ScaI site. The two strands were annealed to generate a heteroduplex that had a 455-bp mismatch in the region of the f1 origin of replication. The circular heteroduplex was linearized by ScaI digestion (duplex I). Duplex DNA identical to the duplex I except for the lack of a mismatch region, was generated by ScaI digestion of pTZ18Rcat (duplex II).
The 305-nucleotide RNA transcript was prepared by in vitro transcription with T7 RNA polymerase (a kind gift of Dave Peabody, University of New Mexico), [32P]UTP (3000 Ci/mmole; New England Nuclear, Boston, MA) and three rNTPs (Pharmacia Biotech, Piscataway, NJ). The template DNA (pTZ18Rcat) was prepared by CsCl centrifugation and linearized at the BamHI site at the 3′ end of the cat gene.
R-loop formation reactions
RecA protein-dependent R-loop reactions were carried out in Tris-HCl buffer (33 mm at pH 7.4). In a standard reaction, duplex I DNA (Fig. 1) at a concentration of 15 μm (bp), was mixed with RecA protein (4 μm; New England Biolabs, Beverly, MA), dithiothreitol (2 mm), and MgCl2 (10 mm). The reaction was then initiated by the addition of [32P]-labeled RNA transcript [0.8 μm (nucleotides)], ATPγS (3.0 mm), and SSB protein (1.5 μm). After 30 min, the reaction was terminated by treatment with SDS (0.4 %) and proteinase K (200 μg/ml) followed by phenol extraction. The reaction product was then electrophoresed in a 1% agarose gel, dried, and subjected to autoradiography using a PhosphorImager (Molecular Dynamics, Sonnydale, CA). From the specific activity, the concentration of 32P-labeled RNA used was estimated to be 0.8 μm (nucleotides); thus, the ratio of RNA to DNA was ∼1:19.
Acknowledgments
We thank Steve Kowalczykowski and Tsuneaki Asai for constructive discussion and critical reading of the manuscript. This work was supported by grant GM22092 from the National Institutes of Health to T.K. and by a grant from the Japan Ministry of Education, Science, and Culture to M.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bates2@fas.harvard.edu; FAX (617) 495-0758.
References
- Asai T, Kogoma T. D-loops and R-loops: Alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J Bacteriol. 1994;176:1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: An RNA-DNA hybrid near oriC. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- Bryant FR, Riddles PW, Lehman IR. Studies of the mechanism of DNA pairing by the RecA protein of Escherichia coli. Cold Spring Harbor Symp Quant Biol. 1984;49:535–539. doi: 10.1101/sqb.1984.049.01.060. [DOI] [PubMed] [Google Scholar]
- Cox MM, Lehman IR. recA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc Natl Acad Sci. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— recA protein-promoted DNA strand exchange. Stable complexes of recA protein and single-stranded DNA formed in the presence of ATP and single-stranded DNA binding protein. J Biol Chem. 1982;257:8523–8532. [PubMed] [Google Scholar]
- Fukuoh A, Iwasaki H, Ishioka K, Shinagawa H. ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J. 1997;16:203–209. doi: 10.1093/emboj/16.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Cadwell GW, Kogoma T. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J. 1995;14:2385–2392. doi: 10.1002/j.1460-2075.1995.tb07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Cadwell GW, Kogoma T. Activation of stable DNA replication in rapidly growing Escherichia coli at the time of entry to stationary phase. Mol Microbiol. 1996;21:953–961. doi: 10.1046/j.1365-2958.1996.591419.x. [DOI] [PubMed] [Google Scholar]
- Itaya M, Crouch RJ. A combination of RNase H (rnh) and recBCD or sbcB mutations in Escherichia coli K12 adversely affects growth. Mol & Gen Genet. 1991;227:424–432. doi: 10.1007/BF00273933. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci. 1980;77:2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DP, Rao BJ, Radding CM. RNA-DNA hybridization promoted by E. coli RecA protein. Nucleic Acids Res. 1992;20:4339–4346. doi: 10.1093/nar/20.16.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- Kogoma T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microb Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Skarstad K, Boye E, von Meyenburg K, Steen HB. RecA protein acts at the initiation of stable DNA replication in rnh mutants of Escherichia coli K-12. J Bacteriol. 1985;163:439–444. doi: 10.1128/jb.163.2.439-444.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Hong X, Cadwell GW, Barnard KG, Asai T. Requirement of homologous recombination functions for viability of the Escherichia coli cell that lacks RNase HI and exonuclease V activities. Biochimie. 1993;75:89–99. doi: 10.1016/0300-9084(93)90029-r. [DOI] [PubMed] [Google Scholar]
- Kogoma T, Barnard KG, Hong X. RecA, Tus protein and constitutive stable DNA replication in Escherichia coli rnhA mutants. Mol & Gen Genet. 1994;244:557–562. doi: 10.1007/BF00583907. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Krupp RA. DNA-strand exchange promoted by RecA protein in the absence of ATP: Implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc Natl Acad Sci. 1995;92:3478–3482. doi: 10.1073/pnas.92.8.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Clow J, Krupp RA. Properties of the duplex DNA-dependent ATPase activity of Escherichia coli RecA protein and its role in branch migration. Proc Natl Acad Sci. 1987;84:3127–3131. doi: 10.1073/pnas.84.10.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer NK, Morrical SW. Initiation of DNA Replication. In: Karam JD, editor. Molecular biology of bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1994. pp. 28–42. [Google Scholar]
- Masukata H, Tomizawa J. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell. 1990;62:331–338. doi: 10.1016/0092-8674(90)90370-t. [DOI] [PubMed] [Google Scholar]
- Mosig G, Colowick N, Gruidl ME, Chang A, Harvey AJ. Multiple initiation mechanisms adapt phage T4 DNA replication to physiological changes during T4's development. FEMS Microbiol Rev. 1995;17:83–98. doi: 10.1111/j.1574-6976.1995.tb00190.x. [DOI] [PubMed] [Google Scholar]
- Shaner SL, Radding CM. Translocation of Escherichia coli recA protein from a single-stranded tail to contiguous duplex DNA. J Biol Chem. 1987;262:9211–9219. [PubMed] [Google Scholar]
- Tsang SS, Muniyappa K, Azhderian E, Gonda DK, Radding CM, Flory J, Chase JW. Intermediates in homologous pairing promoted by recA protein. Isolation and characterization of active presynaptic complexes. J Mol Biol. 1985;185:295–309. doi: 10.1016/0022-2836(85)90405-x. [DOI] [PubMed] [Google Scholar]
- Zaitsev EN, Kowalczykowski SC. The simultaneous binding of two double-stranded DNA molecules by Escherichia coli RecA protein. J Mol Biol. 1999;287:21–31. doi: 10.1006/jmbi.1998.2580. [DOI] [PubMed] [Google Scholar]
- ———. 2000. A novel pairing process promoted by Escherichia coli RecA protein: Inverse DNA and RNA strand exchange. Genes & Dev. (in press). [PMC free article] [PubMed]