Summary
Emerging mechanism for tRNA fragment-based translational regulation expands our understanding of the non-decoding roles of tRNA.
Transfer RNA was discovered by Hoagland and Zamecnik and their coworkers in 1958 (Hoagland et al., 1958). During that era, the aminoacylation of tRNAs in the first step of protein synthesis was shown to establish the rules of the genetic code. Later, tRNAs were sequenced and found to be, typically, comprised of 76 nucleotides that were arranged in a cloverleaf secondary structure which, in turn, folded into a L-shaped molecule with the anticodon at the tip of one arm of the L, and the amino acid attachment site at the tip of the other arm (Figure 1). These structural details have contributed substantially to our understanding of the mechanism of action of tRNA in protein synthesis. Surprisingly, it now seems that the well-researched role of tRNA in protein synthesis is only part of the picture. For several years various groups have noted that fragments of tRNAs are naturally produced and, further, that these fragments are associated with specific biological functions (Thompson and Parker, 2009). In this issue of Molecular Cell, Anderson and coworkers (Ivanov et al., 2011 provides mechanistic insight into the functional expansion of the tRNA world, through the generation of specific fragments.
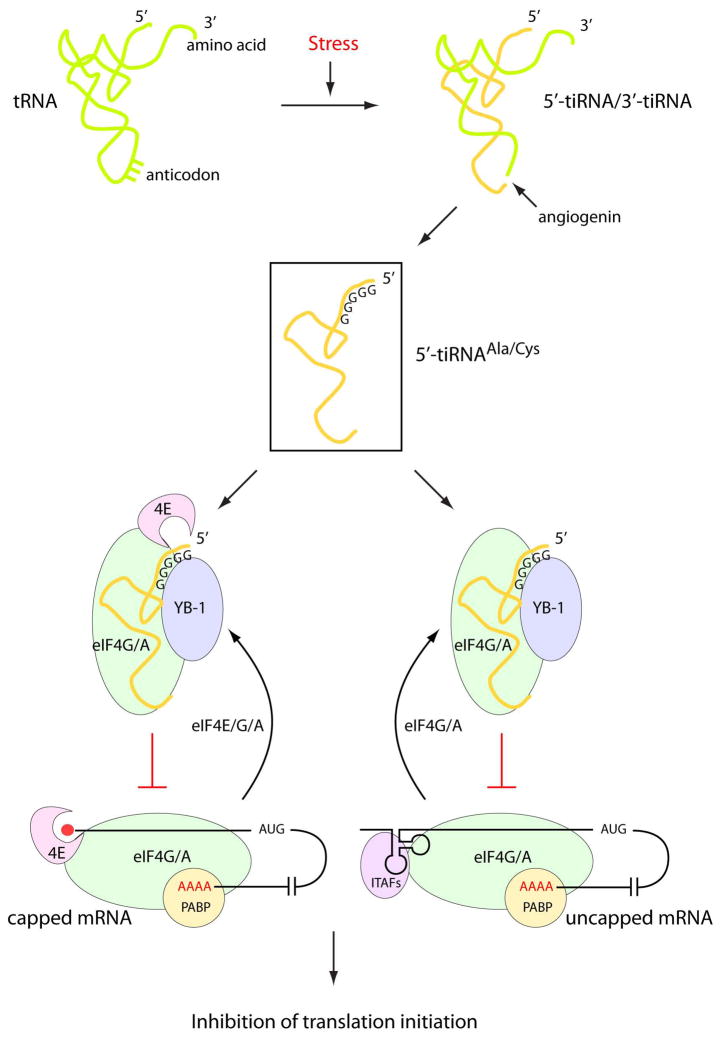
Figure 1. Stress-induced tiRNA-dependent inhibition of translation initiation.
Stress conditions such as oxidation and amino acid starvation induce internalization of extracellular (secreted) angiogenin. When activated, angiogenin cleaves full-length tRNAs at their anticodon loops to give 5′- and 3′-tiRNA fragments of 30–40 nucleotides. A subset of 5′-tiRNAs (5′-tiRNAAla/Cys) that contain an oligo-G motif at the 5′-end cooperates with the translational silencer YB-1 to inhibit translation initiation by recruiting eIF4E/G/A from capped mRNAs or eIF4G/A from uncapped mRNAs.
The fragments of interest are produced by angiogenin, which was discovered by Vallee et al. in 1985 as a secreted protein present in conditioned media of cultured adenocarcinoma cells (Vallee et al., 1985). It was originally characterized as a protein with potent angiogenic activity. Later, and surprisingly, the protein was shown to be a ribonuclease, with substantial sequence similarity to members of the RNase A superfamily (Strydom et al., 1985). Strikingly, stimulation of angiogenesis by angiogenin was dependent on its ribonuclease activity. These observations were particularly interesting because, in 1972, Stein, Moore, and Anfinsen had received the Nobel Prize for their work on pancreatic RNase A. At a time when the tools of modern molecular biology and of recombinant DNA did not exist, they showed that 3 residues—His12, His119, and Lys41—cooperated to form the active site of this monomeric enzyme (Crestfield et al., 1963). That His12, His119, and Lys41 were in close proximity at the active site was borne out by the subsequently determined crystal structure (Wyckoff et al., 1967). Significantly, these same critical residues are present in the active site of angiogenin which, like ribonuclease A, cleaves on the 3′-side of pyrimidines. A key difference between angiogenin and ribonuclease A is the placement in angiogenin of a glutamine in the pyrimidine binding site (Russo et al., 1994). This substitution substantially lowers the ribonucleolytic activity of angiogenin compared to ribonuclease A. This lowering of activity may be essential for the more refined and targeted activity of angiogenin within the cell.
Now it is clear that, under conditions of stress, secreted angiogenin enters cells via receptor-mediated endocytosis and cleaves tRNAs at the well-exposed anticodon loop, to give roughly half-molecules that are designated as 5′- and 3′-tiRNAs (Thompson and Parker, 2009) (Figure 1). These 30–40 nucleotide tiRNAs join the growing list of regulatory non-coding RNAs, such as mi- and pi-RNAs, and smaller tRNA-derived fragments produced by dicer and RNase Z. Stress-induced tRNA cleavage is widely distributed in evolution and, depending on the organism, has been demonstrated with stresses ranging from oxidation to nutrient deprivation. At first glance, the observation that the stress-induced production of tiRNAs resulted in inhibition of protein synthesis was not surprising. After all, cleavage of tRNA should arrest protein synthesis. But it then became clear that the levels of intact tRNAs did not change, even when tiRNAs were produced. Instead, tiRNAs promote the assembly of stress granules in whole cells, which are composed largely of stalled preinitiation complexes, suggesting that the translation initiation machinery is the target of tiRNAs (Emara et al., 2010).
Ivanov et al now show that, consistent with promoting stress granule assembly in vivo, only the 5′-tiRNAs (but not 3′-tiRNAs) inhibit translation in vitro. Moreover, it is only a subset of 5′ti-RNAs that are active. These include 5′-tRNAAla and 5′-tRNACys. The common feature of these two, and not other, tiRNAs is the presence of 4–5 consecutive guanine residues at the 5′-end. In an elegant experiment, the authors activate an inactive 5′-tRNAMet (which lacks the 5′-oligo-G motif) by replacing its first 5 nucleotides with G5. Using in vitro translation and binding assays, they show that 5′-tiRNAAla/Cys inhibits translation initiation by interfering with eIF4G/A binding to uncapped mRNA and, to a lesser extent, with eIF4E/G/A (eIF4F) binding to m7G-capped mRNA (Figure 1). Interestingly, they also show that tiRNAs can displace m7G cap to form a stable complex with eIF4E/G. Lastly, for these tiRNAs to act in the cell, they cooperate with the translational silencer YB-1 to inhibit translation and assembly of stress granules (Figure 1).
These points are clear and well documented by the data presented. However, there is much more to the story that needs to be understood. For example, knocking down YB-1 still leaves the cell vulnerable to production of one-half or more of the stress granules seen without the knock-down. Consistently, for measuring a 5′-tiRNA-induced translation repression with whole cells, the effects of YB-1 are not “all or none”, and are convincing only because of the statistical significance. This partial response also suggests other factors and reactions beyond the tiRNA/YB-1 part of the story. And the structural or mechanistic role of the 5′-oligo-G motif remains an open question. On the later point, a crystal structure of a tiRNAAla/Cys-protein complex would be a major advance. Lastly, because 5′-tiRNAAla/Cys preferentially inhibits translation of uncapped more than capped RNAs in vitro, and because uncapped mRNA can be associated with selective translations of specific mRNAs, for example, those containing an IRES element, a role for the tiRNAs is suggested in reprogramming translation initiation in stressed cells. An important step forward would be to identify the transcripts that are targeted in vivo for tiRNA-dependent regulation of translation.
The delineation of the requirement for the 5′-oligo-G motif raises the question of the role of the other angiogenin-produced non-oligo-G-containing 5′-tiRNAs, and of the complementary 3′-tiRNAs that simultaneously appear. Also, 5′-and 3′-tRNA half molecules do not necessarily quickly dissociate (because of their extensive complementarity) and, if dissociated, can easily re-anneal to form stable structures. This consideration raises the question of what drives apart the two halves of the cleaved tRNA? In addition, tRNAs are bound within cells to a large population of tRNA synthetases and elongation factors. Does angiogenin cleave tRNA when bound to a tRNA synthetase or other tRNA binding proteins? Such binding could suppress the dissociation of the two halves created by cleavage, suggesting that free tRNA may be the more likely substrate in vivo.
In some sense, the further elaboration of functions for tRNAs mirrors that seen with aminoacyl tRNA synthetases. These enzymes also have expanded functions in cell signaling pathways when they are fragmented by alternative splicing or proteolytic cleavage. Both tRNAs and aminoacyl tRNA synthetases appeared at the base of the tree of life, before it split into the 3 great kingdoms. This early origin offered the opportunity for the eons of evolution to co-opt these molecules for other functions, through diverting them from a direct role in translation by cleaving them into pieces. The tiRNA fragments have large numbers of unpaired nucleotides (in the secondary structures proposed by Ivanov et al for tiRNAAla/Cys) and the fragments of tRNA synthetases have exposed new surfaces. These structural transformations of tRNAs (and tRNA synthetases) redirected them to a new repertoire of potential interacting partners. Thus, the functional expansion of tRNAs in evolution could have been driven by stress itself, such as oxidative damage, which led to occasional fragmentation at perhaps the most exposed site—the anticodon. Serendipitously, this fragmentation produced tiRNAs that found novel interactions that were advantageous.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Crestfield AM, Stein WH, Moore S. Properties and conformation of the histidine residues at the active site of ribonuclease. J Biol Chem. 1963;238:2421–2428. [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958;231:241–257. [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;xxx:xxx–xxx. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo N, Shapiro R, Acharya KR, Riordan JF, Vallee BL. Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc Natl Acad Sci U S A. 1994;91:2920–2924. doi: 10.1073/pnas.91.8.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strydom DJ, Fett JW, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Amino acid sequence of human tumor derived angiogenin. Biochemistry. 1985;24:5486–5494. doi: 10.1021/bi00341a031. [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Vallee BL, Riordan JF, Lobb RR, Higachi N, Fett JW, Crossley G, Buhler R, Budzik G, Breddam K, Bethune JL, et al. Tumor-derived angiogenesis factors from rat Walker 256 carcinoma: an experimental investigation and review. Experientia. 1985;41:1–15. doi: 10.1007/BF02005853. [DOI] [PubMed] [Google Scholar]
- Wyckoff HW, Hardman KD, Allewell NM, Inagami T, Johnson LN, Richards FM. The structure of ribonuclease-S at 3.5 A resolution. J Biol Chem. 1967;242:3984–3988. [PubMed] [Google Scholar]