Abstract
Neurons are highly specialized cells whose connectivity at synapses subserves rapid information transfer in the brain. Proper information processing, learning, and memory storage in the brain requires continuous remodeling of synaptic networks. Such remodeling includes synapse formation, elimination, synaptic protein turnover, and changes in synaptic transmission. An emergent mechanism for regulating synapse function is posttranslational modification through the ubiquitin pathway at the postsynaptic membrane. Here, we discuss recent findings implicating ubiquitination and protein degradation in postsynaptic function and plasticity. We describe postsynaptic ubiquitination pathways and their role in brain development, neuronal physiology, and brain disorders.
Keywords: ubiquitin, neuron, synapse, glutamate, circuit, dendrite
INTRODUCTION
Neurons are complex cells whose connectivity at synapses mediates rapid information transfer in the brain. The human brain contains an estimated 100 billion neurons and 100 trillion synapses. Understanding how so many synapses are individually modified to fine-tune neural circuitry remains an enduring challenge for modern biology. Proper information processing and storage in the brain requires a fine balance between circuit plasticity and stability. Furthermore, initial brain development and learning require synapse formation, elimination, and functional plasticity. In contrast, stable information storage requires circuit constancy in the form of stable, enduring synaptic networks. Both the adaptability and stability of synapses is ultimately controlled by the regulated removal, addition, and posttranslational modification of synaptic proteins (Figure 1).
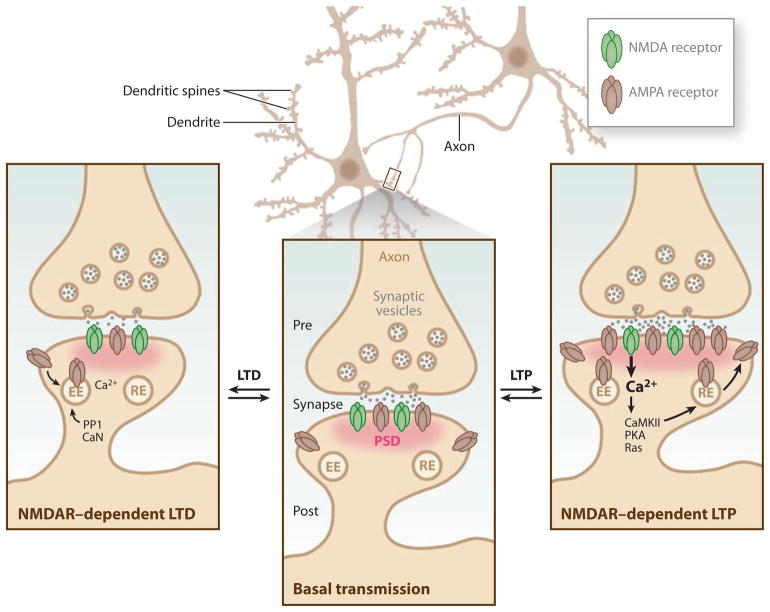
Figure 1.
Bidirectional plasticity at excitatory synapses. Typical synapses in the central nervous system consist of an axon serving as the presynaptic terminal in a junctional contact with the postsynaptic cell. The postsynaptic membrane contains a dense matrix organized in an array of scaffolding molecules known as the postsynaptic density (PSD, pink), which allows for the anchoring and positioning of receptors, such as α-amino-3-hydroxyl-5-methyl-4-isoxazole propionic acid (AMPA) receptors and N-methyl-D-aspartic acid (NMDA) receptors (NMDARs), for efficient activation by presynaptically released glutamate neurotransmitter (middle inset). Learning-related brain plasticity involves the strengthening or weakening of synapses. At excitatory glutamatergic synapses, such synaptic plasticity can be finely tuned by altering the channel properties or abundance of glutamate receptors within the PSD. A prolonged enhancement of synaptic transmission at a synapse is known as long-term potentiation (LTP), whereas a persistent weakening of synaptic transmission at a synapse is known as long-term depression (LTD) (Malenka & Bear 2004). LTP is the most celebrated cellular model for learning and memory in the brain and can be subdivided into two temporal phases that differ in their mechanism of expression. The initial phase of NMDAR-dependent LTP, termed early phase LTP (E-LTP), lasts for ~1–2 hours. E-LTP requires Ca2+ influx through NMDARs and activation of CaMKII, and it is expressed in part by an increase in the number of AMPA receptors trafficked through recycling endosomes (RE). The second phase, known as late phase LTP (L-LTP), requires protein synthesis and gene transcription in the postsynaptic cell to allow for long-lasting enhancement of synaptic transmission for several hours to weeks (Malenka & Bear 2004). Structural remodeling such as increases in PSD size, the growth of new dendritic spines, and the enlargement of preexisting spines is also associated with LTP (Holtmaat & Svoboda 2009). Several forms of LTD contribute to learning-related plasticity at glutamatergic synapses. Various forms of LTD require NMDARs, group I metabotropic glutamate receptors (mGluRs), and endocannabinoid signaling (Malenka & Bear 2004). NMDAR-dependent LTD in CA1 hippocampus is the best-studied form of LTD and, like LTP, its induction requires Ca2+ influx through NMDARs, but it depends on protein phosphatase 1 (PP1) and calcineurin (CaN). The expression of mGluR-LTD requires protein synthesis. In general, LTD is accompanied by a reduction in the number of AMPA receptors that are removed by endocytosis and transported to early endosomes (EE) (Malenka & Bear 2004).
Recent studies indicate that changes in synaptic architecture occur in part via post-translational modification through the ubiquitin pathway. Since its discovery in the late 1970s and early 1980s (Ciechanover et al. 1980, Hershko et al. 1980), the covalent addition of ubiquitin to target proteins has been shown to mediate protein degradation, signal transduction, and membrane trafficking (Haglund & Dikic 2005). Some 30 years later, the ubiquitin pathway has emerged as an important mechanism controlling normal brain functions, including synapse maintenance, regulation, and organization, as well as brain disorders (Yi & Ehlers 2007). In the current review, we highlight recent work on protein ubiquitination at the postsynaptic side of the synapse, its role in brain development and plasticity, and implications for brain disorders. Additional roles for ubiquitin in brain function, neural development, and presynaptic function have been recently reviewed elsewhere (Haas & Broadie 2008, Patrick 2006, Segref & Hoppe 2009, Tai & Schuman 2008, Yi & Ehlers 2007).
PROTEIN MODIFICATION BY UBIQUITIN
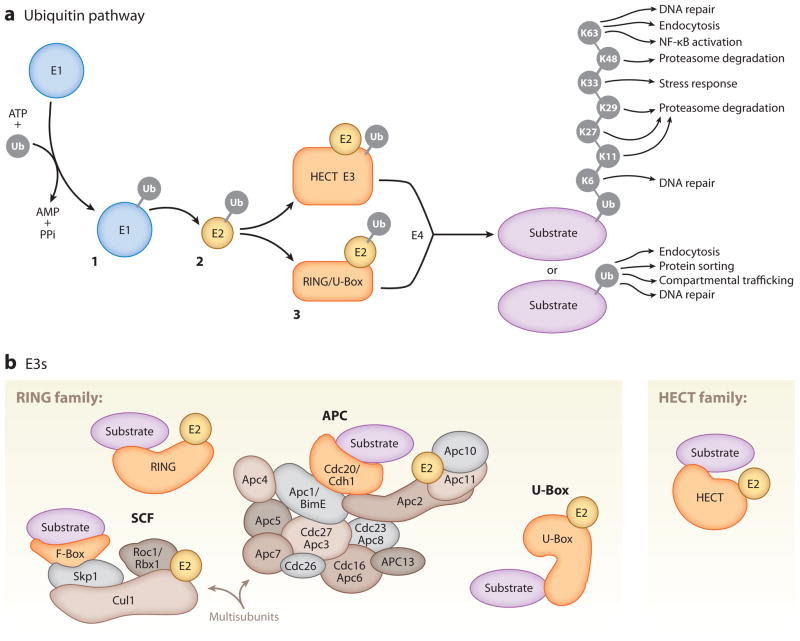
Ubiquitination is a posttranslational modification corresponding to direct conjugation of the 76–amino acid protein ubiquitin to a lysine residue of a protein substrate via an isopeptide bond. The transfer of ubiquitin to a substrate relies on an enzymatic cascade consisting of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), ubiquitin ligases (E3s), and sometimes polyubiquitin ligases (E4s) (Figure 2a; Hershko & Ciechanover 1998, Hoppe 2005). Among the enzymes required for substrate ubiquitination, ubiquitin E3s typically confer substrate specificity, and the coordinated interplay between ubiquitin E2s, E3s, and substrates determines the sites and linkage topology of ubiquitin addition.
Figure 2.
Components of the ubiquitin (Ub) pathway. (a) Substrate ubiquitination requires a three-step enzymatic reaction. In step 1, a ubiquitin-activating enzyme (E1) activates free ubiquitin, a process in which the C-terminal glycine residue of ubiquitin is coupled to a cysteine residue of the E1 via a thioester linkage. In step 2, the activated ubiquitin is transferred to a cysteine residue of a ubiquitin-conjugating enzyme (E2) and linked via a thioester linkage. In step 3, a ubiquitin ligase (E3) transfers ubiquitin to the substrate via an amide isopeptide linkage to the ε-amino group of the substrate lysine (Hershko & Ciechanover 1998). In some cases, a polyubiquitin ligase (E4) coordinates with a ubiquitin E3 to lengthen ubiquitin chains (Hoppe 2005). Ubiquitin can be attached to lysine residues of previously conjugated ubiquitins in multiple polyubiquitin chain configurations (K6, K11, K27, K29, K33, K48, K63) to regulate various functions. (b) Ubiquitin E3s are divided into the RING (really interesting new gene) and HECT (homologous to E6-associated protein C terminus) domain families, among others. The RING domain family is the largest family of ubiquitin E3s. RING ubiquitin E3s function as single subunits or in a multisubunit complex. Illustrated here are the best-studied multisubunit RING ubiquitin E3s, the Skp1/Cullin/F-box (SCF) and anaphase-promoting complex/cyclosome (APC/C) complexes. U-Box proteins are a distinct set of RING-like ubiquitin E3s.
A Diversity of Ubiquitin Ligases
The most abundant and diverse ubiquitin pathway enzymes are the ubiquitin E3s, which in mammalian genomes are encoded by several hundred genes (Deshaies & Joazeiro 2009). There are two main classes of ubiquitin E3s: the RING (really interesting new gene) and HECT (homologous to E6-associated protein C terminus) domain families of E3s (Figure 2b; Deshaies & Joazeiro 2009, Rotin & Kumar 2009). The RING family of ubiquitin E3s is the largest class and contains approximately 600 members. Within this family are multisubunit cullin-RING ubiquitin E3s. One of the best-studied examples is the SCF (Skp1/Cullin/F-box) complex, which utilizes a series of adaptors containing RING finger and cullin domains for substrate binding, ubiquitin E3 activity, and regulation (Deshaies & Joazeiro 2009). Another is the APC/C (anaphase-promoting complex/cyclosome), a 1.5-MDa multicomponent ubiquitin E3 complex that also requires a RING finger and a cullin domain for substrate ubiquitination. Typical substrates of APC/C contain an amino acid recognition motif such as a D-Box or KEN-Box (Peters 2006). The TRIM/RBCC (tripartite-motif/RING domain, one or two B-box motifs and a coiled-coil region) proteins represent a subclass of RING ubiquitin E3s that has a similar RING structure within their TRIM domain (Meroni & Diez-Roux 2005). The PHD (plant homeodomain) finger and LIM (Lin-11/Isl-1/Mec-3) domain subfamilies of RINGs share a similar fold to the RING domain but do not exhibit ubiquitin E3 activity (Deshaies & Joazeiro 2009). The newest addition to the RING family is the U-Box-containing ubiquitin E3s that catalyze substrate ubiquitination through a RING-like structure (Figure 2b; Hatakeyama et al. 2001).
The second main class of ubiquitin E3s is the HECT domain–containing family (Figure 2b), which includes E6-AP/Ube3a, NEDD4, HERC, and other HECT ubiquitin E3s (Rotin & Kumar 2009). HECT family ubiquitin E3s contain a conserved cysteine residue within the HECT domain that accepts ubiquitin from an E2 and is a direct ubiquitin donor. This is in contrast to members of the RING family, in which the RING domain serves as an adaptor for the binding of a ubiquitin-loaded E2 to catalyze the transfer of ubiquitin directly from the E2 to cognate substrates (Figure 2a).
Many Flavors of Ubiquitin Modification
In addition to the diversity of ubiquitin E3s, the ubiquitin moiety itself can be conjugated to substrates in a variety of configurations that mediate distinct cellular functions (Figure 2a). For example, substrate lysines can be covalently modified by a single ubiquitin. Such mono-ubiquitination facilitates endocytosis, initiates DNA repair pathways, and regulates protein sorting at the trans-Golgi network (Haglund & Dikic 2005). Ubiquitin can also be conjugated via its own internal lysines to form polymeric ubiquitin chains on protein targets (Figure 2a). The most prominent type of polyubiquitination occurs through lysine 48 (K48) of ubiquitin, which typically targets substrates for proteasome-dependent degradation. Another form of polyubiquitination is K63-linked chains, which can scaffold signaling complexes such as the NF-κB signaling pathway, regulate DNA repair, and control endo-cytosis (Haglund & Dikic 2005). An additional form of polyubiquitination known as linear ubiquitination has been shown to mediate activation of NF-κB (Tokunaga et al. 2009). Other modes of polyubiquitination occur through K6, K11, K27, K29, and K33, whose functions are less clear (Ikeda & Dikic 2008) but can also lead to proteasomal degradation (Xu et al. 2009).
Deubiquitinases Reverse and Trim Ubiquitin Chains
Protein ubiquitination is a reversible process mediated by a family of ubiquitin proteases also known as deubiquitinases (DUBs). Overall, the number of DUBs is on the order of 100 (Komander et al. 2009). DUBs cleave or trim polyubiquitin chains and aid in priming ubiquitin for the conjugation cascade. There are five distinct subfamilies of DUBs: ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Josephin proteases, and JAB1/MPN/Mov34 metalloenzymes (JAMMs). Most DUBs are cysteine proteases with the exception of the JAMM family, which are Zn2+ metalloproteases. The diversity of DUB family members allows for substrate specificity and for rearrangement or removal of specific ubiquitin configurations (Komander et al. 2009).
UBIQUITINATION IN SYNAPTIC PLASTICITY, LEARNING, AND MEMORY
For decades, the prevailing model for long-term synapse modification has been changes in protein synthesis and stimulus-dependent gene transcription (Flavell & Greenberg 2008, Martin et al. 2000). The contribution of protein turnover and degradation to long-term structural and functional changes at synapses has been much less studied. One of the first demonstrations of a role for protein degradation in regulating synapse function was the finding in Aplysia sensory neurons that the stability of the inhibitory regulatory subunit of the cAMP-dependent protein kinase (PKA) was altered during long-term facilitation (LTF) (Bergold et al. 1990, Chain et al. 1999, Chain et al. 1995, Greenberg et al. 1987, Hegde et al. 1993). Further work revealed that the DUB Ap-Uch was required for both LTF and enhanced PKA degradation by the proteasome (Hegde et al. 1997). Direct inhibition of the proteasome could induce LTF and increase synaptic strength, effects that were correlated with an increase in synapse number between sensory and motor neurons (Zhao et al. 2003).
In mammals, one of the first genetic links between the ubiquitin pathway and synaptic plasticity was the discovery that mice deficient in brain-expressed Ube3a (E6-AP), a HECT domain ubiquitin E3 encoded by the UBE3A gene that is mutated in Angelman syndrome, are seizure prone, have deficits in contextual learning, and exhibit impaired hippocampal long-term potentiation (LTP) (Jiang et al. 1998). More recently, studies have identified widespread synaptic plasticity and learning deficits in mice lacking Ube3a in the brain (van Woerden et al. 2007, Weeber et al. 2003, Yashiro et al. 2009) (see below).
In addition, and likely as a consequence of regulating synaptic plasticity, ubiquitination controls behavior and other higher brain functions. One of the first indications of ubiquitin-dependent control of behavior was the finding that the ubiquitin proteasome system (UPS) mediates LTF of the gill withdrawal reflex in Aplysia (Chain et al. 1995, Hegde et al. 1993, Hegde et al. 1997). Initial studies in the rat showed that bilateral infusion of the proteasome inhibitor lactacystin into CA1 hippocampus causes retrograde amnesia of inhibitory avoidance learning when given within 7 h of training (Lopez-Salon et al. 2001). Inhibitory avoidance training increases the abundance of polyubiquitin conjugates and 26S proteasome proteolytic activity in the hippocampus (Lopez-Salon et al. 2001), which supports learning-dependent activation of the UPS. At glutamatergic synapses in the amygdala, fear conditioning in rats results in an increase in GluA1-containing α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionic acid (AMPA) receptors (Rumpel et al. 2005), a major subclass of excitatory neurotransmitter receptors. Interestingly, pharmacological inhibition of the proteasome augments fear-conditioned learning and increases surface expression levels of GluA1 in amygdala slices (Yeh et al. 2006). Inhibition of the proteasome further prevents the extinction of conditioned fear that is elicited by the N-methyl-D-aspartic acid (NMDA) receptor partial agonist, D-cycloserine, and prevents the associated decrease in surface GluA1 expression (Mao et al. 2008), which suggests that the UPS may reverse or dampen learning-related plasticity. Consistent with this general notion are recent findings that infusion of proteasome inhibitors into CA1 hippocampus immediately following retrieval of contextual fear memory prevents the impairment of contextual memory induced by protein synthesis inhibitors and prevents extinction of fear memory (Lee et al. 2008). In the mouse, both the initial consolidation of allocentric spatial memories and reconsolidation after reactivation requires proteasome activity in CA3 hippocampus (Artinian et al. 2008), which indicates that ubiquitin-dependent protein degradation is a key step for memory consolidation. Interestingly, postsynaptic proteins are ubiquitinated and degraded in the hippocampus after retrieval of contextual fear memory (Lee et al. 2008), which suggests that the UPS alters learning through synapse remodeling. In the crab Chasmagnathus, proteasome inhibition blocks long-term spatial memory consolidation but not short-term memory (Merlo & Romano 2007). These studies highlight the evolutionary conservation of UPS dependency in a variety of forms of learning and behavior in worms, Aplysia, flies, crabs, and rodents.
Further evidence for a role of ubiquitination in learning and memory came from the generation of mice deficient in the ubiquitin E3 APCCdh1. Mice heterozygous for Cdh1, a catalytic subunit of the APC/C complex (Figure 2b), have defects in late-phase LTP (L-LTP) in the hippocampus and are deficient in contextual fear-conditioning (Li et al. 2008). The specific substrates of APC/C required for this phenotype are unknown.
Learning and memory require prolonged alteration of synapses. Both macromolecular synthesis and proteasome activity are required for the maintenance of L-LTP (Dong et al. 2008, Fonseca et al. 2006, Frey et al. 1993), which suggests a tight coupling of protein synthesis and degradation in learning-related plasticity. Notably, blocking either protein synthesis or degradation prevents L-LTP, but simultaneously blocking protein synthesis and protein degradation pathways rescues L-LTP (Dong et al. 2008, Fonseca et al. 2006), which suggests that synthesis-degradation coupling is critical for LTP maintenance. Interestingly, proteasome inhibition enhances the induction of E-LTP in a manner that can partially be blocked by protein synthesis inhibitors and requires the action of NMDA receptors, PKA, and cAMP response element–binding (CREB) (Dong et al. 2008). Taken together, these findings support a model whereby the UPS enhances the induction but inhibits the maintenance of L-LTP.
Deubiquitination is also critical for learning and memory. In mice, the ubiquitin C-terminal hydrolase L3 (UCH-L3), an ortholog of the Aplysia DUB Ap-Uch, is required for working memory (Wood et al. 2005). Interestingly, hippocampal LTP is not affected in mice lacking UCH-L3 (Wood et al. 2005), which suggests that different DUBs have selective functions in distinct paradigms of learning and memory.
In addition to synaptic potentiation, the UPS is important for long-term depression (LTD). During LTD induction at sensorimotor synapses in Aplysia, protein ubiquitination and Ap-Uch levels are increased in a manner dependent on the transcription factor ATF4 (activating transcription factor 4) and the proteasome (Fioravante et al. 2008). Proteasome activity is also required for a decrease in synapse size following induction of LTD (Fioravante et al. 2008). In mammals, inhibition of proteasome activity reduces the magnitude of NMDA receptor–dependent and mGluR-dependent LTD in hippocampal slices (Colledge et al. 2003, Hou et al. 2006). However, in other studies, proteasome inhibition did not alter NMDA receptor-dependent LTD but led to an enhancement of mGluR-dependent LTD (Citri et al. 2009). Although the reasons for the observed differences between these studies is not clear, it is intriguing that the UPS may play a prominent role in some forms of LTD but not others.
It is still unclear how the degradation of specific protein targets can drive the expression of certain forms of memory. Ensembles of post-synaptic proteins are degraded in an activity-dependent manner (Ehlers 2003) that suggests multiple classes of proteins may require degradation following LTP. In the case of LTP, one potential target for degradation is ATF4, which undergoes robust proteasome-dependent degradation following chemical induction of LTP (Dong et al. 2008). Degradation of ATF4, a repressor of CREB, is predicted to promote CREB activation and CREB-induced transcription of BDNF, a growth factor involved in the expression of LTP (Dong et al. 2008). More recent work has solidified the idea that the coupling of protein synthesis and degradation at synapses is required for the expression of molecules critical for learning and memory (Banerjee et al. 2009). Specifically, MOV10, a homolog of the Drosophila RNA-induced silencing complex (RISC), is localized at synapses and is degraded by the proteasome following increases in synaptic activity. MOV10 degradation requires NMDA receptor activity and is required for the translation of proteins such as Ca2+/calmodulin-dependent kinase type II (CaMKII) and LIM kinase 1 (LimK1) (Banerjee et al. 2009).
Although the ubiquitin pathway plays a pivotal role in various states of learning, it has been less clear how the induction of synaptic plasticity is coupled to the ubiquitination of specific molecules essential for remodeling synapses. An important clue came from the finding that changes in synaptic activity lead to bidirectional ubiquitination of ensembles of synaptic proteins involved in remodeling the postsynaptic density (PSD) and that such changes regulate downstream signaling pathways implicated in learning and memory (Ehlers 2003). Subsequent findings confirmed that these ubiquitin-dependent changes in PSD proteins occur during in vivo paradigms of learning (Lee et al. 2008).
Several important questions remain regarding the role of protein degradation in synaptic plasticity. For example, how is proteasome activity itself regulated, and how could the spatial control of proteasome activity target proteins destined for degradation? Insights into these questions have been provided by the demonstration that the proteasome itself is transported into dendritic spines in response to synaptic activity (Bingol & Schuman 2006). The proteasome-associated protein NAC1 may direct this recruitment because mice lacking NAC1 do not exhibit activity-dependent translocation of the proteasome into dendritic spines (Shen et al. 2007). Strengthening the link between protein turnover and synaptic plasticity is the recent finding that proteasome activity is modulated during synaptic activity. As visualized using a rapidly degraded photoactivatable form of green fluorescent protein (paGFPu), blockade of synaptic activity in cultured hippocampal neurons decreases proteasome activity whereas increasing synaptic activity increases proteasome activity (Djakovic et al. 2009), consistent with biochemical observations (Ehlers 2003). Increased proteasome activity is dependent on Ca2+ influx through NMDA receptors and requires L-type voltage-gated Ca2+ channels (VGCCs) and CaMKII activity (Djakovic et al. 2009). Interestingly, CaMKII phosphorylates the AAA ATPase 19S proteasome subunit, Rpt6 (Bingol et al. 2010, Djakovic et al. 2009), which stimulates proteasome activity. Moreover, CaMKII translocation is required for activity-dependent proteasome recruitment and degradation of polyubiquitinated proteins in dendritic spines (Bingol et al. 2010). It is possible that these mechanisms are involved in controlling ensembles of protein substrates at the synapse following changes in activity (Bingol & Schuman 2006, Bingol et al. 2010, Ehlers 2003, Shen et al. 2007).
UBIQUITIN-DEPENDENT REGULATION OF POSTSYNAPTIC ASSEMBLY AND STABILITY
Synapse assembly and stability are critical for establishing and maintaining functional neuronal circuits. Although roles for growth factor and adhesion-based signaling pathways leading to synapse formation are well established (Dalva et al. 2007, Mattson 2008), recent studies indicate that postsynaptic ubiquitination controls synapse composition and contributes to synapse assembly and elimination.
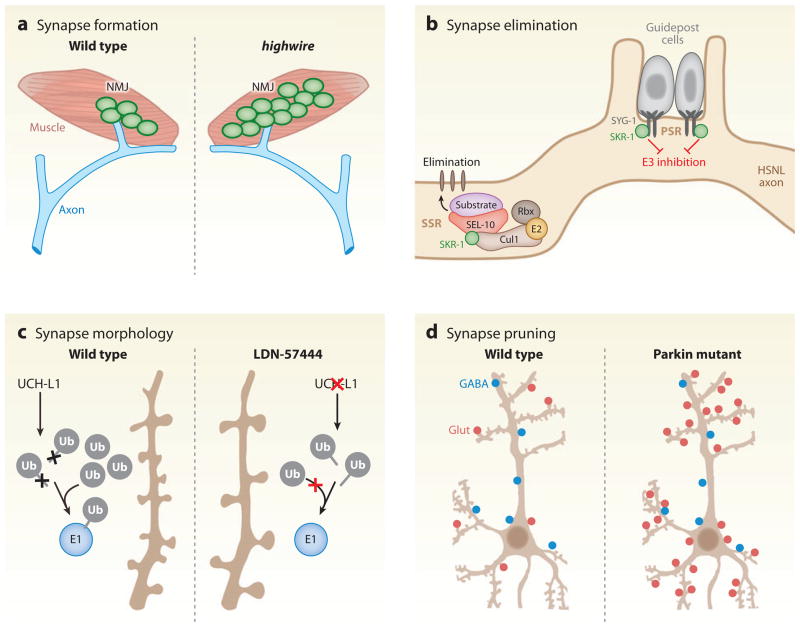
One of the first indications that ubiquitin pathways regulate synaptogenesis came from studies of presynaptic proteins at the glutamatergic neuromuscular junction (NMJ) of Drosophila (DiAntonio et al. 2001). Initially, ubiquitin regulation of the NMJ was suggested by the finding that ubiquitin is highly enriched in motor endplates at the human NMJ (Serdaroglu et al. 1992). Genetic screens of NMJ morphology in Drosophila identified the RING ubiquitin E3 highwire and the DUB fat facets as key regulators of synapse morphology (DiAntonio et al. 2001, Wan et al. 2000). Flies deficient in highwire display synapse overgrowth (Wan et al. 2000), which is phenocopied by overexpression of the DUB fat facets (DiAntonio et al. 2001); this suggests that a delicate balance of ubiquitination and deubiquitination regulates synapse size and morphology (Figure 3a).
Figure 3.
Ubiquitination in synapse regulation. (a) Loss of function of the ubiquitin E3 highwire (hiw) leads to overgrowth of presynaptic terminals at the Drosophila neuromuscular junction (NMJ) (DiAntonio et al. 2001). (b) Sequestration and inhibition of the SCF ubiquitin E3 subunit, SKR-1, by its binding to the synaptic adhesion molecule SYG-1 prevents synapse elimination in the Caenorhabditis elegans hermaphrodite-specific motor neuron (HSNL) axon at the primary synapse region (PSR). During development, synapses in the HSNL axon are eliminated in the secondary synapse region (SSR) owing to the lack of SKR-1 inhibition and corresponding assembly of a functional SKR-1 SCF ubiquitin E3 complex composed of SKR-1, CUL1, SEL-10, and Rbx (Ding et al. 2007). (c) In hippocampal cultures, inhibition of the DUB UCH-L1 by the isatin O-acyl oxime compound, LDN-57444, decreases spine density and increases spine length and width, likely by depletion of free ubiquitin (Cartier et al. 2009). (d) In hippocampal cultures, loss of the RING E3 ligase parkin or expression of Parkinson’s disease–linked mutant forms of parkin increases the number of excitatory synapses (Glut, red circles) with no change in inhibitory synapse number (GABA, blue circles) (Helton et al. 2008).
Interestingly, postsynaptic disruption of the proteasome at the fly NMJ leads to muscle atrophy and gross morphological defects in the sarcomere (Haas et al. 2007b). Furthermore, acute and prolonged postsynaptic proteasome dysfunction leads to an increase in synaptic transmission and postsynaptic accumulation of GluRIIB glutamate receptors in third instar larvae (Haas et al. 2007a). Proteasome inhibition in the postsynaptic muscle cell leads to an increase in both total and ubiquitinated DLG, a postsynaptic MAGUK scaffold protein in the PSD-95 family, indicating that UPS-dependent degradation of DLG may be involved in the maintenance of synaptic transmission at the Drosophila NMJ (Haas et al. 2007a). Such a scenario is analogous to the activity-dependent degradation of PSD-95 (Colledge et al. 2003) and the activity-dependent ubiquitination of Shank, GKAP/SAPAP, and AKAP79/150 (Ehlers 2003, Hung et al. 2010) described at mammalian glutamatergic synapses, which suggests that scaffold ubiquitination and degradation may be a general feature of synapse remodeling conserved across evolution.
Ubiquitin Ligase Control of Synapse Formation
The RING domain ubiquitin E3 PDZRN3A has been implicated in the regulation of NMJ development in mammals through ubiquitination of the receptor tyrosine kinase MuSK (muscle-specific kinase). MuSK activation is required for AChR receptor clustering and the proper formation of the NMJ (DeChiara et al. 1996, Wallace 1989). Overexpression of PDZRN3A in muscle leads to defective NMJ formation in mice and a decrease in postsynaptic MuSK expression levels, which supports the notion that regulation of MuSK by PDZRN3A is required for NMJ formation (Lu et al. 2007).
The identification of numerous multisubunit ubiquitin E3 complexes with the ability to assemble from multiple subunits leads to the conundrum of how their assembly in a variety of unique ubiquitin E3 configurations can degrade specific substrates. An elegant example demonstrating how the spatial assembly of ubiquitin E3s can be utilized to dictate synapse elimination has been provided in Caenorhabditis elegans. In C. elegans, synapse elimination is observed during development at the glutamatergic hermaphrodite-specific motor neuron (HSNL), which forms synapses with the vulval muscles. In the young adult, the secondary synapse region (SSR) in the HSNL is selectively eliminated whereas the primary synapse region (PSR) synapses are retained near the vulva into adulthood (Ding et al. 2007, Shen & Bargmann 2003). The SCF ubiquitin E3 subunits SKR-1 (homolog of vertebrate Skp1) and SCFSEL-10 regulate synapse elimination at HSNL synapses in a spatially restricted manner by an inhibitory interaction with the synaptic adhesion molecule SYG-1 (Ding et al. 2007) (Figure 3b). Specifically, SYG-1 is localized to PSR synapses in the HSNL and binds to SKR-1, which prevents the assembly of an SCF complex containing SKR-1 and SCFSEL-10 at the PSR, whereas at the SSR, a functional SCF E3 ligase is formed, which results in elimination of SSR synapses. Although the relevant ubiquitin ligase functions presynaptically in this case, the ultimate effect includes postsynaptic disassembly. To date, the substrates targeted by the SCFSEL-10 ubiquitin E3 in SSR synapses remain unknown. The use of forward genetics in C. elegans should allow a dissection of both pre- and postsynaptic mechanisms mediating HSNL synapse elimination.
At mammalian glutamatergic synapses, the Rap GTPase activating protein (GAP), SPAR, is an actin-binding protein that is concentrated in the PSD. Overexpression of SPAR leads to irregularly shaped dendritic spines and an increase in dendritic spine head size owing to SPAR’s ability to rearrange the actin cytoskeleton (Pak et al. 2001). Synaptic levels of SPAR are regulated by the serum-inducible serine-threonine kinase/polo-like kinase 2 (SNK/Plk2), which itself is upregulated by neural activity. Induction of SNK/Plk2 leads to ubiquitin-dependent degradation of SPAR by the SCFβTrCP multisubunit ubiquitin E3 (Ang et al. 2008, Pak & Sheng 2003). SPAR binds to βTrCP in a SNK/Plk2-dependent manner. Whereas manipulations that increase SPAR enlarge spines (Pak et al. 2001), overexpression of dominant negative SCFβTrCP did not alter spine morphology (Ang et al. 2008), which suggests additional pathways for SPAR degradation or additional effects of SCFβTrCP. Indeed, in C. elegans, βTrCP (LIN-23) regulates clustering of the GLR-1 glutamate receptor via degradation of β-catenin (see below) (Dreier et al. 2005).
Postsynaptic structures are assembled on dendrites, and dendritic growth and branching occurs in concert with synapse formation (Ahmari et al. 2000, Niell et al. 2004, Washbourne et al. 2002). Several recent studies have implicated ubiquitin-dependent mechanisms in dendritic morphogenesis. For example, the PSD-enriched RING E3 ligase mindbomb1 (Mib1) alters neuronal branching and is regulated by cyclin-dependent kinase 5 (Cdk5), a kinase that is critical for dendritic morphogenesis (Cheung & Ip 2007, Choe et al. 2007). An additional role for ubiquitination in dendritic development was recently provided by the discovery that CaMKIIα promotes the degradation of liprin-α following chronic enhancement of synaptic activity (Hoogenraad et al. 2007). Liprin-α/syd-2 was previously identified as functioning presynaptically to regulate synaptic transmission, synapse structure, and axon guidance (Spangler & Hoogenraad 2007). Overexpression of constitutively active CaMKIIα results in decreased levels of liprin-α that is dependent on liprin-α’s PEST domain. Deletion of this PEST domain impairs dendritic morphogenesis in hippocampal neurons, which suggests that stabilization of liprin-α leads to defects in dendritic morphogenesis (Hoogenraad et al. 2007). The degradation of liprin-α is most likely controlled by the ubiquitin E3 APC/C because RNA interference (RNAi) of APCCdh1 increases liprin-α levels in neurons (Hoogenraad et al. 2007). It is interesting that at the Drosophila NMJ, APC/C mutants have increases in liprin-α levels and postsynaptic glutamate receptors (van Roessel et al. 2004). In hippocampal neurons, post-synaptic liprin-α binds to glutamate receptor interacting protein (GRIP) and is required for proper synaptic targeting of AMPA receptors (Wyszynski et al. 2002), which implicates liprin-α in postsynaptic assembly and plasticity.
The Cdc20-containing multisubunit ubiquitin E3 APC/C also regulates dendritic morphogenesis in postmitotic neurons. Overexpression of APCCdc20 promotes dendritic arbor formation of cerebellar granule neurons, and this effect requires centrosomal localization of APCCdc20 via histone deacetylase 6 (HDAC6) (Kim et al. 2009). One of the critical targets for APCCdc20 is inhibitor of differentiation 1 (Id1), a protein that antagonizes the activities of basic helix-loop-helix transcription factors during development, whose degradation by APCCdc20 is also required for the maintenance of dendritic morphology (Kim et al. 2009).
Deubiquitinase Control of Synapse Formation
In addition to ubiquitin ligases, DUBs have prominent roles in synapse development and regulation. In the ataxia mutant mouse, axJ, loss of function of the ubiquitin-specific protease Usp14 produces profound defects in synaptic transmission along with behavioral deficits including a resting tremor and hindlimb paralysis (Wilson et al. 2002). Usp14 associates with the proteasome, and loss of Usp14 results in reduced levels of monomeric ubiquitin (Anderson et al. 2005), indicating that Usp14 functions to maintain cellular levels of monomeric ubiquitin. These findings, along with recent studies on UCH-L1 (Cartier et al. 2009, Chen et al. 2010) (see below), demonstrate that alterations in the levels of free ubiquitin contribute to synapse regulation and neurological disease. Intriguingly, Usp14 binds the α1 GABAA receptor subunit, and axJ mice lacking Usp14 exhibit increased GABAA receptor levels at Purkinje cell–surface membranes and increased inhibitory postsynaptic currents (IPSCs) (Lappe-Siefke et al. 2009), which suggest that ubiquitin-dependent GABAA receptor turnover at cerebellar synapses contributes to behavioral impairment.
Usp14 is also required for proper muscle mass development and muscular behavior in adult mice (Walters et al. 2008). Although there is no change in motor neuron number in axJ mice, an ultrastructural defect of swelling in motor endplates was observed. Postsynaptic acetylcholine receptors exhibit a plaque-like morphology with a loss of total ubiquitin in synaptic fractions in the spinal cord and sciatic nerve. Notably, this synaptic defect is rescued by genetic restoration of Usp14 (Chen et al. 2009).
The morphology and organization of synapses is also controlled by the brain-specific DUB, UCH-L1. UCH-L1 is among the most abundant proteins in the brain (Wilkinson et al. 1989) and, as stated above, regulates levels of monomeric ubiquitin through processing of ubiquitin precursors and through inhibition of ubiquitin degradation (Setsuie & Wada 2007). A recent study found that NMDA receptor activation induces UCH-L1 activity and increases the availability of free monomeric ubiquitin in cultured neurons (Cartier et al. 2009). Pharmacological inhibition of UCH-L1 activity reduces monomeric ubiquitin levels and results in spine enlargement with a concomitant decrease in spine density and synapse number (Figure 3c). Interestingly, the effects of UCH-L1 inhibition on spine structure are rescued by overexpression of ubiquitin (Cartier et al. 2009), suggesting that the availability of monomeric ubiquitin is critical for the maintenance of synapses. More recently, UCH-L1 has been implicated in proper formation of the NMJ. Mice deficient in UCH-L1 exhibit impaired synaptic transmission at the NMJ that is accompanied by an absence of synaptic vesicles in the presynaptic terminal and denervation of muscles (Chen et al. 2010).
UBIQUITIN-DEPENDENT REGULATION OF GLUTAMATE RECEPTORS AT EXCITATORY SYNAPSES
Ubiquitination and Control of AMPA Receptor Content
AMPA receptors mediate the bulk of charge transfer during fast excitatory transmission, and their abundance is thus a major determinant of synaptic strength (see sidebar on Neurotransmitter Receptors at Excitatory and Inhibitory Synapses) (Kessels & Malinow 2009). In circuits throughout the mammalian brain, the trafficking of AMPA receptors from intracellular compartments to the postsynaptic membrane allows for activity-dependent strengthening of glutamatergic synapses during learning-related synaptic plasticity (Kennedy & Ehlers 2006, Malenka & Nicoll 1999, Newpher & Ehlers 2008). Thus, understanding the mechanisms of AMPA receptor regulation and turnover is critical to understanding normal brain function and the pathology associated with neurological disorders.
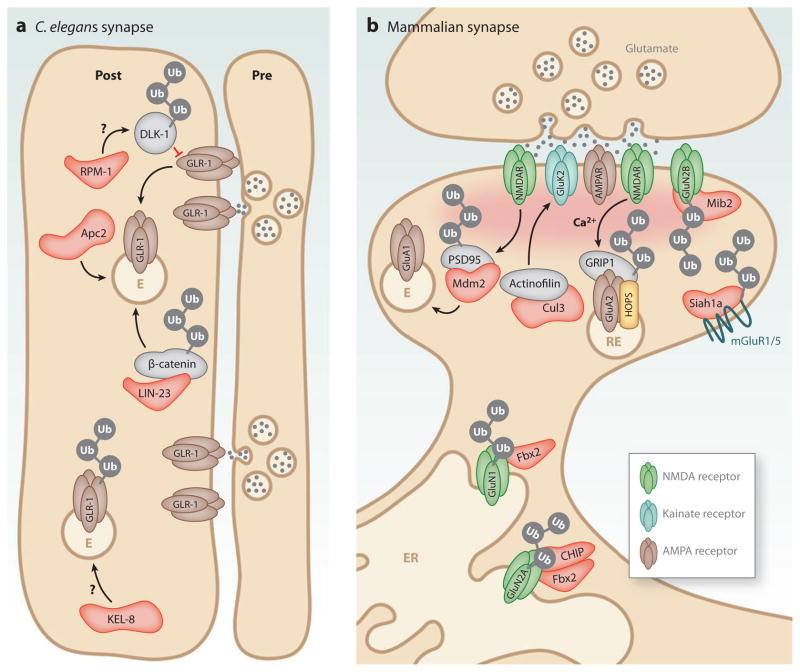
Evidence from several model systems has demonstrated an important role for ubiquitination in AMPA receptor trafficking. In ventral nerve cord interneurons in C. elegans, GLR-1 AMPA receptors mediate nose-touch mechanosensory behavior and spontaneous reversals in locomotion (Hart et al. 1995, Maricq et al. 1995). By visualizing GLR-1 clusters in transgenic C. elegans expressing GLR-1:GFP (Rongo et al. 1998), several genetic studies have identified components of the UPS that regulate GLR-1 receptor abundance (Burbea et al. 2002, Dreier et al. 2005, Juo & Kaplan 2004, Juo et al. 2007, Park et al. 2009, Rongo et al. 1998, Schaefer & Rongo 2006). In particular, ubiquitin itself and subunits of the ubiquitin ligase APC/C negatively regulate GLR-1 abundance at synapses (Burbea et al. 2002, Juo & Kaplan 2004). GLR-1 is ubiquitinated in vivo, and mutations in lysine residues of GLR-1 lead to an increase in receptor abundance and augment locomotor behavior (Burbea et al. 2002). Mutations that disrupt APC/C activity prevent the reduction in synaptic GLR-1 induced by ubiquitin overexpression (Figure 4a; Juo & Kaplan 2004). Interestingly, mutations in APC/C do not alter GLR-1 ubiquitination, which suggests that APC/C functions to regulate the trafficking of GLR-1, perhaps through GLR-1 recycling (Figure 4a; Juo & Kaplan 2004).
Figure 4.
Postsynaptic control of glutamate receptors by ubiquitin E3 ligases. Ubiquitin E3 ligase subunits that have been implicated in glutamate receptor regulation are indicated in red. (a) At C. elegans synapses, ubiquitination of the MAPKKK DLK-1 by SCFRPM-10, ubiquitin-dependent degradation of β-catenin by LIN-23 (C. elegans ortholog of βTrCP), and APC/C ubiquitin E3 activity (Apc2) all lead to a loss of GLR-1-containing AMPA receptors through the endocytic pathway. Ubiquitination by the KEL-8/Cul3 cullin complex also leads to loss of GLR-1 at synapses. E, endosome. (b) At mammalian synapses, Mdm2-dependent ubiquitination of PSD-95 leads to a loss of GluA1-containing AMPA receptors (AMPARs), likely through the endocytic pathway. The ubiquitin-like domain protein Tmub1/HOPS (transmembrane and ubiquitin-like domain-containing 1/Hepatocyte Odd Protein Shuttling) is associated with recycling endosomes (REs) and promotes GluA2 AMPAR recycling in coordination with GRIP1 (glutamate receptor interacting protein 1). The glycosylated NMDA receptor (NMDAR) subunit, GluN1, is ubiquitinated by SCFFbx2 in the endoplasmic reticulum (ER) and is required for surface expression of NMDARs. The NMDAR subunit GluN2A is ubiquitinated by the co-chaperone/ubiquitin ligase C terminus of Hsc-70-interaction protein (CHIP) and SCFFbx2 while in the ER. Ubiquitination of GluN2B by the RING ubiquitin E3 mindbomb2 (Mib2) leads to a loss of GluN2B-containing receptors from the synapse. The ubiquitin RING E3 ligase Siah1a (seven in absentia homolog) is required for ubiquitination of mGluR1 and mGluR5, which leads to their removal from the plasma membrane.
The Wnt signaling pathway also coordinates with the UPS in the regulation of GLR-1 abundance. Degradation of β-catenin by the ubiquitin E3 LIN-23, the worm ortholog of βTrCP, results in decreased GLR-1 abundance at synapses, likely through Wnt pathway target genes (Figure 4a; Dreier et al. 2005). Furthermore, KEL-8, a catalytic subunit of the Cul3-containing cullin-RING ubiquitin E3, localizes in clusters adjacent to GLR-1 and regulates ubiquitin-dependent turnover of GLR-1. Much like APC/C mutants, KEL-8 mutants have increased surface expression of GLR-1, which correlates with increased spontaneous reversal locomotion behaviors. However, a direct interaction between KEL-8 and GLR-1 has not been detected, which suggests that ubiquitination of a novel KEL-8 substrate may be required to regulate GLR-1 abundance (Figure 4a; Schaefer & Rongo 2006).
Beyond its role in axon outgrowth and synapse development (Fulga & Van Vactor 2008), recent studies implicate the ubiquitin E3 ligase RPM-1 in AMPA receptor trafficking in C. elegans. RPM-1 is the worm ortholog of highwire and Phr1 in flies and mice, respectively, and is a component of an SCF-like E3 ligase complex with SCFFSN-10, CUL-1, and SKR-1. Suppressor screens revealed the p38 pathway mitogen-activated protein kinase kinase kinase (MAPKKK) DLK-1 (and its fly ortholog wallenda) to be downstream mediators of RPM-1 signaling critical for synapse development (Collins et al. 2006, Nakata et al. 2005). Interestingly, RPM-1 also regulates postsynaptic AMPA receptor trafficking in C. elegans interneurons (Park et al. 2009). RPM-1 negatively regulates the p38 MAPK pathway by repressing protein levels of DLK-1, which leads to subsequent decreases in GLR-1 endocytosis. Mutations in RPM-1 and FSN-1 lead to accumulation of GLR-1 receptors in neurites (Park et al. 2009). Thus, ubiquitin-dependent regulation of DLK-1 inhibits the activation of the p38 MAPK pathway, which is required for clathrin-mediated endocytosis of GLR-1-containing AMPA receptors (Park et al. 2009). However, direct ubiquitination of DLK-1 by RPM-1 has not been described, which suggests the existence of additional substrates that may be ubiquitinated by RPM-1 to mediate GLR-1 endocytosis (Figure 4a).
NEUROTRANSMITTER RECEPTORS AT EXCITATORY AND INHIBITORY SYNAPSES.
Glutamate receptors are the primary neurotransmitter receptors at excitatory synapses in the mammalian brain. The subtype composition, trafficking, and posttranslational regulation of glutamate receptors are critical for synaptic plasticity (Kessels & Malinow 2009, Lau & Zukin 2007, Newpher & Ehlers 2008, Shepherd & Huganir 2007). The two main types of glutamate receptors are separated according to their mechanism of activation. Ionotropic glutamate receptors (iGluRs) are cation channels that are directly gated by the neurotransmitter glutamate. The three subtypes of iGluRs are AMPA receptors, NMDA receptors, and kainate receptors. The metabotropic glutamate receptors (mGluRs) are G-protein coupled receptors that are divided into three groups (groups 1, 2, and 3) and consist of eight subtypes, mGluR1–8. AMPA receptors contain four subunits (GluA1–4, also referred to as GluR1–4 or GluRA–D), which can homo-or heteromultimerize, and whose differential usage can alter the properties of synaptic transmission and signaling (Palmer et al. 2005). In Drosophila, AMPA receptor orthologs are divided into A-type and B-type (GluRIIA, -IIB) and further include other subunits (GluRIII/IIC, -IID, and -IIE), which are differentially expressed and clustered at the fly neuromuscular synapse (Collins & DiAntonio 2007). In C. elegans, the AMPA receptor orthologs consist of GLR1–8 (Brockie et al. 2001). NMDA receptors are heterotetrameric receptors that assemble from two NR1 subunits (also called GluN1) and two NR2 subunits (also called GluN2) that are derived from four gene products (NR2A–D, also called GluN2A–D). NMDA receptors infrequently contain NR3 subunits, which are derived from two gene products, NR3A–B (also called GluN3A–B) (Kohr 2006).
GABA receptors (GABARs) mediate most inhibitory synaptic transmission in the brain. At inhibitory synapses, the GABAR family can be divided into GABAA and GABAB subclasses, which are ionotropic and metabotropic receptors, respectively. GABAARs are pentameric GABA-gated chloride channels assembled from five different subunits among a large collection (α1–α6, β1–β3, γ1–γ3, δ, ε1–ε3, θ, and π) (Jacob et al. 2008). Most GABAARs in the brain consist of α, β, and γ subunits (Jacob et al. 2008), and subunit composition determines the channel properties and subcellular localization of specific GABAAR heteropentamers. Glycinergic synapses are inhibitory synapses found primarily in the brain stem and spinal cord. Inhibitory transmission at glycinergic synapses is mediated by glycine receptors (GlyRs), which are heteropentameric chloride channels assembled from combinations of multiple α (α1–α4) and a single β subunit (Betz & Laube 2006).
At mammalian synapses, the insertion and removal of AMPA receptors is regulated by the PSD scaffolding molecules SAP97 and PSD-95 (Rumbaugh et al. 2003, Schluter et al. 2006). PSD scaffold proteins are themselves targets of ubiquitin-dependent degradation. For example, PSD-95 was found to be ubiquitinated and degraded by the ubiquitin ligase Mdm2 in response to direct stimulation of NMDA receptors (Figure 4b; Colledge et al. 2003). Furthermore, overexpression of a ubiquitination-defective PSD-95 mutant inhibits AMPA receptor endocytosis (Colledge et al. 2003). PSD-95 is also degraded following direct stimulation of AMPA receptors, and overexpression of PSD-95 reduces AMPA receptor endocytosis (Bingol & Schuman 2004, Colledge et al. 2003). However, ubiquitinated PSD-95 species are rare or absent under many conditions (Bingol & Schuman 2004, Ehlers 2003), which suggests that PSD-95 ubiquitination may occur transiently in response to a restricted set of stimuli. A further proteasome-dependent decrease in the PSD proteins SAP97 and PSD-95 can be observed in the amygdala following extinction of conditioned fear in rats (Mao et al. 2008). This learning paradigm correlates with a proteasome-dependent reduction of surface AMPA receptors, which suggests that loss of critical PSD scaffolds influences AMPA receptor trafficking during fear-conditioned extinction (Mao et al. 2008).
Other targets of proteasome degradation at the PSD include GRIP1, a protein that directly binds to GluA2 (Dong et al. 1997) and is degraded upon stimulation with glutamate (Guo & Wang 2007). Control of proteasome activity and GRIP1 are required for a loss of surface GluA2 expression following glutamate application (Guo & Wang 2007). The degradation of GRIP1 is dependent on NMDA receptor activation and calcium influx (Figure 4b; Guo & Wang 2007). However, the relevant ubiquitin ligase for GRIP1 degradation remains unknown.
In addition to ubiquitin E3 ligases ubiquitin-like (UBL) proteins have been implicated in AMPA receptor trafficking and signaling. Tmub1/HOPS (transmembrane and ubiquitin-like domain-containing 1/Hepatocyte Odd Protein Shuttling) is a UBL-containing integral membrane protein that is associated with recycling endosomes and interacts in a complex with GluA2 and GRIP1. Loss of Tmub1/HOPS leads to decreased recycling of GluA2-containing AMPA receptors and results in reduced amplitudes of AMPA receptor–mediated miniature excitatory post-synaptic currents (mEPSCs) (Figure 4b; Yang et al. 2008). It is not yet clear precisely how the ubiquitin-binding domain of Tmub1/HOPS regulates GluA2 trafficking, although binding to a ubiquitin-modified component in a GluA2/GRIP1 endosome complex seems to be the most likely mechanism.
Control of NMDA Receptor Ubiquitination
During assembly of heteromeric NMDA receptors, GluN1 subunits containing high-mannose glycans are ubiquitinated and degraded through the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway during changes in synaptic activity (Gascon et al. 2007, Kato et al. 2005). This process is regulated by a dendritic spine–localized F-box ubiquitin E3 subunit, SCFFbx2, that binds specifically to high-mannose glycans (Yoshida et al. 2002). Overexpression of SCFFbx2 enhances ubiquitination of glycosylated GluN1 in a manner dependent on its catalytic F-box domain (Figure 4b) (Kato et al. 2005). Furthermore, SCFFbx2 is required for homeostatic regulation of NMDA receptors during chronic changes in synaptic activity (Kato et al. 2005), which suggests that ubiquitination and ERAD contribute to homeostatic synaptic plasticity.
Once assembled, NMDA receptors are targeted to synapses in part through direct interactions with PSD-95 family member scaffold proteins (Kohr 2006), although other interactions may serve to regulate NMDA receptor localization at synapses. For example, NF-L is a neuronal intermediate filament that binds to GluN1-containing NMDA receptors (Ehlers et al. 1998). Interestingly, NF-L antagonizes GluN1 ubiquitination and leads to an increase in NMDA receptor surface expression in heterologous cells (Ratnam & Teichberg 2005). To date, the role of NF-L in GluN1 ubiquitination has not been studied in the brain.
The GluN2 (or NR2) subunits of NMDA receptors are also regulated by ubiquitination. The C terminus of Hsp70-Interacting Protein (CHIP), a U-Box-containing ubiquitin E3 implicated in the turnover of glycoproteins, acts as a co-chaperone with SCFFbx2 to degrade N-linked glycoproteins and regulate the ubiquitination of GluN2A in heterologous cells (Nelson et al. 2006). Cortical neurons cultured from CHIP−/− cells have elevated levels of GluN2B-containing NMDA receptors, which suggests that CHIP controls expression levels of NMDA receptors through the ERAD pathway (Figure 4b; Nelson et al. 2006). Given that SCFFbx2 recognizes both GluN1 and GluN2A in different contexts (Nelson et al. 2006), this E3 ligase may couple with multiple cochaper-one E3s, such as CHIP, to allow for specificity of NMDA receptor subunit degradation.
GluN2B-containing NMDA receptors are ubiquitinated by the RING ubiquitin E3, mindbomb2 (Mib2), which binds to tyrosine-phosphorylated GluN2B (Jurd et al. 2008). Mib2 is localized to the PSD of dendrites in hippocampal neurons and directly ubiquitinates GluN2B in a manner dependent on the non-receptor tyrosine kinase Fyn (Jurd et al. 2008). Overexpression of Mib2 in heterologous cells decreases NMDA receptor–mediated currents, an effect that is rescued by blocking UPS activity in Mib2-overexpressing cells (Figure 4b; Jurd et al. 2008). These findings suggest that Mib2 mediates proteasome-dependent degradation of GluN2B subunits, which may provide a reciprocal mechanism to SCFFbx2 regulation of GluN2A (Nelson et al. 2006). However, the role of Mib2 regulation of GluN2B has not yet been examined in neurons. Interestingly, the related ubiquitin E3 Mib1 is enriched in the PSD and associates with multiple proteins involved in membrane trafficking (Choe et al. 2007), raising the possibility that Mib1 and Mib2 define a family of postsynaptic ubiquitin ligases that control the function and composition of glutamatergic synapses.
In the striatum, synaptic GluN2B surface levels are reduced by amphetamine (Mao et al. 2009). Although amphetamine does not alter the intracellular pool of GluN2B, it does lower the surface levels of GluN2B in PSD-enriched fractions, likely resulting from ubiquitin-dependent remodeling of PSD scaffolds (Mao et al. 2009). These findings are reminiscent of the PSD remodeling that occurs following chronic changes in synaptic activity (Ehlers 2003) and suggest a model whereby chronic amphetamine removes surface GluN2B by enhancing UPS-dependent degradation (Mao et al. 2009). To date, the ubiquitin E3 ligases regulating GluN2B ubiquitination upon amphetamine exposure is unknown, but Mib2 is one potential candidate (Jurd et al. 2008).
Ubiquitination of Kainate Receptors and Metabotropic Glutamate Receptors
Kainate receptors (KARs) are present both pre- and postsynaptically, where they mediate specific aspects of excitatory transmission and synaptic plasticity (Pinheiro & Mulle 2006). The functions of KARs remain largely enigmatic, but recent studies indicate that GluK2-containing KARs (also known as GluR6) are endocytosed and targeted for lysosomal degradation (Martin & Henley 2004). The degradation of GluK2 is mediated by a Cul3-containing SCF ubiquitin E3 complex (Salinas et al. 2006). In this complex, the BTB (bric-a-brac, tram-track, and broad complex)-Kelch domain protein actinofilin serves as a scaffold to link GluK2 to Cul3 for ubiquitination and degradation by the proteasome (Figure 4b; Salinas et al. 2006). However, it is unclear if these ubiquitin E3 subunits mediate the lysosome-dependent degradation of GluK2 (Martin & Henley 2004) or function independently in regulating GluK2 expression.
In addition to ionotropic GluRs, the group I mGluRs, mGluR1 and mGluR5, localize to the postsynaptic membrane, where they mediate slow forms of excitatory transmission and synaptic plasticity (Ferraguti & Shigemoto 2006). The RING ubiquitin E3 Siah1A (seven in absentia homolog) interacts with the C-terminal domains of both mGluR1 and mGluR5, and this interaction is dependent on Ca2+/calmodulin (Ishikawa et al. 1999). Siah1A ubiquitinates multiple lysine residues of mGluR1a and mGluR5 to promote their degradation (Figure 4b; Moriyoshi et al. 2004). However, the role of group I mGluR degradation by Siah1A in regulating synaptic plasticity has yet to be established.
UBIQUITINATION AND POSTSYNAPTIC CONTROL OF INHIBITORY SYNAPSES
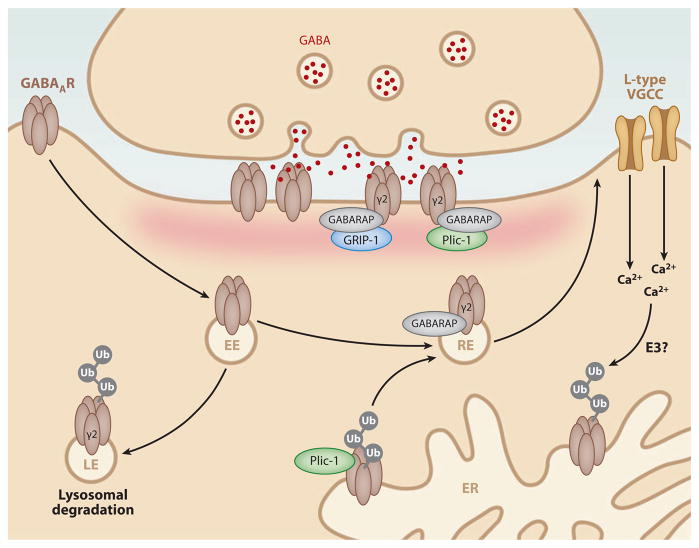
Similar to glutamate receptors (Kennedy & Ehlers 2006, Lau & Zukin 2007, Newpher & Ehlers 2008, Shepherd & Huganir 2007), γ-amino butyric acid (GABA) receptors undergo regulated trafficking and lysosomal degradation. In the case of metabotropic GABAB receptors (GABABRs), degradation is accelerated by blocking GABABR recycling (Grampp et al. 2007, 2008). GABAARs also undergo lysosomal degradation that requires ubiquitination of a motif within the intracellular domain of the GABAA γ2 subunit (Figure 5; Arancibia-Carcamo et al. 2009). Although the relevant GABAA receptor ubiquitin ligases are unknown, the ubiquitin-like protein Plic-1 interacts with GABAA subunits and acts as an adaptor to direct receptor trafficking (Bedford et al. 2001). Plic-1 controls GABAA receptor surface expression and stability but does not affect internalization (Bedford et al. 2001). In addition, Plic-1 increases the stability of ubiquitinated GABAARs in the ER, which suggests that Plic-1 limits ERAD of GABAARs and thus promotes their forward trafficking and ultimate delivery to the synaptic membrane (Figure 5; Saliba et al. 2008). Interestingly, activity blockade decreases the frequency and amplitude of GABAAR-mediated miniature inhibitory post-synaptic currents (mIPSCs), and this effect is correlated with GABAAR ubiquitination and a subsequent loss of ER-associated GABAARs (Saliba et al. 2007). Both GABAAR ubiquitination and decreased mIPSCs are dependent on the UPS and the presence of GABAAR subunit lysine residues (Saliba et al. 2007). However, the mechanism for proteasome-dependent loss of GABAARs following activity blockade is still unclear. One potential mechanism is through the activation of L-type VGCCs. Chronic blockade of L-type VGCCs results in a decrease in synaptic GABAARs, whereas activation of L-type VGCCs alters the turnover of GABAARs in a proteasome-dependent manner(Saliba et al. 2009), which suggests that calcium dynamics regulate GABAAR ubiquitination and UPS-dependent degradation in the ER (Figure 5).
Figure 5.
Ubiquitination at inhibitory synapses. At inhibitory synapses, ubiquitination of the γ2 subunit of GABAA receptors (GABAARs) results in lysosomal degradation of GABAARs. Binding of Plic-1 to GABAARs in the endoplasmic reticulum (ER) leads to stabilization of ubiquitinated GABAARs and promotes their forward trafficking. Interactions of GABAAR with the ubiquitin-like proteins GABARAP and Plic-1 promote recycling and aid in postsynaptic clustering of GABAARs. Interactions of GABARAP with GRIP-1 further aid in the clustering of postsynaptically expressed GABAARs. Ubiquitination and destabilization of GABAARs in the ER are accelerated by Ca2+ signaling through L-type voltage-gated calcium channels (VGCCs) by an unknown ubiquitin E3 ligase. EE, early endosome; LE, late endosome; RE, recycling endosome.
GABAARs are also regulated by the ubiquitin-like protein GABARAP (Kneussel 2002, Wang et al. 1999). GABARAP shares only 7% identity with ubiquitin but contains the conserved ubiquitin-like fold and is conjugated to membrane lipids (Ichimura et al. 2000). GABARAP binds to the γ2 subunit of GABAARs, and this aids in the clustering and anchoring of GABAARs to the PSD (Figure 5; Wang et al. 1999). In addition to GABAARs, GABARAP binds to Plic-1 (Bedford et al. 2001), the inhibitory postsynaptic scaffold protein gephyrin (Kneussel & Betz 2000), and the multi-PDZ (PSD-95/discs-large/ZO-1) protein GRIP1 (Kittler et al. 2004). Through these various interactions, GABARAP is well positioned to regulate GABAAR trafficking and anchoring at inhibitory synapses (Kittler et al. 2004). Much like postsynaptic excitatory synapses (Guo & Wang 2007), the regulation of GRIP1 turnover may also play a role in the regulation of synaptic GABAAR content.
The other major type of inhibitory receptor, the glycine receptor (GlyR), is ubiquitinated following agonist stimulation (Buttner et al. 2001). In Xenopus oocytes, GlyR α1 subunits are conjugated to ubiquitin at the surface of the cell prior to their internalization. Upon internalization, GlyRs are cleaved into multiple fragments by proteases (Buttner et al. 2001). However, the function of GlyR ubiquitination in the nervous system is not yet known, and the relevant ubiquitin ligases that target GlyRs remain to be determined.
UBIQUITIN AND BRAIN DISORDERS
Deficits in proteasomal degradation have been implicated in multiple neurological disorders, including Alzheimer’s disease, Huntington’s disease, amyotrophic lateral sclerosis, Parkinson’s disease, and neurodevelopmental disorders including autism (Lehman 2009). Mutations and deletions in a variety of ubiquitin pathway proteins are associated with neurological and psychiatric diseases (Jiang & Beaudet 2004). Here, we highlight selected brain disorders for which dysfunction of the UPS has been strongly implicated.
Autism Spectrum Disorders and Angelman Syndrome
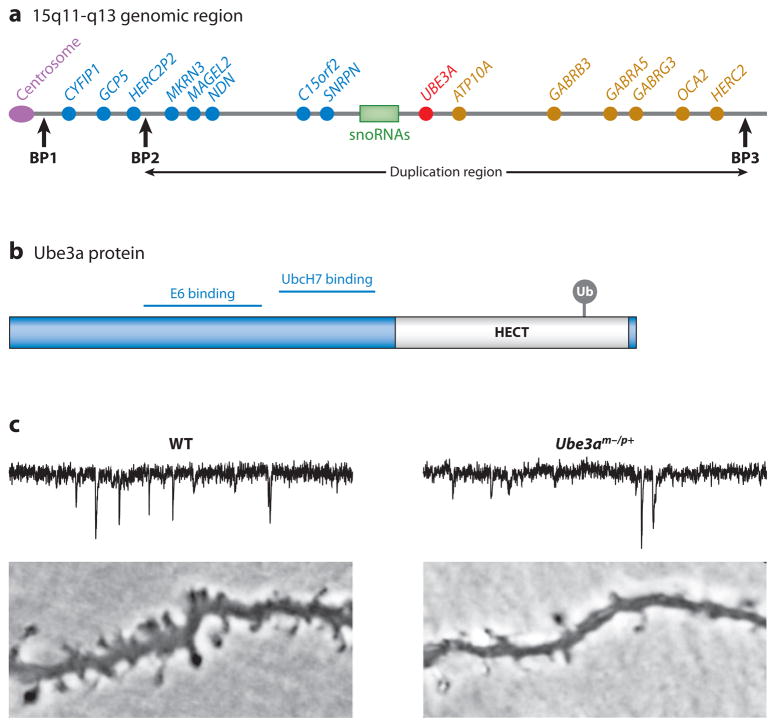
One of the strongest genetic links to autism spectrum disorders (ASD) (~1–5% of cases) are duplications of the 15q11-q13 chromosomal region (Vorstman et al. 2006). The 15q11-q13 region contains UBE3A, a gene that encodes the HECT E3 ligase Ube3a, which has been associated with the autism-related neurodevelopment disorder Angelman syndrome (Figure 6a,b). Maternally inherited loss-of-function mutations in UBE3A cause Angelman syndrome, a severe neurodevelopmental disorder in the autism spectrum characterized by the absence of speech and profound cognitive impairment (Clayton-Smith & Laan 2003). Mice with a maternal deficiency of Ube3a (Ube3am−/p+) exhibit defects in spatial learning and hippocampal LTP (Jiang et al. 1998, van Woerden et al. 2007). Generation of Ube3a yellow fluorescent protein (YFP)-tagged knock-in mice reveal that Ube3a can localize to synapses in the hippocampus (Dindot et al. 2008). Moreover, deletion of Ube3a results in decreased spine density and dendritic branching (Dindot et al. 2008, Lu et al. 2009, Yashiro et al. 2009), which may contribute to the associative learning defects in Ube3a mice.
Figure 6.
The HECT domain E3 ligase Ube3a regulates the number and function of glutamatergic synapses. (a) Location of the UBE3A gene within the human chromosome region 15q11-q13 that is linked to autism. Maternally inherited deletions within the 15q11-q13 region and mutations within Ube3a cause Angelman syndrome, whereas duplications within the 15q11-q13 region are associated with autism. Paternally expressed genes are depicted in blue, whereas maternally expressed genes are depicted in red. Nonimprinted genes are represented in gold. Common breakpoint regions (BP1–3) are represented by the vertical black arrows. Class I deletions (BP1–BP3) and class II deletions (BP2 and BP3) are both associated with Prader-Willi syndrome and Angelman syndrome. (b) Domain structure of the Ube3a protein. Ube3a contains a HECT domain (gray) that is required for the ubiquitin ligase activity of Ube3a. Ubiquitin is conjugated to cysteine 833 within the HECT domain and is required for the ubiquitin E3 activity of Ube3a. Interacting regions for the E2 UbcH7 and the viral oncogenic protein E6 are indicated. (c) Loss of Ube3a impairs synapse development. Traces indicate miniature excitatory postsynaptic currents (mEPSCs) recorded from layer 2/3 pyramidal neurons in visual cortex slices from wild-type (WT) or Ube3am−/p+ juvenile (~P25) mice showing a reduction in mEPSC frequency in Ube3am−/p+ neurons. Lower panels indicate basal dendrites in layer 2/3 pyramidal neurons showing a reduction in dendritic spines in Ube3am−/p+ mice. Figure adapted with permission from Yashiro et al. (2009).
Further evidence has linked Ube3a to brain plasticity by showing that Ube3am−/p+ mice are defective in experience-dependent maturation of the neocortex during episodes of sensory experience (Yashiro et al. 2009). Ube3am−/p+ mice exhibit a decrease in neocortical synapse maturation during development (Figure 6c). Furthermore, layer 4 to layer 2/3 synapses in the primary visual cortex of Ube3am−/p+ mice display a bidirectional impairment in synaptic plasticity that can be alleviated through visual sensory deprivation via dark rearing. Finally, ocular dominance plasticity is defective in Ube3am−/p+ mice (Sato & Stryker 2010, Yashiro et al. 2009), which suggests that a loss of neocortical plasticity in vivo may contribute to the deficits associated with Angelman syndrome. Ube3a is regulated in an activity-dependent manner and controls the expression of the immediate early gene product, activity-regulated cytoskeleton-associated protein (Arc/Arg3.1). Ube3a ubiquitinates Arc/Arg3.1, which results in its degradation, and in turn, leads to a decrease in AMPA receptor endocytosis (Greer et al. 2010).
In addition to duplication of UBE3A, copy number variations in the genes encoding the RING E3 parkin (PARK2), COP1 (RFWD2), and the F-box protein 40 (FBX040) are also linked to autism susceptibility (Glessner et al. 2009). Therefore, ubiquitin pathways and ubiquitin-dependent degradation in the brain are major contributing factors to autism and neurodevelopmental disorders.
Parkinson’s Disease
Mutations in the RING E3 ubiquitin ligase parkin and the DUB UCH-L1 cause early-onset familial forms of Parkinson’s disease (PD), which have the characteristic clinical features of tremor, rigidity, and bradykinesia as well as the classic histopathological loss of midbrain dopaminergic neurons (Leroy et al. 1998, Shimura et al. 2000). Parkin has been isolated from lipid rafts in PSD fractions and has been found to associate with PSD components such as CASK, GluN2B, PSD-95, Homer1a, and CaMKII, which suggests that postsynaptic parkin may be linked to neurotransmission defects (Fallon et al. 2002).
Several studies have found that parkin protects against cell death induced by excitotoxic insult, oxidative stress, mitochondrial dysfunction, and apoptosis (Henchcliffe & Beal 2008). Parkin may also directly affect postsynaptic trafficking by monoubiquitinating PICK1 to regulate the activity of acid-sensing ion channels (ASICs) (Joch et al. 2007). Interestingly, PICK1 is a well-known binding partner of GluA2 (Hanley 2008), which suggests potential cross-talk between parkin, PICK1, and AMPA receptor-mediated synaptic transmission. Consistent with this notion, postsynaptic expression of parkin leads to the pruning and loss of excitatory synapses, whereas postsynaptic shRNA-mediated knockdown of parkin increases AMPA receptor-mediated synaptic transmission and excitatory synapse number in hippocampal neurons (Figure 3d; Helton et al. 2008). Postsynaptic expression of PD-linked mutant forms of parkin increases excitatory synaptic transmission and glutamatergic synapse number, thus phenocopying parkin knockdown and suggesting that enhanced excitatory transmission may be associated with excitotoxic cell death in parkin-linked PD (Helton et al. 2008). The mechanisms underlying parkin-dependent synapse loss are unknown but could involve PICK1 (Joch et al. 2007), mitochondrial regulation (Bueler 2009), or septins (Zhang et al. 2000).
Other Neurological Disorders
Ubiquitin pathway components have been implicated in many other neurological disorders. Huntington’s disease, amyotrophic lateral sclerosis, and frontotemporal dementias are all associated with ubiquitin-positive protein inclusions (Ross & Poirier 2004). Hypoxia leads to the accumulation of ubiquitin conjugates in the PSD, likely owing to defects in protein degradation (Capani et al. 2009). Furthermore, a mutation in the GABAAR α1 subunit that causes early degradation in the ER reduces GABAAR surface expression and causes epilepsy (Gallagher et al. 2007). In addition, a model of ataxia includes abnormal GABA receptor turnover that may be due to the interaction of GABA with the DUB USP14 (Lappe-Siefke et al. 2009). Thus, defining the mechanistic links between the UPS, synapse dysfunction, and neurological disease will be important avenues of future research.
FUTURE PERSPECTIVES
Although the field is rapidly accelerating, we have just begun to understand how ubiquitination and its constituent pathways alter post-synaptic architecture and function (Table 1). Many fundamental questions remain unanswered. For instance, which postsynaptic ubiquitinated substrates are required for learning and memory? How are cohorts of PSD proteins coregulated by ubiquitination during changes in activity? What controls the timing and location of ubiquitination at synapses, and how do these mechanisms couple to classical signaling for synaptic plasticity? How long-lived are various postsynaptic proteins, and how is protein half-life altered during learning-related synaptic plasticity? Although the majority of work has focused on proteasomal degradation, we have little knowledge as to how other types of polyubiquitin chains contribute to synaptic signaling and architecture. Similarly, we know little about other ubiquitin-like modifications such as SUMO, NEDD8, and FAT10 and their role in plasticity. In this regard, it is notable that SUMOylation of the GluK2 KAR is regulated by activity (Martin et al. 2007).
Table 1.
Ubiquitin proteasome system enzymes implicated in neuronal function
| Protein | Neuronal function | Substrates | References |
|---|---|---|---|
| Ubiquitin-conjugating (E2) enzymes | |||
| E2-25K/Hip-2 | Mediating amyloid-β neurotoxicity; control of aggregate formation and cell death in polyglutamine diseases | ? | Song et al. (2003), de Pril et al. (2007) |
| UbcD1 | Dendritic pruning in Drosophila through caspase regulation | DIAP1 | Kuo et al. (2006) |
| Ubiquitin and polyubiquitin ligase (E3/E4) enzymes | |||
| Ube3a/E6-AP | Loss of function of maternal copy leads to Angelman’s syndrome, defects in hippocampal LTP, decreases in dendritic spine density, experience-dependent maturation of neocortex | Arc/Arg3.1 | Dindot et al. (2008), Greer et al. (2010), Jiang et al. (1998), Kishino et al. (1997), Lu et al. (2009), Matsuura et al. (1997), Sato & Stryker (2010), Weeber et al. (2003), Yashiro et al. (2009) |
| F-box protein 40 | Copy number variations lead to autism susceptibility | ? | Glessner et al. (2009) |
| COP1 | Copy number variations lead to autism susceptibility | ? | Glessner et al. (2009) |
| APC/C | Axon growth, dendritic growth, synaptogenesis, L-LTP in mice, glutamate receptor trafficking and abundance in C. elegans, control of antioxidant status | Liprin-α Id1 Pfkfb3 | Burbea et al. (2002), Herrero-Mendez et al. (2009), Juo & Kaplan (2004), Kim et al. (2009), Konishi et al. (2004), Li et al. (2008), van Roessel et al. (2004) |
| highwire/RPM-1/Phr1 | Synapse formation at the Drosophila NMJ, glutamate receptor endocytosis in C. elegans | ? | Bloom et al. (2007), Burgess et al. (2004), DiAntonio et al. (2001), Liao et al. (2004), Nakata et al. (2005), Park et al. (2009), Schaefer et al. (2000), Wan et al. (2000), Wu et al. (2005), Zhen et al. (2000) |
| PDZRN3A | NMJ formation in mice | MuSK | Lu et al. (2007) |
| SKR-1 | Synapse elimination at HSNL synapses in C. elegans | ? | Ding et al. (2007) |
| SCFSEL-10 | Synapse elimination at HSNL synapses in C. elegans | ? | Ding et al. (2007) |
| SCFβTrCP/LIN-23 | Glutamate receptor abundance in C. elegans and regulation of PSD-associated protein SPAR | β-catenin SPAR | Ang et al. (2008), Dreier et al. (2005) |
| mindbomb1 | Neuronal branching | ? | Choe et al. (2007) |
| KEL-8 | Glutamate receptor abundance in C. elegans | ? | Schaefer & Rongo (2006) |
| SCFFSN-10 | Synapse development and glutamate receptor abundance in C. elegans | ? | Liao et al. (2004), Park et al. (2009) |
| Mdm2 | AMPA receptor endocytosis | PSD-95 | Colledge et al. (2003) |
| SCFFbx2 | Regulation of NMDA receptor abundance | GluN1 | Kato et al. (2005), Nelson et al. (2006) |
| CHIP | Enhances parkin ubiquitin ligase activity, NMDA receptor abundance | GluN2A | Imai et al. (2002), Nelson et al. (2006) |
| mindbomb2 | NMDA receptor abundance | GluN2B | Jurd et al. (2008) |
| Cul3 | GluK2 abundance | GluK2 | Salinas et al. (2006) |
| Siah1A | mGluR abundance | mGluR1/5 | Ishikawa et al. (1999), Moriyoshi et al. (2004) |
| Parkin | Controls excitatory synapse number, protects against excitotoxic cell death, regulates ASICs. Copy number variations lead to autism susceptibility; mutations associated with Parkinson’s disease. | PICK1 α-synuclein | Cha et al. (2005), Darios et al. (2003), Glessner et al. (2009), Helton et al. (2008), Henn et al. (2007), Joch et al. (2007), Kitada et al. (1998), Ng et al. (2009), Palacino et al. (2004), Shimura et al. (2001), Staropoli et al. (2003) |
| TRIM3 | Controls dendritic spine size | GKAP | Hung et al. (2010) |
| RNF182 | Brain enriched and upregulated in Alzheimer’s patient brains, regulates cell viability | ATP6V0C | Liu et al. (2008) |
| Mahogunin | Loss of function causes age-dependent neurodegeneration | ? | He et al. (2003), Phan et al. (2002) |
| UFD2 | Loss of function delays Wallerian degeneration | ? | Mack et al. (2001), Zhai et al. (2003) |
| Deubiquitinases | |||
| Ap-Uch/UCH-L3 | Working memory in Aplysia; working memory in mice | ? | Hegde et al. (1993), Wood et al. (2005) |
| fat facets | Synapse formation in Drosophila eye and NMJ; presynaptic membrane dynamics | Epsin 1 | Cadavid et al. (2000), Chen et al. (2003), DiAntonio et al. (2001), Huang et al. (1995), Wan et al. (2000) |
| Usp14 | Muscle mass development and control of motor endplate size, behavioral defects, and increased IPSCs in the ataxia mutant mouse | GABAAR? | Anderson et al. (2005), Chen et al. (2009), Lappe-Siefke et al. (2009), Walters et al. (2008), Wilson et al. (2002) |
| UCH-L1 | Associated with early-onset familial Parkinson’s disease; control of spine size, density, and synapse number; synaptic transmission at the NMJ | ? | Cartier et al. (2009), Chen et al. (2010), Leroy et al. (1998), Saigoh et al. (1999) |
| UBP2 | Overexpression delays Wallerian degeneration | ? | Zhai et al. (2003) |
| Ataxin-3 | Mutated in Machado-Joseph disease | ? | Kawaguchi et al. (1994) |
?, unknown; DIAP1, Drosophila inhibitor of apoptosis; LTP, long-term potentiation; Pfkfb3, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, isoform 3; NMJ, neuromuscular junction; HSNL, hermaphrodite-specific motor neuron; PSD, postsynaptic density; ASIC, acid-sensing ion channel; IPSC, inhibitory postsynaptic current.
The generation of new tools will allow us to identify novel modes of ubiquitin signaling in the brain. Recent developments in the ability to detect chain-specific polyubiquitination (Newton et al. 2008, Wang et al. 2008) and advances in mass spectrometry techniques that enable identification of ubiquitinated substrates and stoichiometries of endogenous ubiquitin chains have been described (Jeram et al. 2009, Newton et al. 2008, Xu et al. 2009) and hold promise for studies of ubiquitination in the brain. Ultimately, ubiquitination may join phosphorylation in the pantheon of posttranslational modifications that mediate postsynaptic plasticity in diverse neural circuits.
Acknowledgments
We thank Juliet Hernandez, Hyunsoo Shawn Je, Matthew Kennedy, Thomas Newpher, Joel Schwartz, Jason Yi, and other members of the Ehlers lab for helpful input and comments on the manuscript. We apologize to those whose work could not be cited owing to space limitations. Work in the lab of M.D.E. is supported by grants from the NIMH and NINDS. A.M.M. is supported by an NRSA postdoctoral fellowship from the NIH. M.D.E. is an Investigator of the Howard Hughes Medical Institute.
Glossary
- Long-term facilitation (LTF)
a form of serotonin-induced LTP in Aplysia that requires cAMP, PKA, and the transcription factor CREB
- Synaptic plasticity
a synapse strength change that typically results from changes in postsynaptic receptor content/function or in neurotransmitter release
- Fear-conditioned learning
a form of learning behavior whereby animals learn to fear a neutral stimulus when paired with noxious or painful stimuli
- Working memory
the temporary storage of information required to carry out complex cognitive tasks. Often referred to as short-term memory
- ER-associated degradation (ERAD)
a protein degradation pathway that targets misfolded proteins in the ER for retrotranslocation, ubiquitination, and degradation by the proteasome
- Homeostatic synaptic plasticity
any of several synaptic mechanisms whereby neurons self-regulate their activity to maintain overall network stability
- Sensory experience
stimuli reaching the brain that derive from the primary senses
- Ocular dominance plasticity
brain plasticity whereby deprivation of visual input from one eye leads to increased responses to input from the nondeprived eye
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Ahmari SE, Buchanan J, Smith SJ. Assembly of presynaptic active zones from cytoplasmic transport packets. Nat Neurosci. 2000;3:445–51. doi: 10.1038/74814. [DOI] [PubMed] [Google Scholar]
- Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, Wilson SM. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–31. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- Ang XL, Seeburg DP, Sheng M, Harper JW. Regulation of postsynaptic RapGAP SPAR by Polo-like kinase 2 and the SCFβ-TRCP ubiquitin ligase in hippocampal neurons. J Biol Chem. 2008;283:29424–32. doi: 10.1074/jbc.M802475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arancibia-Carcamo IL, Yuen EY, Muir J, Lumb MJ, Michels G, et al. Ubiquitin-dependent lysosomal targeting of GABAA receptors regulates neuronal inhibition. Proc Natl Acad Sci USA. 2009;106:17552–57. doi: 10.1073/pnas.0905502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinian J, McGauran AM, De Jaeger X, Mouledous L, Frances B, Roullet P. Protein degradation, as with protein synthesis, is required during not only long-term spatial memory consolidation but also reconsolidation. Eur J Neurosci. 2008;27:3009–19. doi: 10.1111/j.1460-9568.2008.06262.x. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–84. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Bedford FK, Kittler JT, Muller E, Thomas P, Uren JM, et al. GABAA receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat Neurosci. 2001;4:908–16. doi: 10.1038/nn0901-908. [DOI] [PubMed] [Google Scholar]
- Bergold PJ, Sweatt JD, Winicov I, Weiss KR, Kandel ER, Schwartz JH. Protein synthesis during acquisition of long-term facilitation is needed for the persistent loss of regulatory subunits of the Aplysia cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1990;87:3788–91. doi: 10.1073/pnas.87.10.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz H, Laube B. Glycine receptors: recent insights into their structural organization and functional diversity. J Neurochem. 2006;97:1600–10. doi: 10.1111/j.1471-4159.2006.03908.x. [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. A proteasome-sensitive connection between PSD-95 and GluR1 endocytosis. Neuropharmacology. 2004;47:755–63. doi: 10.1016/j.neuropharm.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–48. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIα acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–78. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Bloom AJ, Miller BR, Sanes JR, DiAntonio A. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 2007;21:2593–606. doi: 10.1101/gad.1592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV. The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron. 2001;31:617–30. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- Bueler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson’s disease. Exp Neurol. 2009;218:235–46. doi: 10.1016/j.expneurol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron. 2002;35:107–20. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
- Burgess RW, Peterson KA, Johnson MJ, Roix JJ, Welsh IC, O’Brien TP. Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol Cell Biol. 2004;24:1096–105. doi: 10.1128/MCB.24.3.1096-1105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner C, Sadtler S, Leyendecker A, Laube B, Griffon N, et al. Ubiquitination precedes internalization and proteolytic cleavage of plasma membrane-bound glycine receptors. J Biol Chem. 2001;276:42978–85. doi: 10.1074/jbc.M102121200. [DOI] [PubMed] [Google Scholar]
- Cadavid AL, Ginzel A, Fischer JA. The function of the Drosophila fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development. 2000;127:1727–36. doi: 10.1242/dev.127.8.1727. [DOI] [PubMed] [Google Scholar]
- Capani F, Saraceno GE, Botti V, Aon-Bertolino L, de Oliveira DM, et al. Protein ubiquitination in postsynaptic densities after hypoxia in rat neostriatum is blocked by hypothermia. Exp Neurol. 2009;219:404–13. doi: 10.1016/j.expneurol.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Cartier AE, Djakovic SN, Salehi A, Wilson SM, Masliah E, Patrick GN. Regulation of synaptic structure by ubiquitin C-terminal hydrolase L1. J Neurosci. 2009;29:7857–68. doi: 10.1523/JNEUROSCI.1817-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha GH, Kim S, Park J, Lee E, Kim M, et al. Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proc Natl Acad Sci USA. 2005;102:10345–50. doi: 10.1073/pnas.0500346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain DG, Casadio A, Schacher S, Hegde AN, Valbrun M, et al. Mechanisms for generating the autonomous cAMP-dependent protein kinase required for long-term facilitation in Aplysia. Neuron. 1999;22:147–56. doi: 10.1016/s0896-6273(00)80686-8. [DOI] [PubMed] [Google Scholar]
- Chain DG, Hegde AN, Yamamoto N, Liu-Marsh B, Schwartz JH. Persistent activation of cAMP-dependent protein kinase by regulated proteolysis suggests a neuron-specific function of the ubiquitin system in Aplysia. J Neurosci. 1995;15:7592–603. doi: 10.1523/JNEUROSCI.15-11-07592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Sugiura Y, Myers KG, Liu Y, Lin W. Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc Natl Acad Sci USA. 2010;107:1636–41. doi: 10.1073/pnas.0911516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Polo S, Di Fiore PP, De Camilli PV. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc Natl Acad Sci USA. 2003;100:14908–13. doi: 10.1073/pnas.2136625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PC, Qin LN, Li XM, Walters BJ, Wilson JA, et al. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J Neurosci. 2009;29:10909–19. doi: 10.1523/JNEUROSCI.2635-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ZH, Ip NY. The roles of cyclin-dependent kinase 5 in dendrite and synapse development. Biotechnol J. 2007;2:949–57. doi: 10.1002/biot.200700056. [DOI] [PubMed] [Google Scholar]
- Choe EA, Liao L, Zhou JY, Cheng D, Duong DM, et al. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci. 2007;27:9503–12. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci USA. 1980;77:1365–68. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Soler-Llavina G, Bhattacharyya S, Malenka RC. N-methyl-D-aspartate receptor- and metabotropic glutamate receptor-dependent long-term depression are differentially regulated by the ubiquitin-proteasome system. Eur J Neurosci. 2009;30:1443–50. doi: 10.1111/j.1460-9568.2009.06950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet. 2003;40:87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, DiAntonio A. Synaptic development: insights from Drosophila. Curr Opin Neurobiol. 2007;17:35–42. doi: 10.1016/j.conb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Collins CA, Wairkar YP, Johnson SL, DiAntonio A. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signaling functions at the synapse. Nat Rev Neurosci. 2007;8:206–20. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darios F, Corti O, Lucking CB, Hampe C, Muriel MP, et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12:517–26. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- de Pril R, Fischer DF, Roos RA, van Leeuwen FW. Ubiquitin-conjugating enzyme E2-25K increases aggregate formation and cell death in polyglutamine diseases. Mol Cell Neurosci. 2007;34:10–19. doi: 10.1016/j.mcn.2006.09.006. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Bowen DC, Valenzuela DM, Simmons MV, Poueymirou WT, et al. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–12. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, Goodman CS. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412:449–52. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–18. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Ding M, Chao D, Wang G, Shen K. Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science. 2007;317:947–51. doi: 10.1126/science.1145727. [DOI] [PubMed] [Google Scholar]
- Djakovic SN, Schwarz LA, Barylko B, Demartino GN, Patrick GN. Regulation of the proteasome by neuronal activity and CAMKII. J Biol Chem. 2009;284:26655–65. doi: 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Upadhya SC, Ding L, Smith TK, Hegde AN. Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn Mem. 2008;15:335–47. doi: 10.1101/lm.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, O’Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–84. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- Dreier L, Burbea M, Kaplan JM. LIN-23-mediated degradation of β-catenin regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Neuron. 2005;46:51–64. doi: 10.1016/j.neuron.2004.12.058. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–42. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Ehlers MD, Fung ET, O’Brien RJ, Huganir RL. Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J Neurosci. 1998;18:720–30. doi: 10.1523/JNEUROSCI.18-02-00720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L, Moreau F, Croft BG, Labib N, Gu WJ, Fon EA. Parkin and CASK/LIN-2 associate via a PDZ-mediated interaction and are colocalized in lipid rafts and postsynaptic densities in brain. J Biol Chem. 2002;277:486–91. doi: 10.1074/jbc.M109806200. [DOI] [PubMed] [Google Scholar]
- Ferraguti F, Shigemoto R. Metabotropic glutamate receptors. Cell Tissue Res. 2006;326:483–504. doi: 10.1007/s00441-006-0266-5. [DOI] [PubMed] [Google Scholar]
- Fioravante D, Liu RY, Byrne JH. The ubiquitin-proteasome system is necessary for long-term synaptic depression in Aplysia. J Neurosci. 2008;28:10245–56. doi: 10.1523/JNEUROSCI.2139-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–90. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52:239–45. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Frey U, Huang YY, Kandel ER. Effects of cAMP simulate a late stage of LTP in hippocampal CA1 neurons. Science. 1993;260:1661–64. doi: 10.1126/science.8389057. [DOI] [PubMed] [Google Scholar]
- Fulga TA, Van Vactor D. Synapses and growth cones on two sides of a highwire. Neuron. 2008;57:339–44. doi: 10.1016/j.neuron.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Gallagher MJ, Ding L, Maheshwari A, Macdonald RL. The GABAA receptor α1 subunit epilepsy mutation A322D inhibits transmembrane helix formation and causes proteasomal degradation. Proc Natl Acad Sci USA. 2007;104:12999–3004. doi: 10.1073/pnas.0700163104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon S, Garcia-Gallo M, Renart J, Diaz-Guerra M. Endoplasmic reticulum-associated degradation of the NR1 but not the NR2 subunits of the N-methyl-D-aspartate receptor induced by inhibition of the N-glycosylation in cortical neurons. J Neurosci Res. 2007;85:1713–23. doi: 10.1002/jnr.21309. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–73. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grampp T, Notz V, Broll I, Fischer N, Benke D. Constitutive, agonist-accelerated, recycling and lysosomal degradation of GABA(B) receptors in cortical neurons. Mol Cell Neurosci. 2008;39:628–37. doi: 10.1016/j.mcn.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Grampp T, Sauter K, Markovic B, Benke D. γ-aminobutyric acid type B receptors are constitutively internalized via the clathrin-dependent pathway and targeted to lysosomes for degradation. J Biol Chem. 2007;282:24157–65. doi: 10.1074/jbc.M702626200. [DOI] [PubMed] [Google Scholar]
- Greenberg SM, Castellucci VF, Bayley H, Schwartz JH. A molecular mechanism for long-term sensitization in Aplysia. Nature. 1987;329:62–65. doi: 10.1038/329062a0. [DOI] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, et al. The Angelman Syndrome protein Ube3a regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–16. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Wang Y. Glutamate stimulates glutamate receptor interacting protein 1 degradation by ubiquitin-proteasome system to regulate surface expression of GluR2. Neuroscience. 2007;145:100–9. doi: 10.1016/j.neuroscience.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Haas KF, Broadie K. Roles of ubiquitination at the synapse. Biochim Biophys Acta. 2008;1779:495–506. doi: 10.1016/j.bbagrm.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KF, Miller SL, Friedman DB, Broadie K. The ubiquitin-proteasome system postsynaptically regulates glutamatergic synaptic function. Mol Cell Neurosci. 2007a;35:64–75. doi: 10.1016/j.mcn.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KF, Woodruff E, III, Broadie K. Proteasome function is required to maintain muscle cellular architecture. Biol Cell. 2007b;99:615–26. doi: 10.1042/BC20070019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–59. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JG. PICK1: a multi-talented modulator of AMPA receptor trafficking. Pharmacol Ther. 2008;118:152–60. doi: 10.1016/j.pharmthera.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–20. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- He L, Eldridge AG, Jackson PK, Gunn TM, Barsh GS. Accessory proteins for melanocortin signaling: attractin and mahogunin. Ann NY Acad Sci. 2003;994:288–98. doi: 10.1111/j.1749-6632.2003.tb03192.x. [DOI] [PubMed] [Google Scholar]
- Hegde AN, Goldberg AL, Schwartz JH. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: a molecular mechanism underlying long-term synaptic plasticity. Proc Natl Acad Sci USA. 1993;90:7436–40. doi: 10.1073/pnas.90.16.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, et al. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–26. doi: 10.1016/s0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- Helton TD, Otsuka T, Lee MC, Mu Y, Ehlers MD. Pruning and loss of excitatory synapses by the parkin ubiquitin ligase. Proc Natl Acad Sci USA. 2008;105:19492–97. doi: 10.1073/pnas.0802280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchcliffe C, Beal MF. Mitochondrial biology and oxidative stress in Parkinson disease pathogenesis. Nat Clin Pract Neurol. 2008;4:600–9. doi: 10.1038/ncpneuro0924. [DOI] [PubMed] [Google Scholar]
- Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, et al. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-κB signaling. J Neurosci. 2007;27:1868–78. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Mendez A, Almeida A, Fernandez E, Maestre C, Moncada S, Bolanos JP. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11:747–52. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci USA. 1980;77:1783–86. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10:647–58. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Feliu-Mojer MI, Spangler SA, Milstein AD, Dunah AW, et al. Liprinα1 degradation by calcium/calmodulin-dependent protein kinase II regulates LAR receptor tyrosine phosphatase distribution and dendrite development. Dev Cell. 2007;12:587–602. doi: 10.1016/j.devcel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Hoppe T. Multiubiquitylation by E4 enzymes: “one size” doesn’t fit all. Trends Biochem Sci. 2005;30:183–87. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–54. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Huang Y, Baker RT, Fischer-Vize JA. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–31. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- Hung AY, Sung CC, Brito IL, Sheng M. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One. 2010;5:e9842. doi: 10.1371/journal.pone.0009842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. “Protein Modifications: Beyond the Usual Suspects” review series. EMBO Rep. 2008;9:536–42. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, et al. CHIP is associated with Parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Nash SR, Nishimune A, Neki A, Kaneko S, Nakanishi S. Competitive interaction of seven in absentia homolog-1A and Ca2+/calmodulin with the cytoplasmic tail of group 1 metabotropic glutamate receptors. Genes Cells. 1999;4:381–90. doi: 10.1046/j.1365-2443.1999.00269.x. [DOI] [PubMed] [Google Scholar]
- Jacob TC, Moss SJ, Jurd R. GABAA receptor trafficking and its role in the dynamic modulation of neuronal inhibition. Nat Rev Neurosci. 2008;9:331–43. doi: 10.1038/nrn2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeram SM, Srikumar T, Pedrioli PG, Raught B. Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics. 2009;9:922–34. doi: 10.1002/pmic.200800666. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Beaudet AL. Human disorders of ubiquitination and proteasomal degradation. Curr Opin Pediatr. 2004;16:419–26. doi: 10.1097/01.mop.0000133634.79661.cd. [DOI] [PubMed] [Google Scholar]
- Joch M, Ase AR, Chen CX, MacDonald PA, Kontogiannea M, et al. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol Biol Cell. 2007;18:3105–18. doi: 10.1091/mbc.E05-11-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo P, Harbaugh T, Garriga G, Kaplan JM. CDK-5 regulates the abundance of GLR-1 glutamate receptors in the ventral cord of Caenorhabditis elegans. Mol Biol Cell. 2007;18:3883–93. doi: 10.1091/mbc.E06-09-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo P, Kaplan JM. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol. 2004;14:2057–62. doi: 10.1016/j.cub.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Jurd R, Thornton C, Wang J, Luong K, Phamluong K, et al. Mind bomb-2 is an E3 ligase that ubiquitinates the N-methyl-D-aspartate receptor NR2B subunit in a phosphorylation-dependent manner. J Biol Chem. 2008;283:301–10. doi: 10.1074/jbc.M705580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Rouach N, Nicoll RA, Bredt DS. Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc Natl Acad Sci USA. 2005;102:5600–5. doi: 10.1073/pnas.0501769102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8:221–28. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Ehlers MD. Organelles and trafficking machinery for postsynaptic plasticity. Annu Rev Neurosci. 2006;29:325–62. doi: 10.1146/annurev.neuro.29.051605.112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–50. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–36. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kittler JT, Arancibia-Carcamo IL, Moss SJ. Association of GRIP1 with a GABAA receptor associated protein suggests a role for GRIP1 at inhibitory synapses. Biochem Pharmacol. 2004;68:1649–54. doi: 10.1016/j.bcp.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Kneussel M. Dynamic regulation of GABAA receptors at synaptic sites. Brain Res Rev. 2002;39:74–83. doi: 10.1016/s0165-0173(02)00159-5. [DOI] [PubMed] [Google Scholar]
- Kneussel M, Betz H. Receptors, gephyrin and gephyrin-associated proteins: novel insights into the assembly of inhibitory postsynaptic membrane specializations. J Physiol. 2000;525(Pt. 1):1–9. doi: 10.1111/j.1469-7793.2000.t01-4-00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohr G. NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res. 2006;326:439–46. doi: 10.1007/s00441-006-0273-6. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–30. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–90. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Lappe-Siefke C, Loebrich S, Hevers W, Waidmann OB, Schweizer M, et al. The ataxia (axJ) mutation causes abnormal GABAA receptor turnover in mice. PLoS Genet. 2009;5:e1000631. doi: 10.1371/journal.pgen.1000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8:413–26. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Lee SH, Choi JH, Lee N, Lee HR, Kim JI, et al. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science. 2008;319:1253–56. doi: 10.1126/science.1150541. [DOI] [PubMed] [Google Scholar]
- Lehman NL. The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 2009;118:329–47. doi: 10.1007/s00401-009-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–52. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Li M, Shin YH, Hou L, Huang X, Wei Z, et al. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol. 2008;10:1083–89. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao EH, Hung W, Abrams B, Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–50. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- Liu QY, Lei JX, Sikorska M, Liu R. A novel brain-enriched E3 ubiquitin ligase RNF182 is up regulated in the brains of Alzheimer’s patients and targets ATP6V0C for degradation. Mol Neurodegener. 2008;3:4. doi: 10.1186/1750-1326-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Salon M, Alonso M, Vianna MRM, Viola H, Mello e Souza T, et al. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci. 2001;14:1820–26. doi: 10.1046/j.0953-816x.2001.01806.x. [DOI] [PubMed] [Google Scholar]
- Lu Y, Wang F, Li Y, Ferris J, Lee JA, Gao FB. The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum Mol Genet. 2009;18:454–62. doi: 10.1093/hmg/ddn373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Je HS, Young P, Gross J, Lu B, Feng G. Regulation of synaptic growth and maturation by a synapse-associated E3 ubiquitin ligase at the neuromuscular junction. J Cell Biol. 2007;177:1077–89. doi: 10.1083/jcb.200610060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack TG, Reiner M, Beirowski B, Mi W, Emanuelli M, et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat Neurosci. 2001;4:1199–206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA. Long-term potentiation—a decade of progress? Science. 1999;285:1870–74. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- Mao LM, Wang W, Chu XP, Zhang GC, Liu XY, et al. Stability of surface NMDA receptors controls synaptic and behavioral adaptations to amphetamine. Nat Neurosci. 2009;12:602–10. doi: 10.1038/nn.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao SC, Lin HC, Gean PW. Augmentation of fear extinction by D-cycloserine is blocked by proteasome inhibitors. Neuropsychopharmacology. 2008;33:3085–95. doi: 10.1038/npp.2008.30. [DOI] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signaling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- Martin KC, Barad M, Kandel ER. Local protein synthesis and its role in synapse-specific plasticity. Curr Opin Neurobiol. 2000;10:587–92. doi: 10.1016/s0959-4388(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Martin S, Henley JM. Activity-dependent endocytic sorting of kainate receptors to recycling or degradation pathways. EMBO J. 2004;23:4749–59. doi: 10.1038/sj.emboj.7600483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–25. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Glutamate and neurotrophic factors in neuronal plasticity and disease. Ann NY Acad Sci. 2008;1144:97–112. doi: 10.1196/annals.1418.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo E, Romano A. Long-term memory consolidation depends on proteasome activity in the crab Chasmagnathus. Neuroscience. 2007;147:46–52. doi: 10.1016/j.neuroscience.2007.04.022. [DOI] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of “single protein RING finger” E3 ubiquitin ligases. Bioessays. 2005;27:1147–57. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K, Iijima K, Fujii H, Ito H, Cho Y, Nakanishi S. Seven in absentia homolog 1A mediates ubiquitination and degradation of group 1 metabotropic glutamate receptors. Proc Natl Acad Sci USA. 2004;101:8614–19. doi: 10.1073/pnas.0403042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, et al. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–20. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nelson RF, Glenn KA, Miller VM, Wen H, Paulson HL. A novel route for F-box protein-mediated ubiquitination links CHIP to glycoprotein quality control. J Biol Chem. 2006;281:20242–51. doi: 10.1074/jbc.M602423200. [DOI] [PubMed] [Google Scholar]
- Newpher TM, Ehlers MD. Glutamate receptor dynamics in dendritic microdomains. Neuron. 2008;58:472–97. doi: 10.1016/j.neuron.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–78. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, et al. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J Neurosci. 2009;29:11257–62. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niell CM, Meyer MP, Smith SJ. In vivo imaging of synapse formation on a growing dendritic arbor. Nat Neurosci. 2004;7:254–60. doi: 10.1038/nn1191. [DOI] [PubMed] [Google Scholar]
- Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–73. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, et al. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–22. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- Palmer CL, Cotton L, Henley JM. The molecular pharmacology and cell biology of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Pharmacol Rev. 2005;57:253–77. doi: 10.1124/pr.57.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EC, Glodowski DR, Rongo C. The ubiquitin ligase RPM-1 and the p38 MAPK PMK-3 regulate AMPA receptor trafficking. PLoS One. 2009;4:e4284. doi: 10.1371/journal.pone.0004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick GN. Synapse formation and plasticity: recent insights from the perspective of the ubiquitin proteasome system. Curr Opin Neurobiol. 2006;16:90–94. doi: 10.1016/j.conb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Phan LK, Lin F, LeDuc CA, Chung WK, Leibel RL. The mouse mahoganoid coat color mutation disrupts a novel C3HC4 RING domain protein. J Clin Invest. 2002;110:1449–59. doi: 10.1172/JCI16131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro P, Mulle C. Kainate receptors. Cell Tissue Res. 2006;326:457–82. doi: 10.1007/s00441-006-0265-6. [DOI] [PubMed] [Google Scholar]
- Ratnam J, Teichberg VI. Neurofilament-light increases the cell surface expression of the N-methyl-D-aspartate receptor and prevents its ubiquitination. J Neurochem. 2005;92:878–85. doi: 10.1111/j.1471-4159.2004.02936.x. [DOI] [PubMed] [Google Scholar]
- Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–59. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Rumbaugh G, Sia GM, Garner CC, Huganir RL. Synapse-associated protein-97 isoform-specific regulation of surface AMPA receptors and synaptic function in cultured neurons. J Neurosci. 2003;23:4567–76. doi: 10.1523/JNEUROSCI.23-11-04567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpel S, LeDoux J, Zador A, Malinow R. Postsynaptic receptor trafficking underlying a form of associative learning. Science. 2005;308:83–88. doi: 10.1126/science.1103944. [DOI] [PubMed] [Google Scholar]
- Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, et al. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- Saliba RS, Gu Z, Yan Z, Moss SJ. Blocking L-type voltage gated Ca2+ channels with dihydropyridines reduces γ-aminobutyric acid type a receptor expression and synaptic inhibition. J Biol Chem. 2009;284:32544–50. doi: 10.1074/jbc.M109.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba RS, Michels G, Jacob TC, Pangalos MN, Moss SJ. Activity-dependent ubiquitination of GABAA receptors regulates their accumulation at synaptic sites. J Neurosci. 2007;27:13341–51. doi: 10.1523/JNEUROSCI.3277-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba RS, Pangalos M, Moss SJ. The ubiquitin-like protein Plic-1 enhances the membrane insertion of GABAA receptors by increasing their stability within the endoplasmic reticulum. J Biol Chem. 2008;283:18538–44. doi: 10.1074/jbc.M802077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas GD, Blair LA, Needleman LA, Gonzales JD, Chen Y, et al. Actinfilin is a Cul3 substrate adaptor, linking GluR6 kainate receptor subunits to the ubiquitin-proteasome pathway. J Biol Chem. 2006;281:40164–73. doi: 10.1074/jbc.M608194200. [DOI] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc Natl Acad Sci USA. 2010;107:5611–16. doi: 10.1073/pnas.1001281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–56. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Schaefer H, Rongo C. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol Biol Cell. 2006;17:1250–60. doi: 10.1091/mbc.E05-08-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter OM, Xu W, Malenka RC. Alternative N-terminal domains of PSD-95 and SAP97 govern activity-dependent regulation of synaptic AMPA receptor function. Neuron. 2006;51:99–111. doi: 10.1016/j.neuron.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Segref A, Hoppe T. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Rep. 2009;10:44–50. doi: 10.1038/embor.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serdaroglu P, Askanas V, Engel WK. Immunocytochemical localization of ubiquitin at human neuro-muscular junctions. Neuropathol Appl Neurobiol. 1992;18:232–36. doi: 10.1111/j.1365-2990.1992.tb00785.x. [DOI] [PubMed] [Google Scholar]
- Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Int. 2007;51:105–11. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shen H, Korutla L, Champtiaux N, Toda S, LaLumiere R, et al. NAC1 regulates the recruitment of the proteasome complex into dendritic spines. J Neurosci. 2007;27:8903–13. doi: 10.1523/JNEUROSCI.1571-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Bargmann CI. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003;112:619–30. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–43. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–5. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, et al. Ubiquitination of a new form of α-synuclein by parkin from human brain: implications for Parkinson’s disease. Science. 2001;293:263–69. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- Song S, Kim SY, Hong YM, Jo DG, Lee JY, et al. Essential role of E2-25K/Hip-2 in mediating amyloid-β neurotoxicity. Mol Cell. 2003;12:553–63. doi: 10.1016/j.molcel.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Spangler SA, Hoogenraad CC. Liprin-α proteins: scaffold molecules for synapse maturation. Biochem Soc Trans. 2007;35:1278–82. doi: 10.1042/BST0351278. [DOI] [PubMed] [Google Scholar]
- Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–49. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9:826–38. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol. 2009;11:123–32. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell. 2004;119:707–18. doi: 10.1016/j.cell.2004.11.028. [DOI] [PubMed] [Google Scholar]
- van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, et al. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of αCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10:280–82. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry. 2006;11:18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- Wallace BG. Agrin-induced specializations contain cytoplasmic, membrane, and extracellular matrix-associated components of the postsynaptic apparatus. J Neurosci. 1989;9:1294–302. doi: 10.1523/JNEUROSCI.09-04-01294.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters BJ, Campbell SL, Chen PC, Taylor AP, Schroeder DG, et al. Differential effects of Usp14 and Uch-L1 on the ubiquitin proteasome system and synaptic activity. Mol Cell Neurosci. 2008;39:539–48. doi: 10.1016/j.mcn.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, Goodman CS. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–29. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW. GABAA-receptor-associated protein links GABAA receptors and the cytoskeleton. Nature. 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- Wang H, Matsuzawa A, Brown SA, Zhou J, Guy CS, et al. Analysis of nondegradative protein ubiquity-lation with a monoclonal antibody specific for lysine-63-linked polyubiquitin. Proc Natl Acad Sci USA. 2008;105:20197–202. doi: 10.1073/pnas.0810461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washbourne P, Bennett JE, McAllister AK. Rapid recruitment of NMDA receptor transport packets to nascent synapses. Nat Neurosci. 2002;5:751–59. doi: 10.1038/nn883. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, et al. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23:2634–44. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KD, Lee KM, Deshpande S, Duerksen-Hughes P, Boss JM, Pohl J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;246:670–73. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, et al. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–25. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Brensinger CM, Guo W, Abel T. Ubiquitin C-terminal hydrolase L3 (Uchl3) is involved in working memory. Hippocampus. 2005;15:610–21. doi: 10.1002/hipo.20082. [DOI] [PubMed] [Google Scholar]
- Wu C, Wairkar YP, Collins CA, DiAntonio A. Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J Neurosci. 2005;25:9557–66. doi: 10.1523/JNEUROSCI.2532-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski M, Kim E, Dunah AW, Passafaro M, Valtschanoff JG, et al. Interaction between GRIP and liprin-α/SYD2 is required for AMPA receptor targeting. Neuron. 2002;34:39–52. doi: 10.1016/s0896-6273(02)00640-2. [DOI] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Takagi H, Konishi Y, Ageta H, Ikegami K, et al. Transmembrane and ubiquitin-like domain-containing protein 1 (Tmub1/HOPS) facilitates surface expression of GluR2-containing AMPA receptors. PLoS One. 2008;3:e2809. doi: 10.1371/journal.pone.0002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12:777–83. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SH, Mao SC, Lin HC, Gean PW. Synaptic expression of glutamate receptor after encoding of fear memory in the rat amygdala. Mol Pharmacol. 2006;69:299–308. doi: 10.1124/mol.105.017194. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol Rev. 2007;59:14–39. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–42. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Wang J, Kim A, Liu Q, Watts R, et al. Involvement of the ubiquitin-proteasome system in the early stages of Wallerian degeneration. Neuron. 2003;39:217–25. doi: 10.1016/s0896-6273(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA. 2000;97:13354–59. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hegde AN, Martin KC. The ubiquitin proteasome system functions as an inhibitory constraint on synaptic strengthening. Curr Biol. 2003;13:887–98. doi: 10.1016/s0960-9822(03)00332-4. [DOI] [PubMed] [Google Scholar]
- Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–43. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]