Abstract
Ah receptor (AhR) is a ligand-activated transcription factor that mediates pleiotropic effects of environmental pollutants such as 2,3,7,8-tetrachlorodibenzo-p-dioxin on host animals. In addition to induction of drug-metabolizing enzymes, the liganded AhR complex was found to activate gene expression of a factor designated AhR repressor (AhRR), which inhibits AhR function by competing with AhR for dimerizing with Arnt and binding to the XRE sequence. Thus, AhR and AhRR form a regulatory circuit in the xenobiotic signal transduction pathway and provide a novel mechanism of regulation of AhR function that may determine tissue-specific sensitivity to environmental pollutants.
Keywords: AhR, Arnt, bHLH–PAS, TCDD, XRE
AhR (arylhydrocarbon receptor, or dioxin receptor) has been known to mediate pleiotropic biological effects of various environmental contaminants, mainly polycyclic aromatic hydrocarbons usually represented by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). These biological effects include teratogenesis, tumor promotion, thymic atrophy, epithelial hyperplasia, hepatotoxicity, and induction of drug-metabolizing enzymes (Poland and Knutson 1982; Swanson and Bradfield 1993; Hankinson 1995; Sogawa and Fujii-Kuriyama 1997). AhR is usually present in cytoplasm in association with Hsp90 (Perdew 1988; Pongratz et al. 1992). Subsequently, upon high-affinity binding of inducing chemicals, the liganded AhR translocates to nuclei, where it switches the partner from Hsp90 to Arnt (AhR nuclear translocator) and binds the cognate enhancer sequence, XRE, upstream of the target genes for CYP1A1, GST, and others to activate their expressions (Fujisawa-Sehara et al. 1987; Telakowski-Hopkins et al. 1988). Recently, involvement of AhR in TCDD-induced teratogenesis, such as cleft palate and hydronephrosis in fetal development and cytotoxicity in adult animals, has been demonstrated by using AhR knockout mice (Fernandez-Salguero et al. 1996; Mimura et al. 1997).
Structurally, AhR and Arnt belong to a superfamily of bHLH transcription factors that include MyoD and Myc (Murre et al. 1989). A characteristic common domain of AhR and Arnt, designated PAS [conserved sequence among Per (Jackson et al. 1986), Arnt/AhR (Hoffman et al. 1991), and Sim (Nambu et al. 1991)], which abuts on the carboxyl terminus of the bHLH, defines a growing family of bHLH–PAS factors among the bHLH superfamily.
These transcription factors with bHLH motif form homo- and/or heterodimers with themselves or other members of the same family, to bind the cognate binding site upstream or downstream of their target genes, resulting in activation of gene expression (Murre et al. 1989). Because many of the bHLH transcription factors are involved in physiologically and developmentally important functions such as cell proliferation and differentiation, their transcription activities are negatively regulated by competitive heterodimer formation with inhibitory bHLH proteins. Two Myc-related proteins, Mad (Ayer et al. 1993) and Mxi1 (Zervos et al. 1993), dimerize with Max, and these heterodimers bind the same core sequence (CACGTG) as Myc/Max heterodimer. Thereby, Mad and Mxi1 interfere with Myc function either by sequestering Max or by direct competition for the DNA target sequence. During differentiation of certain myeloid cell lines in vitro, relative changes in the intracellular concentration of Myc and Mad (or Mxi1) rapidly modulate the expression of a set of genes responsive to these transcription factors (Ayer and Eisenman 1993; Larsson et al. 1994). In another group of bHLH transcription factors such as MyoD and E12/E47, inhibitory proteins, Ids, which lack a basic region adjacent to the HLH, are able to dimerize with a member of bHLH proteins including MyoD and E12/E47, resulting in inhibition of their transcription activation activity via sequestration into dimers that cannot bind DNA (Christy et al. 1991; Neuhold and Wold 1993). It has been reported that Id inhibits muscle differentiation by associating with E12 and prevents it from forming the active MyoD/E12 heterodimer (Benezra et al. 1990; Jen et al. 1992). During terminal differentiation, the Id levels decrease, suggesting that Id can act as an inhibitor of differentiation.
Although two kinds of suppressive forms of bHLH transcription factors change in concentration in association with cell proliferation and differentiation, the mechanism of their gene expression remains unknown.
During investigation of AhR and Arnt of a third group of bHLH transcription factors, we isolated cDNA clones that encode a polypeptide with high similarity to the sequence of the bHLH/PAS of AhR. This polypeptide was found to repress the transcription activity of AhR by competing with AhR in forming a heterodimer with Arnt and binding with the XRE sequence, and is thus designated AhRR or AhR repressor. Furthermore, the expression of AhRR is induced by the AhR/Arnt heterodimer through binding to the enhancer sequence XRE, upstream of the AhRR gene; thus the AhR function is regulated by the feedback inhibition of AhRR. A similar mechanism has been suggested recently: the circadian rythmic regulation of the mammalian clock system, consisting of the same bHLH–PAS factors, that is, mClock, BMAL1, and mPer1 (Gekakis et al. 1998).
Results and Discussion
During the screening of a mouse genomic library with AhR cDNA used as a hybridization probe, we isolated a genomic clone that has a sequence with high similarity to a part of AhR cDNA encoding the bHLH region (Ema et al. 1992). Subsequently, we isolated cDNA clones from a mouse small intestine cDNA library with the genomic DNA fragment showing high similarity to the AhR used as probe. The longest insert of the isolated cDNA clones was estimated to be 4.5 kb, and the determined sequence contains a long ORF of 2103 nucleotides, encoding a polypeptide of 701 amino acids. By comparison with other bHLH–PAS proteins, the encoded sequence shows the highest degree of sequence similarity to AhR in the sequence of the bHLH and PAS-A regions (Fig. 1). However, the sequence carboxy-terminal to PAS-A is quite variable. Notably, the PAS-B sequence, which functions as a ligand binding site and an interaction interface with Hsp90 in AhR, is missing in the deduced sequence and the sequence of the carboxy-terminal half, which corresponds to the transactivation domain for AhR (Sogawa et al. 1995), differs greatly from that of AhR (we designated the isolated factor AhRR for the reasons described below).
Figure 1.
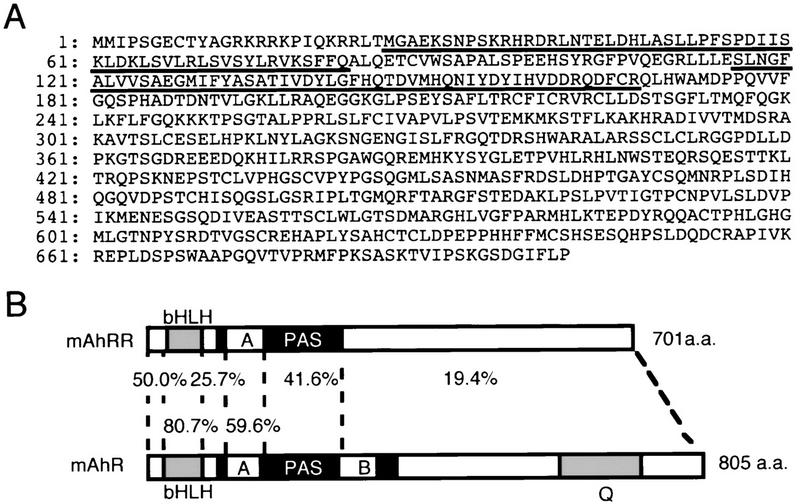
Predicted mouse AhRR primary structure and sequence comparison with that of AhR. (A) Predicted amino acid sequence of mAhRR, in single-letter symbols. bHLH (26–82 amino acids) and PAS-A regions (116–167) are underlined. (B) Domain structures of mAhRR and mAhR. Percentages of identity between mAhRR and mAhR are indicated.
Close similarity between AhRR and AhR in the bHLH and PAS-A regions led us to investigate, by immunoprecipitation assay, whether AhRR interacts with Arnt. When incubated with Arnt, AhRR was coimmunoprecipitated efficiently by an anti-Arnt antibody in a manner independent of the presence of a ligand, 3-methylcholanthrene (3MC) (Fig. 2B, lanes 3,4), whereas interaction between AhR and Arnt was significantly enhanced (twofold) by 3MC (Fig. 2B, lanes 8,9) as reported previously (Hirose et al. 1996). The degree of enhanced interaction between in vitro-synthesized AhR and Arnt by 3MC was variable, but the enhancement was reproducibly observed. This interaction between AhRR or AhR and Arnt was confirmed by the mammalian two-hybrid system (Dang et al. 1991). A fusion gene encoding the VP16 activation domain (AD) and the bHLH–PAS region of AhRR was transfected into 293T cells together with a fusion gene encoding GAL4–DBD and the bHLH–PAS region of Arnt, and the pG3E–Luc reporter gene. As a control experiment, a fusion gene of AhR bHLH–PAS and VP16 AD was used as prey. Whereas the AhR fusion gene enhanced the reporter gene expression in response to 3MC (Fig. 2C, lane 3), the AhRR fusion gene activated the luciferase expression in a manner independent of the inducer (Fig. 2C, lane 4). These results indicated that AhRR interacted constitutively with Arnt, whereas interaction between AhR and Arnt is ligand-dependent. In contrast to AhR, AhRR was not bound with Hsp90, as revealed by the immunoprecipitation assay (data not shown), and the expressed fusion protein composed of AhRR and green fluorescent protein (GFP) in COS7 cells was found to be constitutively localized in the nuclei (Fig. 2A).
Figure 2.
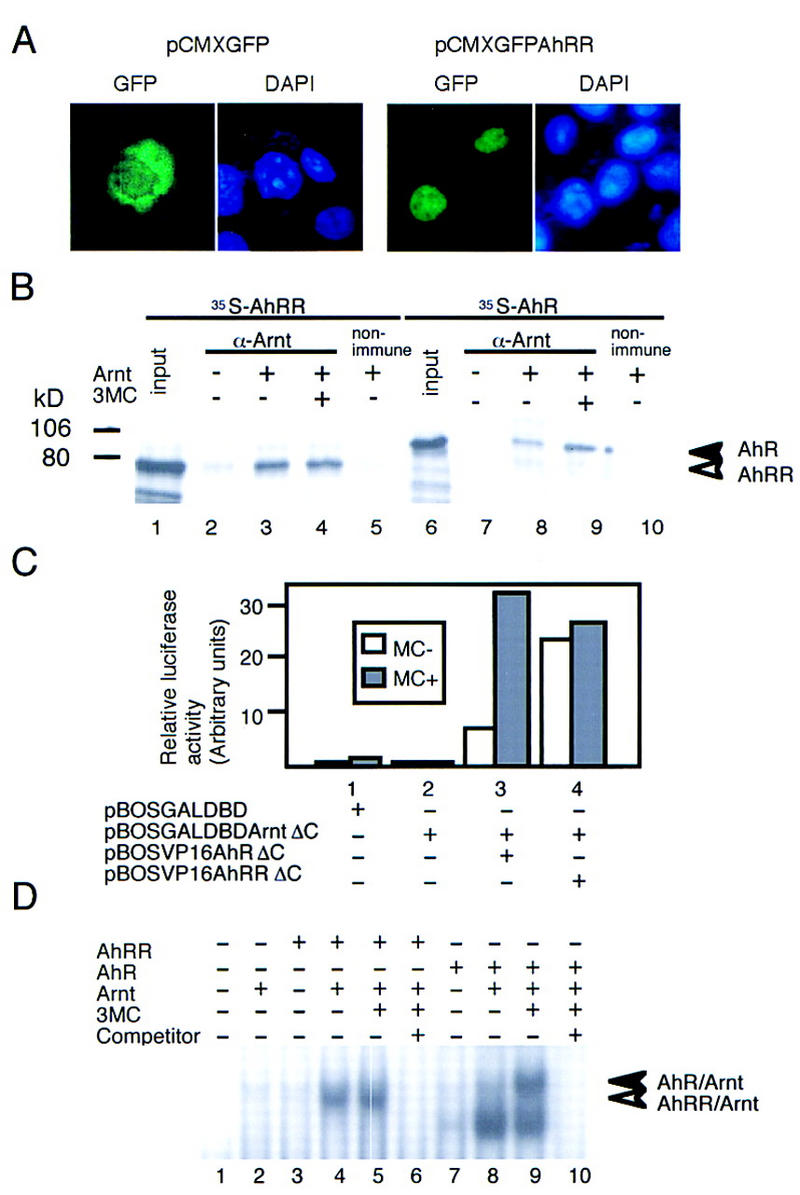
Subcellular localization of AhRR, heterodimer formation of AhRR with Arnt, and its binding to XRE sequence. (A) Subcellular localization of GFP–AhRR fusion protein. To construct pCMX–GFP–AhRR, an EcoRI–SalI fragment of AhRR was ligated with the SalI–BamHI site of pCMX–GFP–hGR (Ogawa et al. 1995). GFP (left) or GFP–AhRR fusion protein (right) was expressed in COS7 cells and visualized as described (Ogawa et al. 1995). (B) Coimmunoprecipitation of AhRR and AhR with an anti-Arnt antibody in the presence or absence of 3MC. Open and closed arrowheads indicate coprecipitated AhRR and AhR, respectively. (C) Interaction between AhRR or AhR and Arnt revealed by the mammalian two-hybrid method. A fusion gene encoding GAL4–DBD and Arnt–bHLH–PAS as bait and those encoding AhR or AhRR bHLH–PAS and VP16 AD were constructed. Various prey/bait combinations were cotransfected into 293 cells with pG3E–Luc; interaction of AhR or AhRR with Arnt was assessed by measuring the expressed luciferase activity. 3MC (1 μm) was used as inducer. (D) In vitro interaction of AhRR/Arnt heterodimer with XRE sequence. (Lanes 6,10) Nonlabeled XRE (250-fold) was used as competitor. Open and closed arrowheads indicate AhRR–Arnt–XRE and AhR–Arnt–XRE complexes, respectively.
Because the basic sequence on the amino terminus of the AhRR bHLH domain is closely related to that of AhR, gel mobility shift assay (GMSA) was performed to determine whether the AhRR/Arnt heterodimer is able to bind the XRE sequence as the AhR/Arnt heterodimer does. As shown in Figure 2D, a mixture of in vitro-synthesized AhRR and Arnt constitutively gave a specific retarded band with XRE that migrated slightly faster than that produced by the AhR/Arnt heterodimer in the presence of 3MC (Fig. 2D, lane 4). The specificity of the XRE binding activity of these heterodimers was confirmed by competitive GMSA (Fig. 2D, lanes 6,10). The reason for the constitutive shifted bands below the inducer-specific band (Fig. 2D, lanes 8,9) remains unknown.
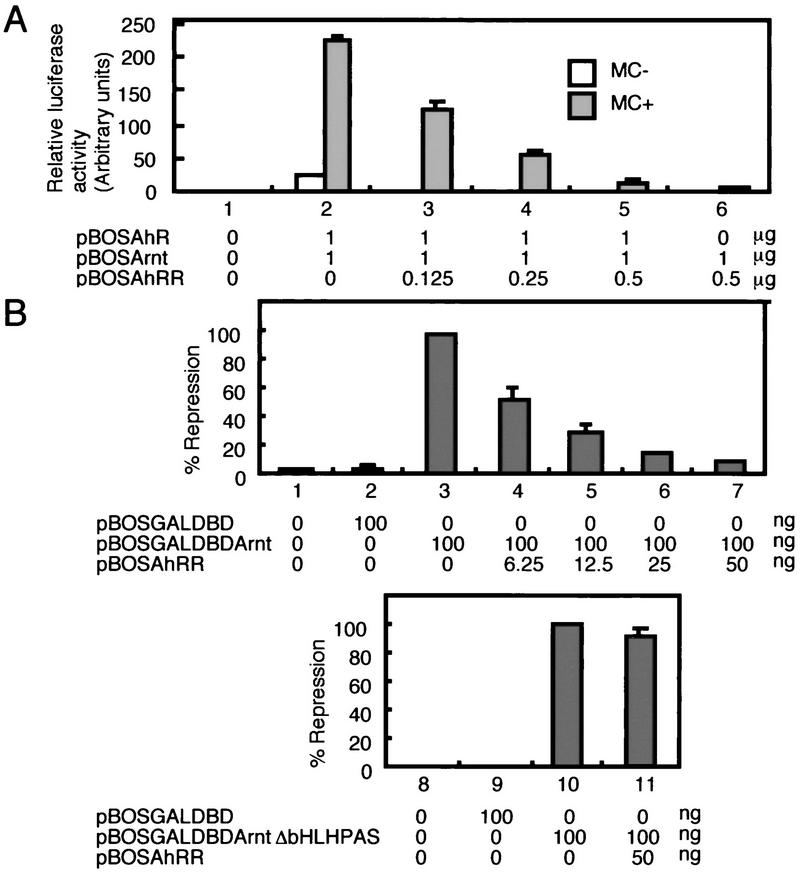
We examined transactivation activity of the AhRR/Arnt heterodimer on the XRE-driven pX4TK–Luc reporter gene in HeLa cells. Cotransfection of AhRR and Arnt expression vectors showed no enhancement, or even suppression, in the basal luciferase expression from the reporter gene (Fig. 3A, lane 6), whereas simultaneous expression of AhR and Arnt stimulated a high level of luciferase expression in an inducer-dependent manner (Fig. 3A, lane 2). Next, we investigated the effects of AhRR on the transactivation activity of the AhR/Arnt heterodimer. Transfection of AhRR expression vector into HeLa cells, together with the expression vectors of AhR and Arnt and the reporter gene pX4TK–Luc, repressed luciferase expression originally induced by AhR and Arnt plasmids in a dose-dependent fashion (Fig. 3A, lanes 3–5). This repression was reversed by further addition of AhR expression vector (data not shown). These results indicate that AhRR functions as a competitive repressor of AhR. Inhibition of the AhR/Arnt function by AhRR can be explained by two possible mechanisms: competition between AhRR and AhR for recruitment of Arnt, and/or competition between these protein complexes for binding the XRE sequence. AhRR only moderately inhibited transactivation by the HIF-1α/Arnt heterodimer (data not shown). This is probably due to the fact that the AhRR/Arnt heterodimer cannot compete with HIF-1α/Arnt for binding to the HRE sequence, although it cannot be ruled out that the affinity of Arnt for HIF-1α is higher than that for AhRR. ΔbAhRR, which lacks the basic DNA-binding domain and therefore forms a heterodimer with Arnt without the XRE-binding activity, gave milder inhibition than AhRR on an XRE-driven reporter gene (data not shown). These results support the conclusion that the efficient repression of AhR function by AhRR requires the two competitive ways of inhibition. Transactivation activity of Arnt per se was exhibited by GAL–DBD–Arnt on the UAS sequence in the promoter (Fig. 3B, lane 3), and this activity was inhibited by addition of AhRR expression plasmid (Fig. 3B, lanes 4–7). Binding of AhRR with Arnt is essential for the inhibitory effect of AhRR, because the activity of GAL–DBD–ArntΔbHLH–PAS, which lacks the bHLH–PAS binding domain for AhRR was not repressed by AhRR (Fig. 3B, lane 11). A small fragment of ∼150 amino acids in the carboxy-terminal half of AhRR was found to be sufficient for inhibitory activity (data not shown). It was shown that the expression of AhRR also inhibited the inducible expression of the endogenous CYP1A1 gene in Hepa-1 cells in response to 3MC (data not shown). In summary, AhRR showed an inherent ability to repress the transactivation activity of Arnt and, in addition, competed for XRE binding upon dimerization with Arnt. The inhibition mechanism of AhRR resembles that of Mad or Mxi1 rather than that of Id. Expression of GAL–DBD–AhRR has also been found to repress the basal transcription of pG3E–Luc (J. Mimura and Y. Fujii-Kuriyama, unpubl.). It is now under investigation whether the inhibition acivity of AhRR is mediated by a corepressor like Mad and Mxi1.
Figure 3.
Repression of transactivation activity of AhR and Arnt heterodimer by AhRR. (A) Enhanced expression of luciferase activity by AhR and Arnt in the presence of 3MC was repressed by AhRR. pX4TK–Luc was cotransfected into HeLa cells with AhR and Arnt expression plasmids, and the transfected cells expressed luciferase activity in response to treatment with 3MC for 44 hr at 37°C. (B) Repression of Arnt transactivation activity by cotransfection with increasing amounts of AhRR expression plasmid. 293T cells were cotransfected with pG3E–Luc reporter plasmid, and increasing amounts of AhRR expression vector and either pBOS–GAL–DBD–Arnt (left) or pBOS–GAL–DBD–ArntΔbHLH–PAS (right) plasmids. Expressed luciferase activities were determined following 44 hr of transfection.
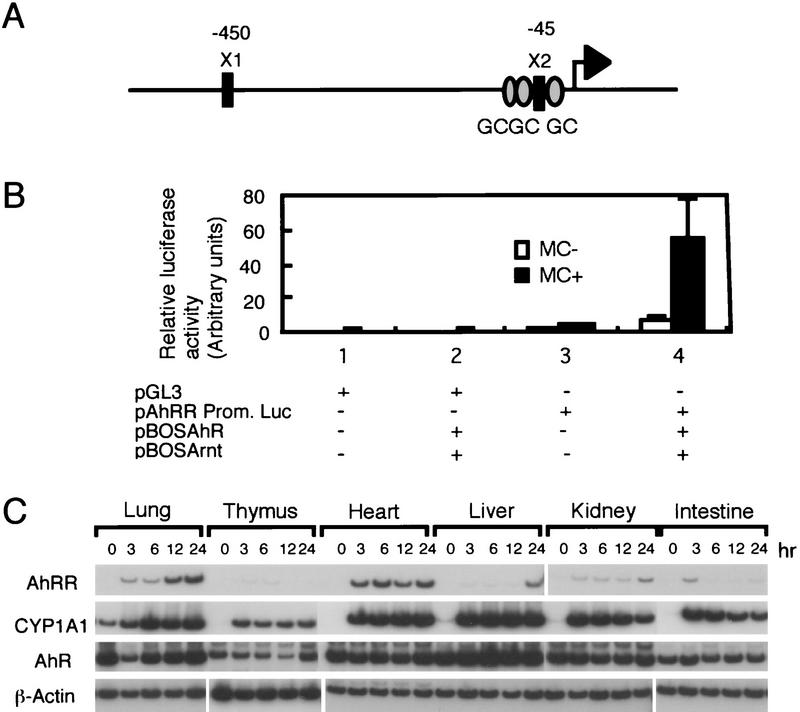
Our next step was to investigate the expression of the AhRR gene and isolated its genomic DNA. Interestingly, sequence analysis revealed that the 5′-flanking sequence of the gene carries two and three copies of the XRE and GC box, respectively, as shown in Figure 4A, which suggests that AhRR gene expression is regulated by the AhR/Arnt heterodimer in response to 3MC.
Figure 4.
Promoter activity of AhRR gene in response to 3MC. (A) Structure of the mAhRR gene promoter. (X1 and X2) Relative location of two XRE sequences in the regulatory region upstream of the AhRR gene. The AhRR gene is a TATA box-less gene; GC boxes are indicated by shaded ovals. (B) Activation of the mouseAhRRgene promoter in response to 3MC. pGL3 or pAhRR-Prom–Luc was cotransfected into HeLa cells and various combinations of AhR and Arnt expression vectors, and the transfected cells were cultured for 44 hr in the presence or absence of 3MC. The promoter activity of the AhRR gene was assessed by measuring expressed luciferase activities. (C) Inducible expression of AhRR mRNA in mouse various tissues in response to 3MC. Male mice (C57BL/6J) were injected intraperitoneally with 3MC in sesame oil (80 mg/kg body weight), Total RNAs from various tissues were subjected to RT–PCR analysis.
When the 5′-flanking sequence of the AhRR gene was fused to the luciferase gene and transfected into HeLa cells along with the expression plasmids of AhR and Arnt, expression of luciferase activity was markedly activated by the administration of 3MC (Fig. 4B, lane 4), indicating that the XRE in the AhRR gene is functional as inducible enhancer. These experiments established AhRR as a novel target gene of AhR in addition to genes encoding drug metabolizing enzymes. Against this background we investigated inducible expression of AhRR mRNA in various tissues of mice in response to 3MC by RT–PCR. In untreated animals, essentially no expression of the mRNA was detected, in liver, heart, lung or other tissues. Upon treatment with 3MC, however, AhRR mRNA levels were induced in these tissues, although the mode of expression was different from tissue to tissue. As shown in Figure 4C, heart and lung showed most abundant AhRR mRNA expression, whereas liver, thymus, kidney, and intestine expressed relatively small amounts of mRNA. Interestingly, tissues such as liver and thymus, which are known to be most susceptible to the toxic effects of dioxin (Poland and Knutson 1982; Fernandez-Salguero et al. 1996) appear to express AhRR mRNA poorly in response to inducers. Expression of the AhRR gene and susceptibility of various tissues to dioxin are now under detailed investigation.
It has long been known that superinduction of TCDD-induced CYP1A1 mRNA was generated in cultured cells such as Hepa-1c1c7 by treatment with inhibitors of protein synthesis such as cycloheximide (Israel et al. 1985), that the AhR function is down-regulated after a short interval of treatment with the inducers, and this down-regulation is blocked by inhibitors of protein synthesis (Lusska et al. 1992). The interesting phenomena of superinduction and block of down-regulation caused by protein synthesis inhibitors have been postulated to be due to inhibited synthesis of a short-lived repressor of AhR function, the entity of which remains elusive. The present data suggest that this repressor is most likely AhRR. Recently, a similar regulatory circuit has been suggested with a mammalian circadian system carried out by the same bHLH–PAS proteins as AhR, such as Clock, BMAL1 and mPer, although their transcriptional activities have not been demonstrated clearly. An elaborate regulatory circuit of AhR function may support the fact that it is involved in cell cycle regulation, as suggested previously (Ma and Whitlock 1996; Weiss et al. 1996).
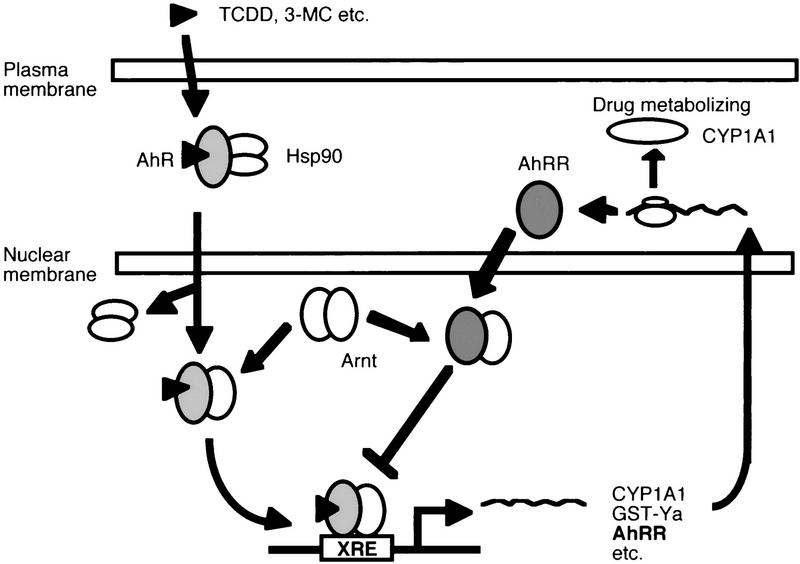
In conclusion, we have identified AhRR as a target gene of the AhR, providing a novel mechanism of feedback inhibition of the receptor function in that a transcription factor induces directly the expression of its repressor gene through binding to the cognate regulatory sequence of the gene. As shown in Figure 5, this regulatory circuit involves activation of AhR by xenobiotics to induce expression of the AhRR gene and many others through binding to the XRE as a heterodimeric complex with Arnt, and induced levels of AhRR inhibit in a tissue-specific manner the AhR function by competing with AhR for Arnt and XRE binding activity.
Figure 5.
Mechanism of negative feedback regulation of AhR function by AhRR. Ligand-activated AhR/Arnt heterodimer transactivates the expression of target genes (including AhRR gene) driven by the XRE sequence. ExpressedAhRRmRNA is translated into AhRR protein, and the resulting AhRR inhibits AhR function by competing with AhR for heterodimerizing with Arnt and binding with the XRE sequence. Consequently, inhibition of translation of AhRR mRNA by inhibitors of protein synthesis results in superinduction of mRNAs by TCDD or 3MC.
Materials and methods
Isolation of mouse AhRR cDNA
A mouse genomic library was screened with a mAhR cDNA to obtain a genomic DNA clone. The genomic fragment encoding the bHLH region was used as a probe to screen a mouse cDNA library under stringent hybridization conditions to isolate AhRR cDNA. The cDNA sequence has been registered in DDBJ (accession no. AB015140).
Immunoprecipitation assay
AhRR and AhR proteins were synthesized in vitro with a TnT-coupled transcription–translation kit (Promega) in the presence of [35S]methioine (Amersham). Arnt protein was synthesized with nonlabeled methionine. Labeled AhRR (5 μl) or AhR (5 μl) was mixed with nonlabeled Arnt (5 μl) and incubated for 2 hr at 30°C in the absence or presence of 3MC (1 μm). The reaction mixtures were immunoprecipitated with anti-Arnt or nonimmune sera and then adsorbed on protein A–Sepharose. Immunoprecipitates were eluted by adding 2× SDS sample buffer (20 μl), boiled for 5 min, and subjected to SDS-PAGE. The radioactivity was visualized with the Image Analyzer BAS 1000 (Fuji film).
Plasmid construction
For construction of pBOSAhR, we first produced a mAhR cDNA fragment that contains the Kozak sequence (Kozak 1987) by PCR, using pBSK–mAhR as a PCR template and the following pair of primers: 5′-GTCGAAGCTTCCGCCACCATGGCCAGCAGCGGCGCC-3′ (sense) and 5′-AGTCGGACGAATAGGTTTC-3′ (antisense). The PCR product was inserted into the blunt-ended XbaI site of pBluescript vector to generate an XbaI site at the 3′ end of cDNA [pBSK–AhR(K)]. pBOSAhR was produced by inserting the blunt-ended HindIII–XbaI fragment of pBSK–AhR(K) into the blunt-ended XbaI site of pEFBOS vector (Mizushima and Nagata 1990). To obtain pBOS–Arnt, the mouse Arnt cDNA fragment was excised by EcoRI–BamHI digestion of pBSK–mArnt (Numayama-Tsuruta et al. 1997), blunt-ended by Klenow and was then ligated with the blunt-ended XbaI site of pEFBOS. The blunt-ended SmaI–HindIII fragment (2.4 kb) of AhRR cDNA was inserted into the blunt-ended XbaI site of pEFBOS to produce pBOS–AhRR. To construct VP16 fusion constructs, we first generated VP16 cDNA by PCR with pSRα–GAL4–DBD–VP16 used as a template, and the resulting fragment was inserted into pBSK–NLS, which also contained the HindIII–BamHI fragment of pENL (Mimura et al. 1997) in pBluescript vector to obtain pNV. The blunt-ended HindIII–EcoRV fragment of pBSK–AhR(K) and the EcoRI–SmaI fragment of AhRR cDNA were inserted into the EcoRV and EcoRI–EcoRV sites of pNV to produce pNV–AhRΔC and pNV–AhRRΔC, respectively. Excision of HindIII–SalI or NotI–SalI from these pNV constructs and subsequent ligation of these fragments (blunt-ended) with the SmaI site of pEFBOS(Stop) (containing an XbaI nonsense linker at the 5′ end of the polyadenylation signal) gave pBOS–VP16–AhRΔC and pBOS–VP16–AhRRΔC, respectively. For construction of pBOS–GAL–DBD–ArntΔC, pBOS–GAL–DBD–Arnt, and pBOS–GAL–DBD–ArntΔbHLH–PAS, we first produced pGBT–ArntΔC, pGBT–Arnt, and pGBT–ArntΔbHLH–PAS by inserting blunt-ended NcoI–PvuII, NcoI–BamHI, and PvuII–BamHI fragments of pBSK–Arnt into the blunt-ended EcoRI site of the pGBT9 vector, respectively. pBOS–GAL–DBD–ArntΔC, pBOS–GAL–DBD–Arnt, and pBOS–GAL–DBD–ArntΔbHLH–PAS were constructed by excising the HindIII–SalI fragments from pGBT–ArntΔC, pGBT–Arnt, and pGBT–ArntΔbHLH–PAS and, subsequently, subcloning these fragments into the blunt-ended XbaI site of pEFBOS vector, respectively.
pG3E–Luc was produced by inserting three copies of the GAL4 binding site and E1b TATA sequence excised from pG5EC vector (Sogawa et al. 1995) into the SmaI site of pGL3 vector (Clontech). pX4TK–Luc was constructed by subcloning four copies of synthesized XRE1 (Kubota et al. 1991) and the TK promoter of pBLCAT2 (BamHI–XhoI fragment) into the XhoI site of pGL3. pAhRR-Prom–Luc was produced by subcloning the blunt-ended HindIII–SacII fragment in the promoter region of the AhRR genomic clone into the SmaI site of pGL3.
GMSA
A double-stranded oligonucleotide XRE probe (Matsushita et al. 1993) was end-labeled with [γ-32P]ATP (Amersham). In vitro-translated AhRR or AhR (5 μl) was mixed with in vitro-translated Arnt (5 μl) or reticulocyte lysate without mRNA and incubated for 2 hr at 30°C in the absence or presence of 3MC (1 μm). The reaction mixtures were diluted by adding 10 ml of 2× binding buffer [200 mm HEPES–KOH (pH 7.9), 1 m KCl, 2 mm EDTA, 60 mm MgCl2, 20 mm DTT, 0.2 mg/ml salmon sperm DNA]. After 15 min incubation at 25°C, the labeled XRE probe (2 × 104 cpm) was added and incubated at 25°C for another 15 min. Protein–DNA complexes were resolved by 4.5% PAGE and subjected to autoradiography.
RT–PCR
Total RNAs (3 μg) from various tissues were used for cDNA synthesis (20 μl), and an aliquot (2 μl) of synthesized cDNA was amplified in a total volume of 20 ml containing 150 mm dNTP, 0.2 units of Taq polymerase, and 0.12 μg of each primer and [α-32P]dCTP. Samples were amplified by repeated cycles of 94°C for 45 sec, 60°C for 45 sec, and 72°C for 1 min. Amplifications of 30 and 28 cycles were applied for AhRR, AhR, CYP1A1, and β-actin, respectively. PCR products were separated on 4% polyacrylamide gels and subjected to autoradiography. PCR primers for amplification of AhR, CYP1A1, and β-actin cDNA were described previously (Mimura et al. 1997), and those for AhRR were 5′-GGCTTACCATGGGCGCTGAG-3′ (sense) and 5′-CCACCAGAGCGAAGCCATTGAG-3′ (antisense). Mice were treated in accordance with institutional guidelines.
Acknowledgments
We thank Dr. L. Poellinger for critical reading of this manuscript and advice. Our thanks also go to Drs. K. Umesono, S. Nagata, H. Handa, M. Katsuki and M. Green for pCMXGFPhGR, pEFBOS, pSRαGAL–4DBD–VP16, pENL, and pG5EC, respectively. This work was supported in part by Grants-in-Aid for Scientific Reseach of priority area from the Ministry of Education, Culture, Sports, and Science of Japan, by funds for Research for the Future Program of the Japan Society for Promotion of Science, and from Sankyo Co.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ykfujii@mail.cc.tohoku.ac.jp; FAX 81-22-217-6594.
References
- Ayer DE, Eisenman RN. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes & Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- Ayer DE, Kretzner L, Eisenman RN. Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D. An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci. 1991;88:1815–1819. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Barrett J, Villa-Garcia M, Resar LM, Kato GJ, Fearon ER. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol Cell Biol. 1991;11:954–962. doi: 10.1128/mcb.11.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Sogawa K, Watanabe N, Chujoh Y, Matsushita N, Gotoh O, Funae Y, Fujii-Kuriyama Y. cDNA cloning and structure of mouse putative Ah receptor. Biochem Biophys Res Commun. 1992;184:246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol Appl Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- Fujisawa-Sehara A, Sogawa K, Yamane M, Fujii-Kuriyama Y. Characterization of xenobiotic responsive elements upstream from the drug-metabolizing cytochrome P-450c gene: A similarity to glucocorticoid regulatory elements. Nucleic Acids Res. 1987;15:4179–4191. doi: 10.1093/nar/15.10.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijo Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt) Mol Cell Biol. 1996;16:1706–1713. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EC, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- Israel DI, Estolano MG, Galeazzi DR, Whitlock J., Jr Superinduction of cytochrome P1-450 gene transcription by inhibition of protein synthesis in wild type and variant mouse hepatoma cells. J Biol Chem. 1985;260:5648–5653. [PubMed] [Google Scholar]
- Jackson FR, Bargiello TA, Yun SH, Young MW. Product of per locus of Drosophila shares homology with proteoglycans. Nature. 1986;320:185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program in vivo association of Id with E2A proteins. Genes & Dev. 1992;6:1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Kubota M, Sogawa K, Kaizu Y, Sawaya T, Watanabe J, Kawajiri K, Gotoh O, Fujii-Kuriyama Y. Xenobiotic responsive element in the 5′-upstream region of the human P-450c gene. J Biochem. 1991;110:232–236. doi: 10.1093/oxfordjournals.jbchem.a123562. [DOI] [PubMed] [Google Scholar]
- Larsson LG, Pettersson M, Oberg F, Nilsson K, Lüscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells: opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- Lusska A, Wu L, Whitlock J., Jr Superinduction of CYP1A1 transcription by cycloheximide. Role of the DNA binding site for the liganded Ah receptor. J Biol Chem. 1992;267:15146–15151. [PubMed] [Google Scholar]
- Ma Q, Whitlock JP., Jr The aromatic hydrocarbon receptor modulates the Hepa 1c1c7 cell cycle and differentiated state independently of dioxin. Mol Cell Biol. 1996;16:2144–2150. doi: 10.1128/mcb.16.5.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita N, Sogawa K, Ema M, Yoshida A, Fujii-Kuriyama Y. A factor binding to the xenobiotic responsive element (XRE) of P-4501A1 gene consists of at least two helix-loop-helix proteins, Ah receptor and Arnt. J Biol Chem. 1993;268:21002–21006. [PubMed] [Google Scholar]
- Mimura J, Yamashita K, Nakamura K, Morita M, Takagi TN, Nakao K, Ema M, Sogawa K, Yasuda M, Katsuki M, Fujii-Kuriyama Y. Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells. 1997;2:645–654. doi: 10.1046/j.1365-2443.1997.1490345.x. [DOI] [PubMed] [Google Scholar]
- Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Boltimore D. A new DNA binding and dimerization motif in immunogloblin enhancer binding, doughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton K, Jr, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Neuhold LA, Wold B. HLH forced dimers: Tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell. 1993;74:1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- Numayama-Tsuruta K, Kobayashi A, Sogawa K, Fujii-Kuriyama Y. A point mutation responsible for defective function of the aryl-hydrocarbon-receptor nuclear translocator in mutant Hepa-1c1c7 cells. Eur J Biochem. 1997;246:486–495. doi: 10.1111/j.1432-1033.1997.00486.x. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Inouye S, Tsuji FI, Yasuda K, Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc Natl Acad Sci. 1995;92:11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdew GH. Association of the Ah receptor with the 90-kDa heat shock protein. J Biol Chem. 1988;263:13802–13805. [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Pongratz I, Mason GG, Poellinger L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J Biol Chem. 1992;267:13728–13734. [PubMed] [Google Scholar]
- Sogawa K, Iwabuchi K, Abe H, Fujii-Kuriyama Y. Transcriptional activation domains of the Ah receptor and Ah receptor nuclear translocator. J Cancer Res Clin Oncol. 1995;121:612–620. doi: 10.1007/BF01197779. [DOI] [PubMed] [Google Scholar]
- Sogawa K, Fujii-Kuriyama Y. Ah receptor, a novel ligand-activated transcription factor. J Biochem. 1997;122:1075–1079. doi: 10.1093/oxfordjournals.jbchem.a021864. [DOI] [PubMed] [Google Scholar]
- Swanson HI, Bradfield CA. The AH-receptor: Genetics, structure and function. Pharmacogenetics. 1993;3:213–230. doi: 10.1097/00008571-199310000-00001. [DOI] [PubMed] [Google Scholar]
- Telakowski-Hopkins CA, King RG, Pickett CB. Glutathione S-transferase Ya subunit gene: Identification of regulatory elements required for basal level and inducible expression. Proc Natl Acad Sci. 1988;85:1000–1004. doi: 10.1073/pnas.85.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss C, Kolluri SK, Kiefer F, Göttlicher M. Complementation of Ah receptor deficiency in hepatoma cells: negative feedback regulation and cell cycle control by the Ah receptor. Exp Cell Res. 1996;226:154–163. doi: 10.1006/excr.1996.0214. [DOI] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]