Abstract
The PRDI-BF1/Blimp-1 protein is a transcriptional repressor required for normal B-cell differentiation, and it has been implicated in the repression of β-interferon (IFN-β) and c-myc gene expression. Here, we show that PRDI-BF1 represses transcription of the IFN-β promoter and of an artificial promoter through an active repression mechanism. We also identified a minimal repression domain in PRDI-BF1 that is sufficient for transcriptional repression when tethered to DNA as a Gal4 fusion protein. Remarkably, this repression domain interacts specifically with hGrg, TLE1, and TLE2 proteins, all of which are members of the Groucho family of transcriptional corepressors. In addition, the hGrg protein itself can function as a potent repressor when tethered to DNA through the Gal4 DNA-binding domain. We also find that the amino-terminal glutamine-rich domains of hGrg and TLE1 are sufficient to mediate dimerization of the two Groucho family proteins. Proteins containing only this domain can function as a dominant-negative inhibitor of PRDI-BF1 repression, and can significantly increase the IFN-β promoter activity after virus induction. We conclude that PRDI-BF1/Blimp-1 represses transcription by recruiting a complex of Groucho family proteins to DNA, and suggest that such corepressor complexes are required for the postinduction repression of the IFN-β promoter.
Keywords: PRDI-BF1 Blimp-1 protein, transcriptional repressor, B-cell differentiation, Groucho proteins, IFN-β promoter
β-Interferon (IFN-β) is not synthesized usually in normal growing tissue cultures or in animals, but its production is highly inducible by virus or double-stranded RNA (for review, see DeMaeyer and DeMaeyer-Guignard 1988). The level of IFN-β mRNA in human fibroblast cells peaks ∼6–12 hr after induction and then decreases rapidly. This transient induction of the IFN-β gene is caused by transcriptional activation followed by postinduction repression of transcription (for review, see Maniatis et al. 1992). It has been shown that the postinduction shutoff requires protein synthesis as well as the PRDI (positive regulatory domain I) site within the IFN-β gene promoter (Whittemore and Maniatis 1990a).
Two or more copies of the PRDI elements are transiently virus-inducible and are turned off at the same time as the intact IFN-β promoter (Whittemore and Maniatis 1990a,b). The PRDI-BF1 protein, which was first identified by virtue of its ability to bind to the PRDI site (hence, the name PRDI-binding factor), can repress transcription of the human IFN-β gene in cotransfection experiments (Keller and Maniatis 1991). The PRDI-BF1 gene is virus-inducible, and the level of PRDI-BF1 mRNA is detectable 4 hr after virus induction. It increases for the next 20 hr (Keller and Maniatis 1991). The level of PRDI-BF1 mRNA reaches a maximum at a time at which the IFN-β gene is turned off after virus induction. Thus, PRDI-BF1 is a postinduction repressor of the IFN-β gene (Keller and Maniatis 1991).
The PRDI-BF1 gene also plays an essential role in the differentiation of B cells into plasma cells (Turner et al. 1994). The PRDI-BF1 mouse homolog, also known as B-lymphocyte induction maturation protein-1 (Blimp-1), is specifically expressed late in B-cell differentiation and continues to be expressed in plasma cells. Ectopic expression of Blimp-1 in a mature B-lymphoma cell line results in phenotypic changes characteristic of B cells that differentiate into plasma cells, including the induction of J-chain message and immunoglobulin secretion, up-regulation of Syndecan-1, and an increase in cell size and granularity (Turner et al. 1994). Interestingly, when the Blimp-1 gene is expressed at earlier stages of B-cell development, it induces apoptosis (Lin et al. 1997; Messika et al. 1998). Thus, the mouse Blimp-1 gene seems to act as a checkpoint gene in the course of B-cell differentiation: It ensures that fully activated B cells proceed to the plasma cell stage and helps to eliminate the immature and partially activated B cells (Messika et al. 1998). The human PRDI-BF1 gene has likewise been implicated as a regulator of B-cell differentiation, as deletion of the PRDI-BF1 locus is found in several human malignancies, particularly in B-cell non-Hodgkin lymphoma (B-NHL) (Mock et al. 1996).
At least one of the functions of PRDI-BF1/Blimp-1 in B-cell differentiation appears to be repression of the c-myc proto-oncogene (Turner et al. 1994; Lin et al. 1997). The general function of the c-myc gene is to promote cellular proliferation, thus preventing terminal differentiation. In particular, the regulation of c-myc during B-cell development correlates with transitions in B-cell differentiation. Expression of c-myc is induced upon stimulation by antigens and maintained at high levels in proliferating cells, but is absent in quiescent or terminally differentiated plasma cells (Hoffman-Liebermann and Liebermann 1991; Melchers 1997). Therefore, it is not surprising that deregulation of c-myc gene expression is a common feature of virtually all plasma cell tumors and Burkitt lymphomas (Potter and Marcu 1997). Thus, activation and repression of c-myc expression correlates with B-cell proliferation and differentiation, respectively.
Recently, Blimp-1 was shown to repress c-myc transcription by binding to the plasma repression factor (PRF) site located 290 bp upstream from the P1 transcriptional start site of the c-myc gene (Kakkis and Calame 1987; Kakkis et al. 1989; Lin et al. 1997). This site is identical to the previously identified PRDI-BF1 binding site in the human IFN-β promoter (Keller and Maniatis 1991). At present, the mechanism of repression by PRDI-BF1/Blimp-1 is not well understood. In the case of the IFN-β promoter, PRDI-BF1 binds to a sequence that is also recognized by members of the interferon regulatory factor (IRF) family of transcriptional activators (Miyamoto et al. 1988; Wathelet et al. 1998). Thus, PRDI-BF1 is thought to act at least in part by interfering with the binding of critical activator proteins to the IFN-β enhancer (Keller and Maniatis 1991). In the case of the c-myc promoter Blimp-1 does not prevent the binding of an activator. Rather, Blimp-1 forms a stable complex with the YY1 protein bound to an adjacent site (Kakkis et al. 1989; Riggs et al. 1993; Lin et al. 1997). How this interaction leads to repression is not understood.
Here, we investigate the mechanism of repression by PRDI-BF1/Blimp-1 at the IFN-β promoter and a simple promoter containing multiple binding sites for the PRDI-BF1/Blimp-1 protein. Using PRDI-BF1 deletions and Gal4 hybrid proteins we show that a 58-amino acid sequence in PRDI-BF1 is both necessary and sufficient for PRDI-BF1-mediated repression. This repression domain interacts specifically with hGrg and other members of the Groucho family of transcriptional corepressors. Furthermore, we show that hGrg functions as a potent repressor when fused to the Gal4 DNA-binding domain, and that the conserved Q domain in the Groucho proteins can act as a dominant-negative mutant and relieve the PRDI-BF1 repression in cultured cells. Remarkably, the hGrg dominant-negative mutant can delay the postinduction repression of IFN-β transcription significantly. Based on these observations, we propose that PRDI-BF1/Blimp-1 represses transcription by recruiting the Groucho family corepressors to adjacent promoters.
Results
PRDI-BF1 is a long-range repressor
Previous experiments showed that PRDI-BF1 can repress IFN-β promoter activity in cotransfection experiments. The mechanism for this repression was thought to be steric hindrance, because the PRDI site is also recognized by activator proteins that play critical roles in the activation phase of the IFN-β expression and it is conceivable that the binding of PRDI-BF1 to this site can displace activators. This model, however, has not been tested directly.
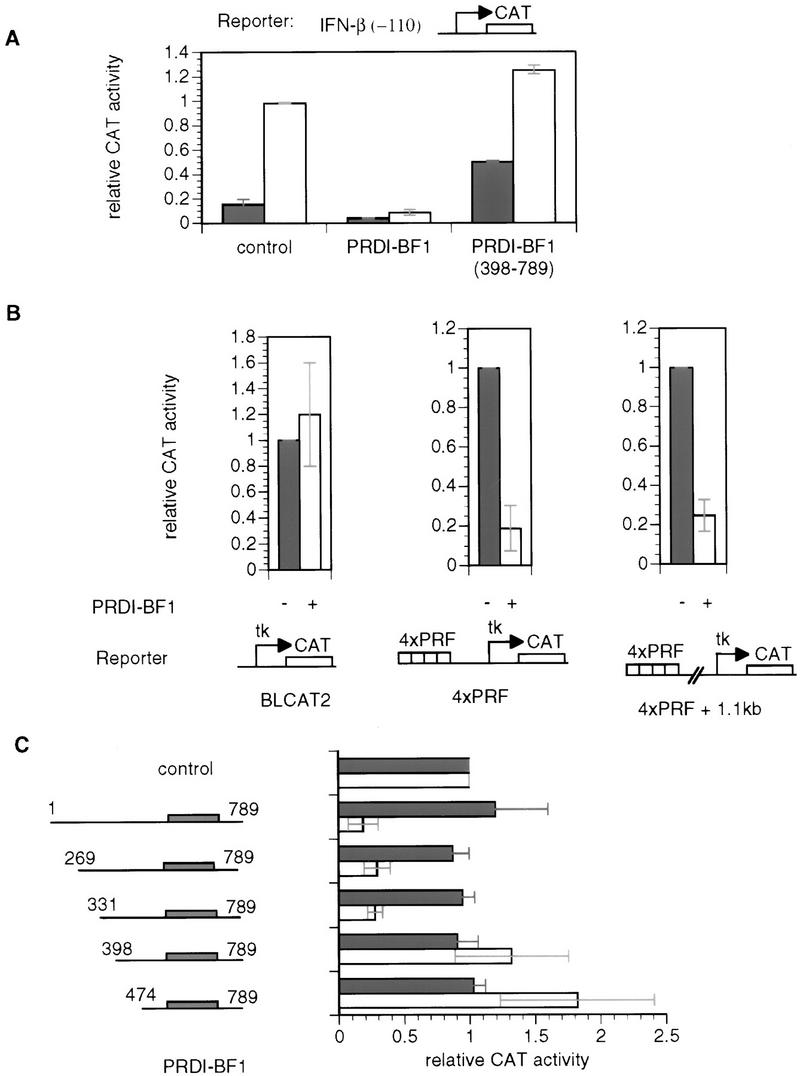
PRDI-BF1 is a 789-amino-acid protein bearing five carboxy-terminal zinc finger DNA-binding motifs (Keller and Maniatis 1991). The carboxy-terminal domain of PRDI-BF1, including the zinc finger motifs, is sufficient for specific DNA binding (Keller and Maniatis 1991). If PRDI-BF1 represses transcription through steric hindrance, the carboxy-terminal DNA-binding domain of PRDI-BF1 should be sufficient for repression. This possibility was tested by carrying out cotransfection experiments in HeLa cells. The PRDI-BF1 full-length cDNA or a carboxy-terminal truncation of PRDI-BF1 that retains the DNA-binding domain was cloned into a mammalian expression vector (pcDNA3, Invitrogen) and then cotransfected with a reporter plasmid containing the intact IFN-β promoter driving the expression of the bacterial chloramphenicol acetyltransferase (CAT) gene (Fig. 1A). As a control, the mammalian expression vector lacking the PRDI-BF1 insert was cotransfected with the reporter gene. Consistent with the previous results, expression of the full-length PRDI-BF1 gene resulted in a dramatic inhibition of the IFN-β promoter activity both before and after virus induction. Surprisingly, expression of the carboxy-terminal DNA-binding domain of PRDI-BF1 failed to repress the activity of the reporter. Rather, it elevated the level of the IFN-β promoter activity to >threefold before virus induction, and also slightly increased the virus-induced level of transcription. Thus, the DNA-binding domain of PRDI-BF1 is not sufficient for the repression, suggesting that the PRDI-BF1 repression is not mediated through a steric hindrance.
Figure 1.
PRDI-BF1 is an active repressor. (A) PRDI-BF1 represses transcription from the natural IFN-β promoter, and this repression requires the amino terminus of the protein. The histogram shows the CAT activity produced in HeLa cells transfected with 5 μg of the reporter gene containing the −110 IFN-β promoter fused to the CAT gene, cotransfected with 1 μg of the indicated expression vector, 4 μg of the pcDNA3 vector, and 2 μg of pCMV–lacZ. The cells were either untreated (dark bars) or infected with Sendai virus (open bars) 24 hr after transfection, and harvested 16 hr later. (Control) pcDNA3 vector. CAT activities in this and the following experiments are normalized to the activity of the cotransfected pCMV–lacZ gene. (B) PRDI-BF1 represses transcription of the tk promoter when bound to sites located 1000 nucleotides from the start site of transcription. HeLa cells were transfected with 2 μg of pCMV–lacZ control plasmid, 6 μg of pXM, 3 μg of reporter, and 1 μg of effector pcDNA3 (−) or pcDNA3–PRDI-BF1 (+). The control reporter BLCAT2 contains a fragment (−109 to +55 bp) of the herpes simplex virus tk promoter driving the expression of the bacterial CAT gene. The 4×PRF reporter contains four copies of the PRF element inserted adjacent to the tk promoter. In the case of the 4×PRF + 1.1-kb reporter a 1.1-kb λDNA fragment was inserted between the tk promoter and the PRF elements. The data shown are representative of three independent assays. For each reporter, the CAT activity for PRDI-BF1 was normalized to that of the negative control pcDNA3. (C) Amino acids 331–398 of PRDI-BF1 functions as a transcriptional repression domain. The PRDI-BF1 sequences tested in each assay are illustrated at left. Horizontal lines represent PRDI-BF1 sequences of the full-length protein and various deletion mutants as illustrated. Shaded boxes denote the zinc finger DNA-binding domains of PRDI-BF1. PRDI-BF1 constructs (1 μg of each) were transfected into HeLa cells with the same control reporter and PRF-containing reporter (4×PRF) as in B. The data shown are representative of three independent assays, and the CAT activity for all PRDI-BF1 constructs was normalized by the CAT activity of the cells transfected with the control pcDNA3–Flag vector and the respective reporter BLCAT2 (dark bars) or 4×PRF–BLCAT2 (open bars).
The above result implies that PRDI-BF1 is an active repressor that can repress transcription on its own, probably by recruiting corepressor proteins through its amino-terminal region. If this is true, one would expect that PRDI-BF1 may repress transcription regardless of the distance between the core promoter and the PRDI-BF1-binding site. To determine whether PRDI-BF1 could function at a distance from the promoter, HeLa cells were transfected with a plasmid expressing full-length PRDI-BF1 along with three different reporter plasmids. The control reporter pBLCAT2 contains the herpes simplex virus thymidine kinase (tk) promoter driving the expression of CAT gene. The second reporter contains four copies of the PRF element from the mouse c-myc gene inserted upstream of the herpes virus tk promoter. The third reporter contains a 1.1-kb insert of λ DNA between the four PRF sites and the promoter. Sequence analysis verified that the λ DNA insert does not contain a fortuitous PRDI-BF1-binding site.
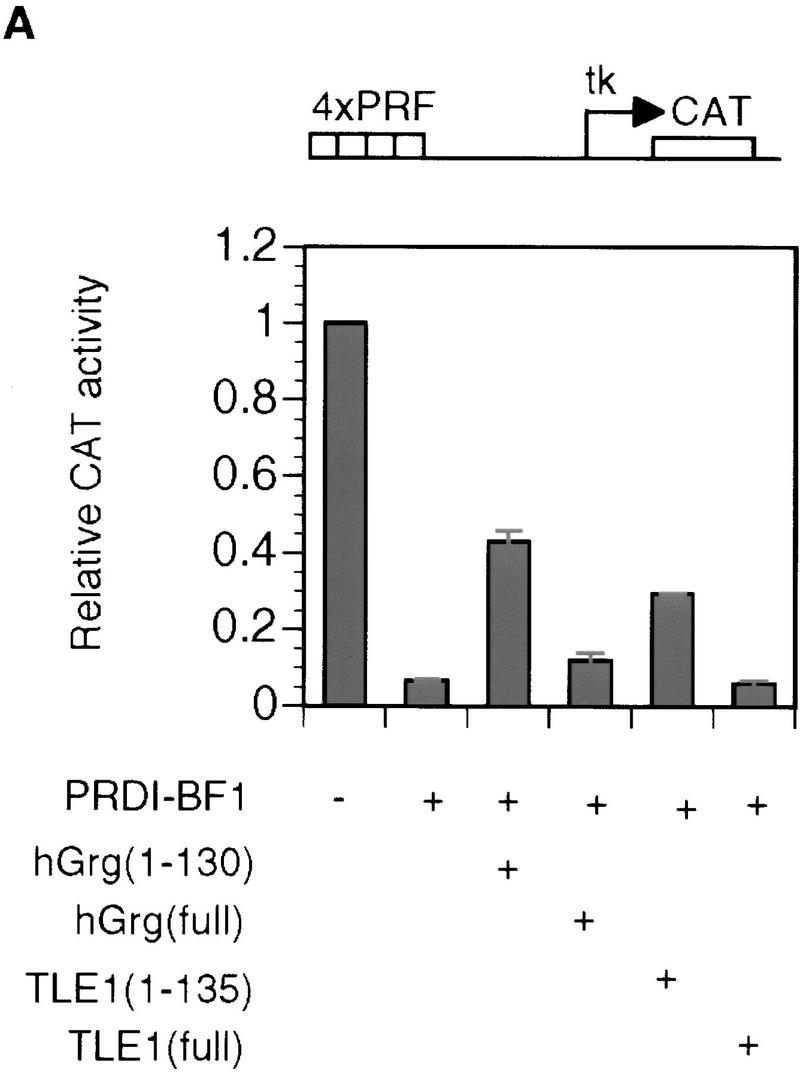
As shown in Figure 1B, PRDI-BF1 repressed transcription regardless of the distance of the PRF sites from the promoter. This result confirms that PRDI-BF1 is an active repressor, and suggests that its amino-terminal region contains a repression domain that is necessary and sufficient for repression.
Identification and characterization of a PRDI-BF1 repression domain
To identify the repression domain of PRDI-BF1, systematic amino-terminal truncations of the protein were constructed and overexpressed in HeLa cells along with a reporter bearing four PRF sites (Fig. 1B). Successive truncations of ∼65 amino acids were constructed and tested. The first two truncations, PRDI-BF1(269–789) and (331–789), exhibited >threefold repression of the reporter gene, which is comparable to the repression by the full-length PRDI-BF1 protein. By contrast, subsequent truncations actually exhibited 1.5- to 2-fold activation (Fig. 1C). Western blotting assays were performed to confirm that each construct was expressed at the same level (data not shown). Thus, the region of PRDI-BF1 between 331 and 398 amino acids is necessary for transcriptional repression. Remarkably, the corresponding sequence in the mouse Blimp-1 gene was shown recently to be required for the induction of apoptosis in immature B cells by Blimp-1 (Messika et al. 1998). Thus, PRDI-BF1/Blimp-1 protein appears to induce apoptosis by repressing transcription of genes required for protection of cells from programmed cell death.
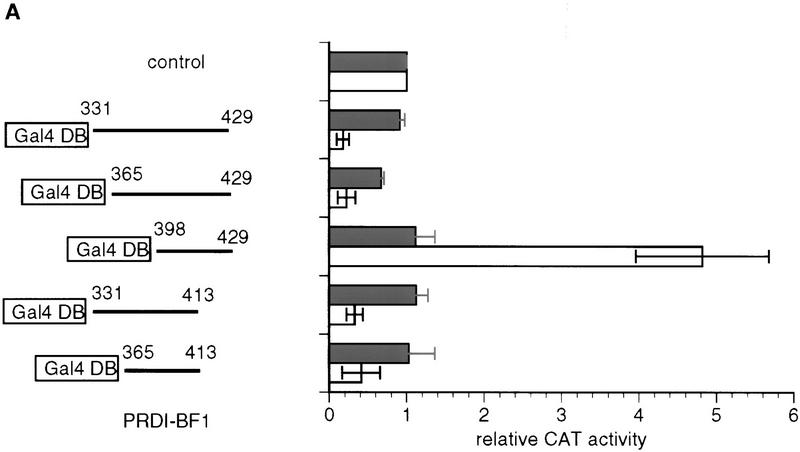
To test whether the repression domain of PRDI-BF1 is sufficient to repress transcription, this sequence and the adjacent 30-amino-acid residues (331–429) were fused to a Gal4 DNA-binding domain. As shown in Figure 2A, this fusion protein was capable of repressing the expression of a reporter containing five Gal4-binding sites upstream of the tk promoter which drives the expression of the CAT gene (G5BLCAT2; Fig. 2A). Thus, the PRDI-BF1 repression domain is sufficient for transcriptional repression.
Figure 2.
The PRDI-BF1 repression domain functions when fused to the Gal4-DNA-binding domain. (A) The ability of various Gal4–PRDI-BF1 fusion proteins to repress transcription was tested with a reporter that bears five Gal4 DNA-binding sites upstream of the tk promoter (G5BLCAT2). The BLCAT2 (see Fig. 1B) reporter was used as a control. Horizontal lines represent various PRDI-BF1 amino- and carboxy-terminal truncations cloned into the pBXG expression vector, which contains a DNA sequence encoding the Gal4 DNA-binding domain, Gal4(1–147) (boxed). PRDI-BF1 constructs (1 μg) were transfected into HeLa cells with either the control reporter BLCAT2 (dark bars) or G5BLCAT2 (open bars). The data shown are representative of three independent assays, and the CAT activities for all Gal4–PRDI-BF1 constructs were normalized by the CAT activity of the cells transfected with the control vector pECE and the respective reporter G5BLCAT2 or BLCAT2. (B) The sequence of a portion of the repression domain in PRDI-BF1. The sequence of the homologous region in Blimp-1 is also shown. The amino acid residues shared by the two proteins are listed between the two sequences. Two stretches of proline-rich regions, PRI and PRII, are indicated by brackets. (C) The proline-rich region of PRDI-BF1 is compared to similar region in WT1. Residues shared by PRDI-BF1 and WT1 protein are listed between the two sequences.
To further localize the PRDI-BF1 repression domain, various amino- and carboxy-terminal truncation mutants were fused to the Gal4 DNA-binding domain. When these PRDI-BF1 constructs were transfected into HeLa cells, between two- and fivefold repression was observed for all of the constructs, with the exception of PRDI-BF1(398–429), which activated transcription by almost fivefold above the control vector (Fig. 2A). This observation suggests that there is an activation domain in the region from amino acids 398–429 in PRDI-BF1 that is not detected when the repression domain is also present. Although the minimal sequence required to observe repression lies between amino acids 365 and 398, the minimal sequence required for maximal repression lies between amino acids 331 and 429 (Fig. 2A).
We note that the extended repression domain of PRDI-BF1 contains two proline-rich sequences, PRI and PRII, that are highly conserved between PRDI-BF1 and its murine homolog Blimp-1 (Fig. 2B). Similar proline-rich regions have been identified in the Wilm’s tumor protein (WT1), which also functions as a transcriptional repressor (Fig. 2C). The proline-rich region between amino acids 13–34 of WT1 lies outside of a minimal 40-amino acid repression domain, but the proline-rich region is required for maximal repression in vivo (Madden et al. 1991; Wang et al. 1995). Although a proline-rich region is shared by many transcriptional repressors, the functional significance of the sequence is not understood (Hanna-Rose and Hansen 1996).
PRDI-BF1 interacts with the human Groucho-related gene
Previously characterized long-range repressors have been shown to function through recruitment of corepressor proteins (Gray and Levine 1996). We therefore carried out a yeast two-hybrid screen with the repression domain of PRDI-BF1 (amino acids 331–429) as bait in an effort to identify a potential corepressor protein. A plasmid library of fusions between a transcriptional activation domain and cDNAs from human bone marrow cells (Clontech) was used as prey in the two-hybrid screen. Five positive clones were isolated and all were found to correspond to the human Groucho-related gene (hGrg), also named human amino-terminal enhancer of split (hAES) (Mallo et al. 1993; Miyasaka et al. 1993). To demonstrate that the two-hybrid PRDI-BF1/hGrg interaction is specific, the hGrg plasmid was reintroduced into yeast cells along with Gal4–PRDI-BF1 fusion constructs that contain or lack the repression domain of PRDI-BF1. Yeast cells containing hGrg and Gal4–PRDI-BF1(398–429; lacking the repression domain) were unable to grow in the selective medium, indicating that no interaction between hGrg and the PRDI-BF1 fragment occurred. In contrast, yeast transformed with Gal4–PRDI-BF1(331–429) and hGrg grew in selective medium, and the colonies were blue in the X-gal assay (data not shown).
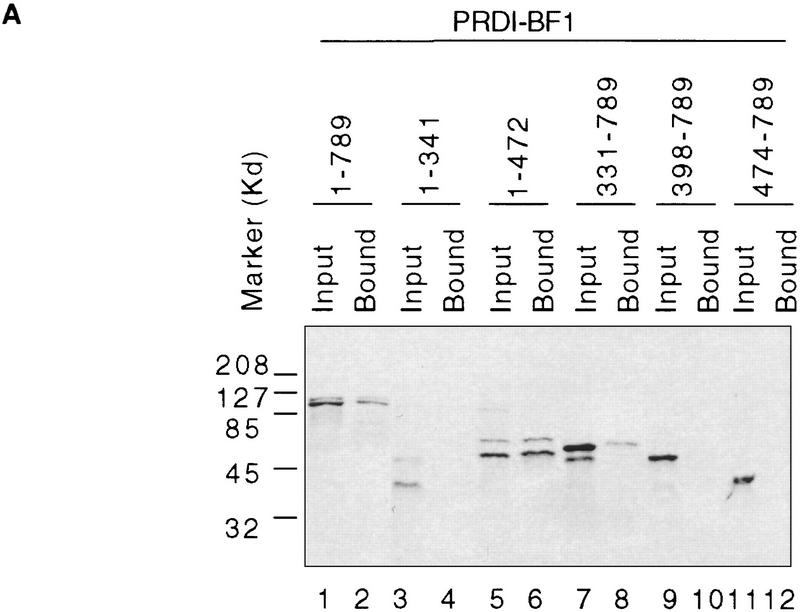
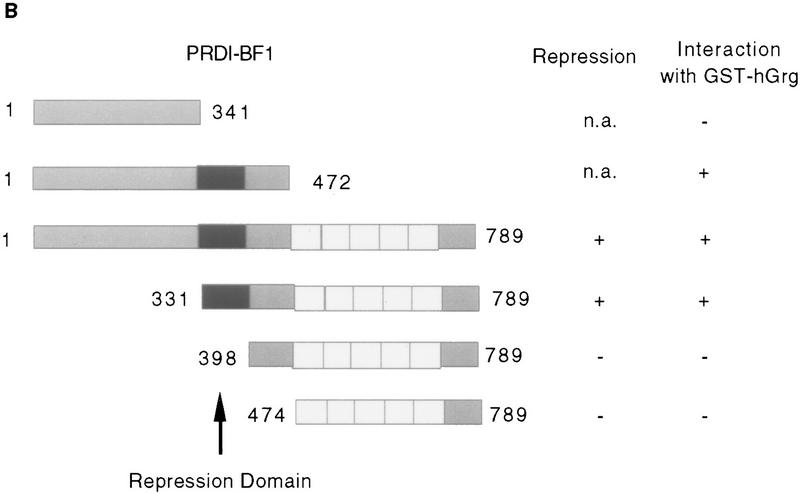
To further investigate the interactions between hGrg and PRDI-BF1 we performed in vitro protein–protein interaction assays using GST fusion proteins. Various PRDI-BF1 mutant proteins were synthesized and labeled with [35S]methionine by in vitro transcription/translation, and incubated with a GST–hGrg fusion protein immobilized on glutathione–agarose beads. Whereas the full-length PRDI-BF1 protein, as well as truncations that retain the repression domain of PRDI-BF1, interacted with GST–hGrg, truncation mutants in which the repression domain was deleted failed to interact (Fig. 3). Thus, the PRDI-BF1/hGrg interaction requires the repression domain of PRDI-BF1.
Figure 3.
The PRDI-BF1 protein interacts specifically with the human Groucho-related protein (hGrg) in vitro. (A) Radiolabeled PRDI-BF1 full-length protein or various truncations of PRDI-BF1 were incubated with GST–hGrg protein immobilized on agarose beads. After washing the beads, the bound PRDI-BF1 proteins and one-fifth of the input were analyzed on a 10% SDS–polyacyrlamide gel and visualized by autoradiography. The protein size marker (in kD) is shown at left. (B) Correlation between the repressive function of various PRDI-BF1 truncations and their interaction with GST–hGrg protein is shown. (−) Lack of repression or protein–protein interactions; (+) presence of repressive activity or protein–protein interactions; (n.a.) repressive function was not studied.
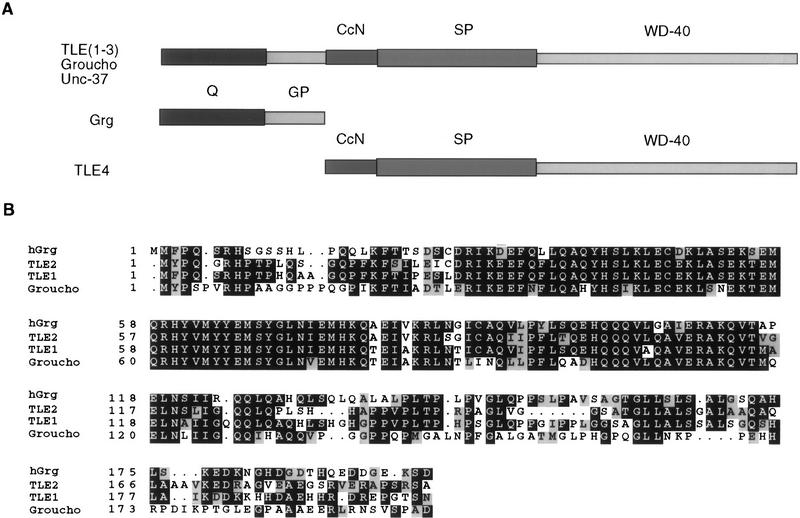
The hGrg protein encodes a 197-amino-acid nuclear protein that belongs to a family of highly conserved proteins named for one of its members, the Drosophila Groucho protein (Hartley et al. 1988). The Groucho family includes three types of proteins (for review, see Fisher and Caudy 1998). The larger proteins such as Groucho and its mammalian homologs, the TLE proteins [transducin-like enhancer of split (TLE1-3)], share five domain structures: the amino-terminal Q domain that is glutamine-rich, the GP domain that is rich in glycine and proline residues, a CcN domain that includes a casein kinase II site/cdc2 kinase site/nuclear localization sequence motif, an SP domain that is rich in serine and proline residues, and carboxy-terminal WD-40 repeats (Stifani et al. 1992) (Fig. 4A). Three of these domains, the Q, CcN, and WD-40 domains, are the most highly conserved. A shorter protein, the human TLE4, contains all the domains except for the amino-terminal Q domain. The shortest proteins in the Groucho family, including hGrg and its murine homolog mGrg, share only the first two regions from the amino terminus (Mallo et al. 1993). Significant homology is observed in the Q domain between hGrg and other Groucho proteins except for TLE4 (Fig. 4B). This region has been shown to participate in the dimerization of Groucho family proteins (Pinto and Lobe 1996).
Figure 4.
Organization of Groucho family proteins. (A) Domain structures of three forms of the Groucho family proteins. The Q domain is rich in glutamine. The GP domain is proline- and glycine-rich. The CcN domain contains target sites for casein kinase II and cdc2 kinase, and a nuclear localization signal. The SP domain is rich in serine and proline. The carboxy-terminal domain contains four WD-40 repeats. Among these, the Q domain, the CcN domain, and the WD-40 domain are the most conserved. (B) An amino acid sequence alignment of the hGrg, TLE1, TLE2, and Groucho proteins. The entire lengths of hGrg and the amino-terminal 200 amino acids of proteins are shown. Identical residues to hGrg shared by any of the three other proteins are marked by black boxes, and the similar amino acids marked by shaded boxes. Dots denote the alignment gaps.
PRDI-BF1 interacts with the Q domain of Groucho family proteins
Based on the extensive homology shared by hGrg and other members of the Groucho family (except for TLE4), it is likely that PRDI-BF1 also interacts with other Groucho proteins, such as TLE1 and TLE2. To test this hypothesis, GST pull-down assays were carried out. The amino-terminal 269 amino acid residues of TLE1 and 315 amino acid residues of TLE2 were used to create GST fusion proteins (Fig. 5A). Labeled PRDI-BF1 proteins with truncations in the amino or caboxy terminus were incubated with the GST–TLE1 and GST–TLE2 proteins immobilized on glutathione–agarose beads. Figure 5, B and D, shows that both TLE1 and TLE2 interacted specifically with the PRDI-BF1 truncations containing the repression domain, whereas no protein–protein interactions were detected between TLE proteins and PRDI-BF1 truncations lacking the repression domain. The GST protein was used in parallel as a negative control and did not interact with any of the PRDI-BF1 proteins. These findings therefore confirm that PRDI-BF1 can associate in vitro with TLE1 and TLE2.
Figure 5.
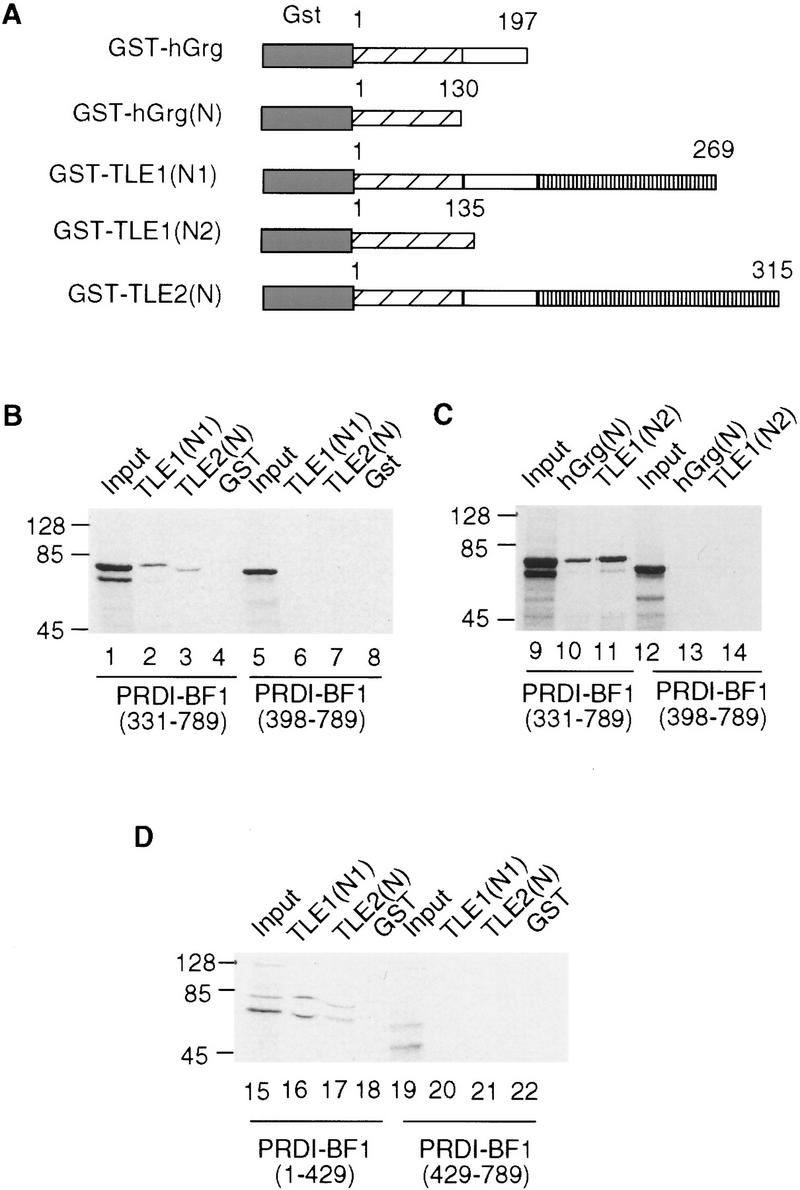
Groucho family proteins interact with each other and with PRDI-BF1. (A) Diagram of the GST/hGrg, GST/TLE1, and GST/TLE2 fusion constructs used in B–D. The GST domain was fused to various fragments of Groucho proteins, with the numbers indicating the starting and ending amino acids. (B,D) Groucho family proteins TLE1 and TLE2 bind to the repression domain of PRDI-BF1 in vitro. (C) The Q domain of hGrg and TLE1 is sufficient to bind to PRDI-BF1. PRDI-BF1 truncations (lanes 1–22) were translated and radiolabeled in vitro, and incubated with immobilized GST fusion proteins or GST protein, as indicated at the top. After the beads were washed, one-fifth of the input (top) and bound proteins were analyzed on a 10% SDS–polyacrylamide gel and visualized by autoradiography. The protein size marker (in kD) is listed at left. The abnormality of PRDI-BF1(331–789) migration in lane 3 is probably caused by the comigrating GST/TLE2(N) protein, which has a similar molecular weight.
The Q domain is the most homologous region shared by all three PRDI-BF1 interacting proteins, and may therefore mediate TLE/PRDI-BF1 interactions. To test this possibility, truncations of hGrg and TLE1 containing only the Q domain were fused to the GST protein, and incubated with the in vitro-labeled PRDI-BF1 proteins (Fig. 5A). Consistent with our prediction, these GST fusion proteins interacted with the PRDI-BF1 truncation that retains the repression domain, that is, PRDI-BF1(331–789), but not with the one that lacks the repression domain, that is, PRDI-BF1(398–789) (Fig. 5C).
Transcription repression by DNA-bound hGrg and TLE1 proteins
Many members of the Groucho family can function as corepressors. For example, genetic experiments showed that the Groucho protein is required for Hairy-mediated repression during Drosophila embryonic segmentation (Paroush et al. 1994). The Groucho protein can interact directly with a conserved WRPW motif in the Hairy protein, and this protein–protein interaction is important for the function of Hairy as a repressor in vivo (Jimenez et al. 1997). Groucho and TLE proteins do not have a recognizable DNA-binding domain, but they can repress transcription directly if tethered to DNA through a Gal4 DNA-binding domain, and the region in Groucho that is sufficient to mediate repression has been localized to its amino-terminal 269 amino acids (Fisher et al. 1996). However, it is not known whether the short form of Groucho proteins, that is, hGrg and its murine homolog mGrg, can function as transcriptional corepressors.
To test this possibility, and to further delineate the region in TLE1 that is necessary for repression, full-length hGrg, full-length TLE1, and truncations of hGrg and TLE1 were fused to the Gal4 DNA-binding domain (Fig. 6A). When these constructs were overexpressed in HeLa cells along with the reporter G5BLCAT2, strong inhibition was observed with the full-length hGrg fusion construct and all the TLE1 fusion constructs (Fig. 6B). Thus, the full-length hGrg can function as a potent repressor when tethered to DNA, and the amino-terminal 135 amino acids, i.e., the Q domain, are sufficient for TLE1-mediated repression. These observations are consistent with the hypothesis that the Groucho family proteins are part of a corepressor complex, that can mediate transcriptional repression when tethered to the promoter. The fact that the Q domain also functions to repress transcription as a Gal4 fusion protein suggests that this domain can dimerize with the endogenous Groucho family proteins and recruit a corepressor complex to the promoter.
Figure 6.
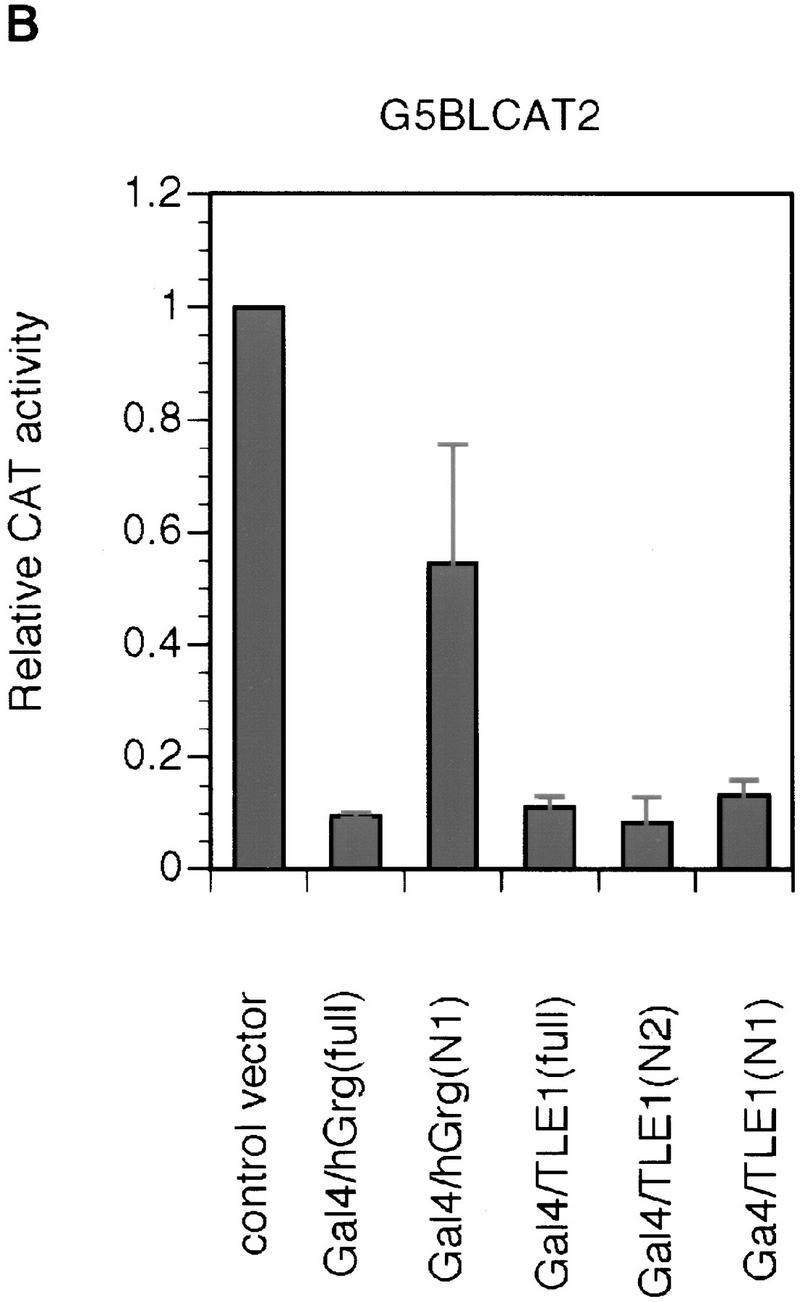
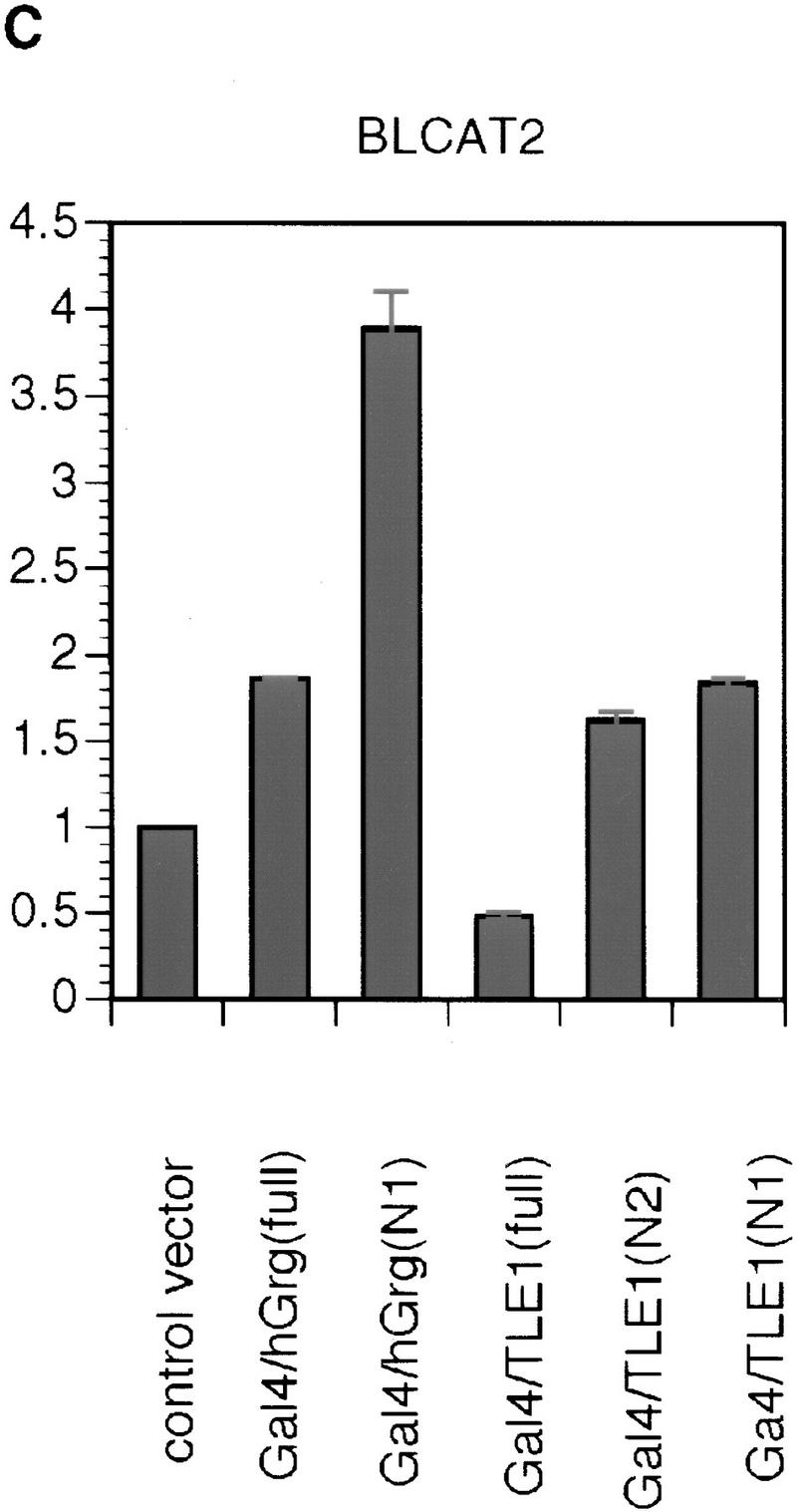
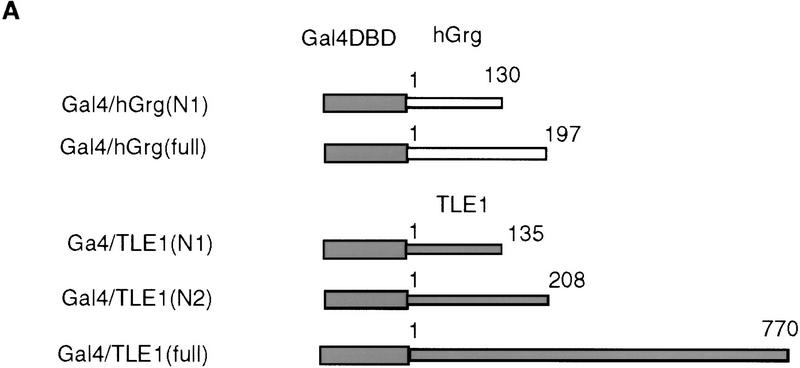
Repression by hGrg and TLE1 proteins fused to the Gal4 DNA-binding domain. (A) Diagram of the Gal4/hGrg and Gal4/TLE1 fusion constructs. (B,C) The full-length hGrg and the amino-terminal Q domains of TLE1 are sufficient for transcriptional repression. HeLa cell transfections similar to those in Fig. 2A were carried out with 0.1 μg of a control vector (pECE) or the indicated Gal4 fusion expression vector, along with a reporter G5BLCAT2 (B) or BLCAT2 (C). Relative CAT activities of the transfections are compared in the histogram.
Given the extensive homology shared by hGrg and TLE1 in their Q domain (see Fig. 4B), it is surprising to find that the hGrg truncation containing amino acids 1–130 does not repress transcription to the same extent as the TLE1 amino-terminal truncation when fused to the Gal4 DNA-binding domain. This might be caused by the 4-amino-acid region at the carboxyl terminus of the Q domain, which is missing in the hGrg truncation but present in the TLE1 truncation. Alternatively, this phenomenon could be explained by a nonspecific activation effect of this particular Gal4–hGrg fusion protein, as it also appeared to activate the reporter that does not have the Gal4-binding sites (Fig. 6C).
The Groucho family proteins dimerize through the amino-terminal Q domain
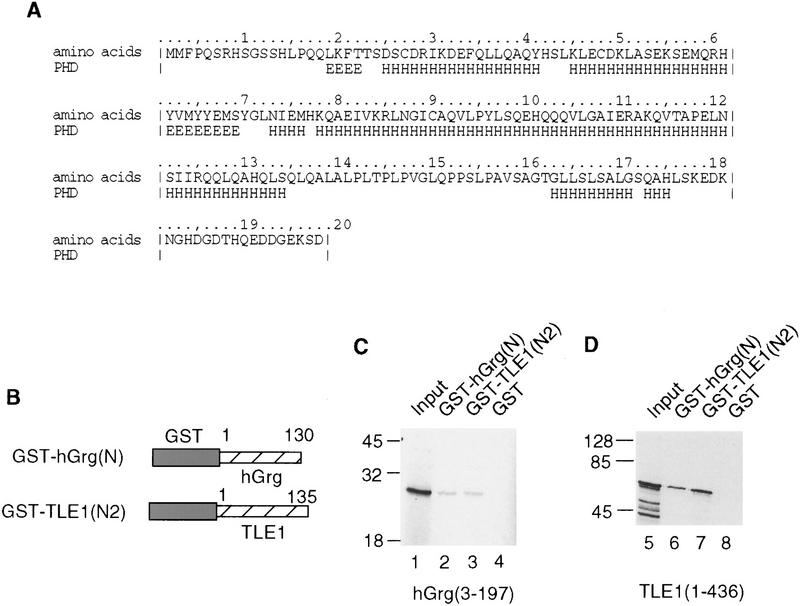
We have performed a secondary-structure prediction analysis of the entire hGrg protein using the PHD server, which predicts protein secondary structure based on information about evolutionary conservation derived from multiple sequence alignments (Rost et al. 1994). We found that the Q domain is predicted to have two α helical structures, one located between amino acids 25 and 69, and the other from amino acid 73 to 135 (Fig. 7A). The first α helix is likely to be involved in the formation of a coiled–coil structure, as predicted by the PairCoil program, which uses an algorithm based on pairwise residue correlation (Berger et al. 1995). The sequence from amino acids 30–59 scored a probability of 0.628, with the cutoff for scoring a coiled coil being 0.50. Further analysis with the MultiCoil program shows that this region is more likely to form three-stranded coils than two-stranded coils (Wolf et al. 1997). Thus, the hGrg protein very likely forms oligomers through its Q domain.
Figure 7.
The Q domains of hGrg and TLE1 are sufficient for dimerization. (A) A secondary structure prediction of the entire hGrg sequence using the PHD server. Below the amino acid sequence is the PHD secondary prediction for each residue. (E) Residues likely involved in extended structure (strand); (H) residues involved in forming helix structure; (blank spaces) residues likely to form loop structure or unpredictable. (B) Diagram of the GST–hGrg(N) and GST–TLE1(N2) fusion constructs used in C and D. (C,D) The Q domains of hGrg and TLE1 are sufficient to mediate homo- and heterodimerization. hGrg protein (lanes 1–4) or TLE1 truncations (lanes 5–8) were translated and radiolabeled in vitro, and incubated with immobilized GST–hGrg(N), GST–TLE1(N2) or GST protein, as indicated at the top. After the beads were washed, one-fifth of the input (top) and bound proteins were analyzed on a 10% SDS–polyacrylamide gel and visualized by autoradiography. The protein size marker (in kD) is listed at left.
Consistent with this possibility, the Q domain of the mouse Grg proteins was shown previously to mediate the formation of homo- and heterodimers (Pinto and Lobe 1996). However, neither the first nor the second α helix alone was sufficient for dimerization, and the minimal region for optimal dimerization includes amino acids 1–162 (Pinto and Lobe 1996). Based on this result, and the results of the secondary structure prediction for hGrg, we speculate that the two α helices cooperate with each other to mediate optimal oligomerization, and thus the carboxy-terminal boundary of the minimal dimerization domain should be around amino acid 130.
To test this hypothesis, GST fusion proteins that contain the amino-terminal 130 amino acids of hGrg or the amino-terminal 135 amino acids of TLE1 were mixed with the in vitro-translated and radiolabeled full-length hGrg protein or the truncated TLE1 protein containing amino acids 1–436 (Fig. 7B). Both GST fusion proteins bound to either hGrg or TLE1 proteins, indicating that the Q domain is sufficient to promote homo- and heterodimerization of the Groucho family proteins (Fig 7C,D).
Truncations of hGrg and TLE1 protein relieve PRDI-BF1 repression
Previous studies suggested that Groucho and TLE proteins form large multiprotein complexes in cells (Palaparti et al. 1997). Although the exact composition of these complexes is unclear, there is evidence suggesting that they may contain multimeric forms of Groucho or TLEs (Palaparti et al. 1997). Because the amino-terminal Q domain in hGrg and TLE1 is the minimal region that is sufficient for dimerization between these proteins, truncations containing only this domain may exhibit dominant-negative effect in vivo, because they can dimerize with the endogenous hGrg or TLE proteins but form nonfunctional multimers because of lack of other domains in these hGrg or TLE1 truncations. We therefore made use of the hGrg and TLE1 truncations hGrg(1–130) and TLE1(1–135), which contain only the amino-terminal Q domain in each protein, to test their role in the PRDI-BF1 repression in vivo. HeLa cells were transfected with a CAT reporter plasmid that bears four PRF sites along with a control vector or a PRDI-BF1-expressing plasmid (Fig. 8A). PRDI-BF1 exhibited a 13-fold repression of the reporter, and this repression was reduced to only two- to threefold when the hGrg and TLE1 truncations were also included in the transfection. In contrast, cotransfection of the full-length hGrg- or TLE1-expressing plasmids did not affect the PRDI-BF1-dependent repression. The failure to observe an augmentation of PRDI-BF1 repression by overexpressing full-length hGrg or TLE1 is most likely caused by the high level of endogenous Groucho proteins in HeLa cells (Stifani et al. 1992). As a control, HeLa cells were also transfected with a CAT reporter without PRF sites along with a control vector or the same hGrg- or TLE1-expressing plasmids as above (Fig. 8B). The hGrg or TLE1 truncations did not affect the transcription of this reporter. The full-length hGrg and TLE1, for unknown reasons, activated the levels of reporter activity slightly. We conclude that the Q domains of hGrg and TLE1 function as dominant-negative inhibitors of the PRDI-BF1 dependent repression in HeLa cells. These observations provide additional evidence that Groucho proteins are required for repression by PRDI-BF1.
Figure 8.
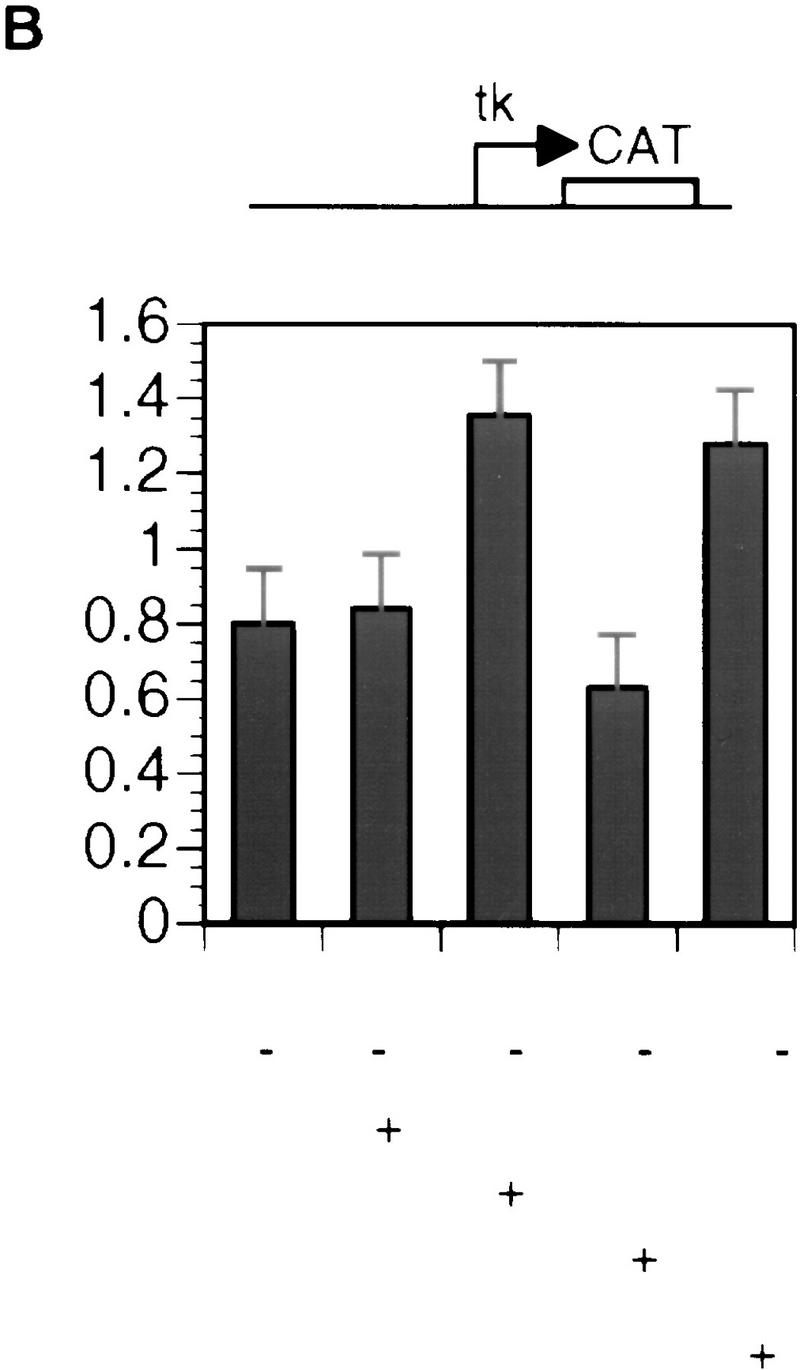
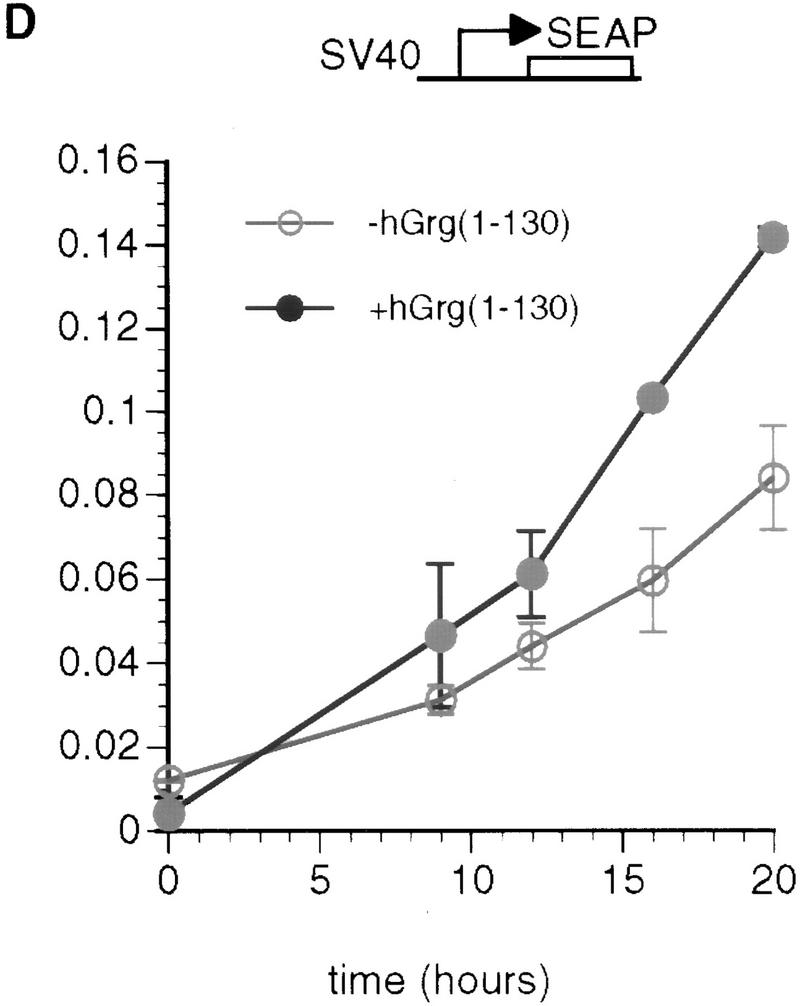
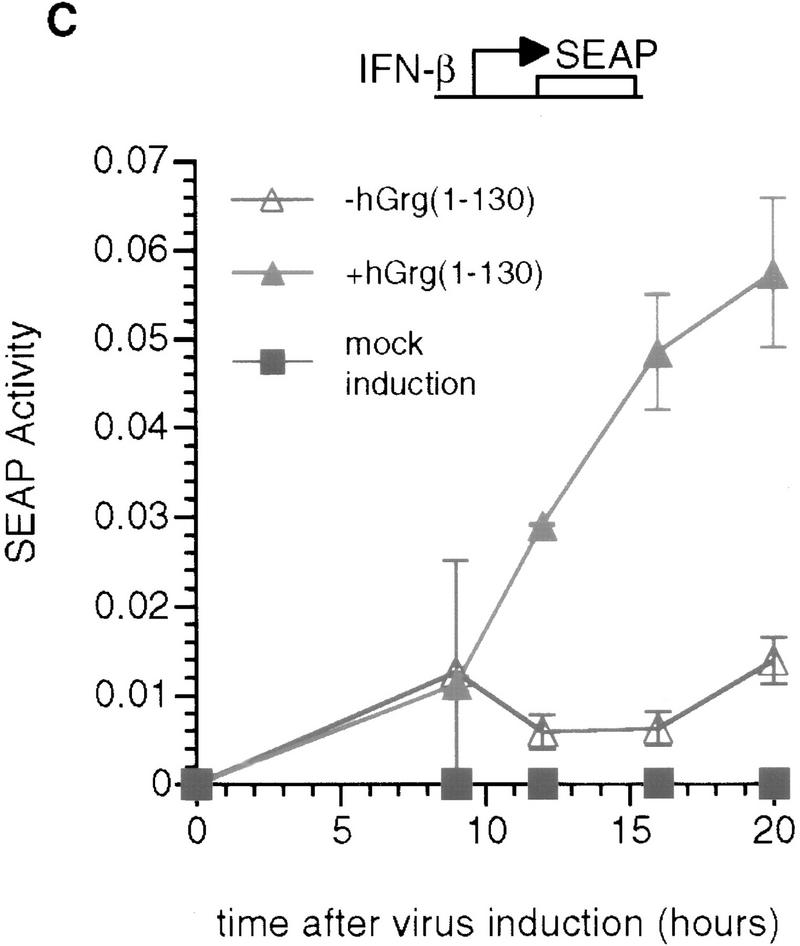
The Q domains of hGrg and TLE1 act as dominant-negative inhibitors of PRDI-BF1 repression. (A,B) Histograms showing CAT activities produced in HeLa cells transfected with a the reporter gene and various hGrg and TLE constructs. One microgram of a control vector (pcDNA3) or a PRDI-BF1 full-length expression vector was cotransfected as indicated by − or +, respectively. One microgram of the expression plasmid containing the Q domain of hGrg [hGrg (1–130)], the full-length hGrg, the Q domain of TLE1 [TLE1(1–135)], or the full-length TLE1 was cotransfected as indicated at the bottom of the graph. (A) Reporter gene containing four tandem PRF sites upstream of the tk promoter driving the CAT gene. (B) A control reporter containing only the tk promoter driving the CAT gene. (C,D) A dominant-negative inhibitor hGrg(1–130) relieves the postinduction repression of the IFN-β promoter. HeLa cells were transfected with 5 μg of a reporter plasmid, indicated at the top, cotransfected with 2 μg of a mammalian expression vector containing [+hGrg(1–130); ▵] or lacking [−hGrg(1–130); ▴] the hGrg(1–130) gene. Twenty-four hours after transfection, the cells were either infected with Sendai virus or untreated (mock induction, █), and SEAP activities in the culture medium were measured at various times following virus infection. (C) Reporter containing the −110 IFN-β promoter driving the expression of the SEAP gene. (D) Reporter containing the herpes virus SV40 promoter driving the SEAP gene (Clontech) (○) −hGrg. The cells were untreated, and SEAP activities were measured at the corresponding time points to those in C.
Dominant-negative hGrg mutants relieve the postinduction repression of the IFN-β promoter
Previous studies showed that the human IFN-β promoter is induced transiently by virus. Its mRNA level peaks at ∼6–12 hr after virus induction, then rapidly decreases (Whittemore and Maniatis 1991a,b). The PRDI-BF1 protein has been implicated as a postinduction repressor of the IFN-β gene because of its DNA-binding properties and the fact that its virus-induced accumulation in cells correlates with the shutoff of the IFN-β transcription after virus induction (Keller and Maniatis 1991). To test whether the PRDI-BF1 interactions with Groucho family proteins play a role in the repression of the IFN-β promoter after virus induction, we performed transient transfection experiments. We constructed a reporter gene in which the intact IFN-β promoter drives the secreted alkaline phosphotase (SEAP) gene, then cotransfected this reporter with a mammalian expression vector (pcDNA3) containing the dominant-negative hGrg mutant, hGrg(1–130). As a control, the SEAP reporter gene was cotransfected with the pcDNA3 vector. The cells were infected by sendai virus 24 hr after transfection, and the SEAP activity present in the cell culture medium was measured at various time after infection. Because SEAP protein is quite stable in the medium, the rate of the accumulation of SEAP activity corresponds to the promoter activity, assuming that the SEAP mRNA is degraded rapidly.
As shown in Figure 8C, the reporter SEAP expression was highly induced by virus, but was not detectable in the mock induction. In the absence of hGrg(1-130), the SEAP activity peaked at ∼9 hr after addition of virus, then was maintained at a constant level, consistent with the fact that the IFN-β promoter was shut off by this time. However, in the presence of hGrg(1–130), the SEAP activity did not plateau 9 hr after induction, but continued to increase over the next several hours. The rate of SEAP accumulation did not drop until 20 hr after virus induction. Thus, hGrg(1–130) dramatically delayed or eliminated the postinduction shutoff of the IFN-β promoter. As a control, hGrg(1–130) was cotransfected with a reporter containing the SV40 promoter driving expression of the SEAP gene. Only slight activation of this promoter by hGrg(1–130) was observed (Fig. 8D). These results suggest that PRDI-BF1 and its association with Groucho family corepressors play an important role in the shutoff of the IFN-β transcription after virus induction.
Discussion
PRDI-BF1 represses transcription by recruiting Groucho family proteins
Transcriptional repressors utilize a variety of mechanisms to inhibit transcription and thereby regulate cellular growth. In this study, we provide a number of lines of evidence that the PRDI-BF1 repressor functions by recruiting Groucho family corepressor proteins to DNA. Our results indicate that PRDI-BF1 is a long-range repressor, effectively mediating repression when its binding site is >1 kb upstream from the promoter. We have mapped the repression domain of PRDI-BF1 to a minimal 58-amino-acid region that is sufficient to repress transcription when fused to a Gal4 DNA-binding domain. We also demonstrated that PRDI-BF1 interacts through its repression domain specifically with proteins of the Groucho family, in particular hGrg, TLE1, and TLE2. These Groucho family proteins were shown to function as repressors when fused to the Gal4 DNA-binding domain, and the amino-terminal dimerization domains of hGrg and TLE1 act as dominant-negative inhibitors of the PRDI-BF1 repression in HeLa cells. Taken together, these observations provide strong evidence for the corepressor recruitment model of PRDI-BF1 repression.
One of the target genes for the PRDI-BF1 protein is the human IFN-β promoter. PRDI-BF1 is thought to inhibit the transcription of the promoter after virus induction, thereby contributing to the transient expression profile of the IFN-β (Keller and Maniatis 1991). Although previous studies were consistent with the conclusion that PRDI-BF1 acts by a steric interference mechanism (Keller and Maniatis 1991), we show here that PRDI-BF1 repression not only depends on the binding of PRDI-BF1 to the PRDI site of the IFN-β promoter, it also requires a repression domain in the amino terminus of PRDI-BF1, which recruits a corepressor complex containing the Groucho family proteins. This repression domain, interestingly, was discovered independently by Messika et al. (1998) to be required for the ability of PRDI-BF1/Blimp-1 to induce apoptosis in immature B cells. We also find that overexpression of the PRDI-BF1 protein in HeLa cells is sufficient to cause cell death, and this phenomenon depends on the repression domain of PRDI-BF1 (B. Ren, K. Chee, and T. Maniatis, unpubl.). Thus, PRDI-BF1/Blimp-1 appears to repress transcription of genes whose expression protects cells from undergoing apoptosis directly or indirectly.
It is highly possible that a corepressor-recruiting mechanism is used by PRDI-BF1/Blimp-1 protein at its other target genes, for example, the mouse c-myc promoter. In this case, the Blimp-1 binding site is located 290 bp upstream of the P1 promoter of the c-myc gene. Overexpression of Blimp-1 in a B-cell line causes repression of the c-myc gene transcription (Lin et al. 1997). Based on the high homology between PRDI-BF1 and Blimp-1 proteins, and the observation that PRDI-BF1 represses transcription regardless of the distance between its binding site and the promoter, we propose that Blimp-1 represses the c-myc gene by recruiting the Groucho family corepressors. Work is in progress to determine whether the dominant-negative inhibitor hGrg(1–130) can relieve the repression of c-myc gene transcription by Blimp-1.
Mechanisms of Groucho family protein function
At present, relatively little is known about the mechanisms by which Groucho family proteins function as eukaryotic corepressors (Fisher and Caudy 1998). Repressors that functionally depend on Groucho proteins include the products of genes that participate in a number of regulatory pathways. They include the Drosophila Hairy-related proteins, Runt domain proteins, Engrailed, Dorsal, and their mammalian homologs. These repressors recruit the Groucho proteins through specific interactions between their repression domains and various regions of Groucho proteins.
The structural resemblance of Groucho and TLE proteins to Tup1, a general transcription repressor in yeast, suggests that the two proteins may function by similar mechanisms (Keleher et al. 1992; Johnson 1995). Both Groucho family proteins and Tup1 lack DNA-binding domains and must therefore be recruited to DNA through interactions with transcriptional regulators. Tup1 and Groucho proteins share a conserved amino-terminal glutamine-rich dimerization domain, a serine–proline-rich central region, and carboxy-terminal tandem WD-40 repeats (Williams and Trumbly 1990; Stifani et al. 1992). The Tup1 protein, together with the Ssn6 protein, constitute a multimeric corepressor complex that is recruited to many different promoters by specific DNA-binding regulatory proteins (Keleher et al. 1992; Komachi et al. 1994; Treitel and Carlson 1995; Tzamarias and Struhl 1995; Varanasi et al. 1996). Tup1/Ssn6 complex mediates transcriptional repression through at least two mechanisms. The findings that mutations in Tup1 result in derepression and perturbation of nucleosome positioning on DNA, and that the repression domain of Tup1 directly interacts with the amino-terminal tails of histones H3 and H4, suggest that the Tup1/Ssn6 complex interacts with nucleosomal histone proteins to modulate chromatin structure (Cooper et al. 1994; Edmondson et al. 1996). Significantly, the Tup1–histone interactions occur only with underacetylated histones (Edmondson et al. 1996). Histone acetylation reduces the net positive charge of the histones, presumably rendering them less effective in associating with negatively charged DNA to form an ordered chromatin structure (Grunstein 1997). The Tup1/Ssn6 complex also appears to function by interacting directly with components of the general transcription machinery (Herschbach et al. 1994; Redd et al. 1997). In support of this model, deletions or mutations in Srb10, Srb11, SIN4, and Rox3, which are all associated with the yeast RNA polymerase II holoenzyme, cause a partial loss of repression at Tup1-dependent promoters (Chen et al. 1993; Wahi and Johnson 1995; Kuchin and Carlson 1998).
Because of their structural and functional similarities to Tup1 as transcriptional repressors, the Groucho family proteins may mediate repression through similar mechanisms. It has been shown that TLE proteins are associated with chromatin in Jurkat cell extracts (Palaparti et al. 1997). In addition, TLE proteins can specifically interact with the amino-terminal tail of histone H3, the same region shown to be important for the interaction between yeast histone H3 and Tup1. Mutations of the amino-terminal tail of yeast histone H3 were shown to cause derepression of the a-type-specific genes in α mating-type cells, as well as depression of the haploid-specific genes in diploid cells (Edmondson et al. 1996; Huang et al. 1997). Thus, Groucho proteins may regulate transcription by promoting formation of a repressive chromatin configuration in the vicinity of their target sites. Alternatively, it is also possible that they contact components in the general transcription machinery directly and inhibit transcription.
Although previous studies have shown that Groucho family proteins can form oligomers (Pinto and Lobe 1996), here we provide the first evidence that the multimerization of Groucho family proteins is required for their in vivo functions. A dominant-negative inhibitor, the hGrg(1–130), retains the ability to dimerize with other Groucho family proteins. However, because it lacks other functional domains, it may form nonfunctional complexes with the endogenous Groucho family proteins, and thus disrupt the repression of IFN-β transcription by PRDI-BF1.
Recent studies have identified a number of corepressors that can function by histone deacetylation. These include Sin3/mSin3, N-CoR, SMRT, and Rb, each required for transcription repression by a different set of transcription factors (Weintraub et al. 1992, 1995; Ayer et al. 1995; Downes et al. 1996; Zamir et al. 1996). All of these proteins are associated in vivo with a protein complex possessing histone deacetylase activity (DePinho 1998; Wolffe 1997), which are believed to alter chromatin structure by removing the acetyl groups from amino termini of core histones (Grunstein 1997). It remains to be shown whether these histone–deacetylase-containing complexes represent a group of transcriptional corepressors that are distinct from the Groucho protein-containing complexes.
Groucho family proteins play important roles in development
The Drosophila Groucho gene was first identified as a component of the Notch signal transduction pathway, which is critical for cell-fate determination in both invertebrates and vertebrates (Hartley et al. 1988). Subsequently Groucho proteins were shown to function as corepressors for the Hairy family of bHLH transcription factors, including Hairy, Deadpan, and the Enhancer of Split (Esl), proteins that act in Drosophila embryonic segmentation, sex determination, and neurogenesis, respectively (Paroush et al. 1994; Jimenez et al. 1997). Groucho is also required for the Drosophila dorsal–ventral patterning, as it participates in the conversion of the transcription activator dorsal to a repressor at the ventral repression regions (VRRs) located in dorsal fate-determining genes, such as zerknullt (zen) and decapentaplegic (dpp) (Dubnicoff et al. 1997). In all of these cases, direct protein–protein interactions were detected between the Groucho protein and the repressors. Groucho binds to the WRPW motif that is shared by all the Hairy family proteins, and this interaction is mediated by the internal region of Groucho (Paroush et al. 1994; Fisher et al. 1996). Groucho’s interaction with dorsal protein requires the Rel homology domain. However, this interaction is not likely to be sufficient for Groucho recruitment in vivo, as the dorsal-mediated repression requires additional repression elements in the proximity of the dorsal-binding sites (Courey and Huang 1995; Dubnicoff et al. 1997).
The role of Groucho as a transcription corepressor appears to be highly conserved in mammals. The mammalian homologs of Groucho, including TLE1 and TLE2, have been shown to interact with the mammalian Hairy-related genes such as hairy-like enhancer of split-1 (HES-1) through the WRPW motif (Fisher et al. 1996; Grbavec and Stifani 1996). HES-1 and TLE genes are expressed in overlapping tissues during development, suggesting similar functions of the Groucho family proteins in Drosophila and in mammals (Dehni et al. 1995).
The short form of Groucho proteins, that is, Grg has thus far been found only in humans, mice, and rats. Expression of the mouse Grg protein is ubiquitous in embryos after gestation, and is widespread in adults (Mallo et al. 1993). The function of the mouse Grg protein was studied by gene knockout experiments (Mallo et al. 1995). Mice homozygous for a Grg null mutation are viable and appear normal upon birth, but later exhibit various degrees of growth deficiency. The cause of this growth retardation phenotype is not clear. However, it is also possible that phenotypic consequence of the Grg null mutation may be complemented by TLE proteins, which have widespread expression patterns in both embryos and adults (Stifani et al. 1992; Miyasaka et al. 1993; Dehni et al. 1995).
Materials and methods
Plasmid construction
To construct the reporter plasmid 4×PRF_BLCAT2, a double-stranded PRF oligonucleotide was synthesized (5′-CTAGCGTACAGAAAGGGAAAGGA-3′) and (5′-CTAGTCCTTTCCCTTTCTGTACG-3′), self-ligated, digested with NheI and SpeI (New England Biolabs), and the products were separated by gel electrophoresis. A 100-bp DNA fragment was cloned into the XbaI site of the vector BLCAT2. The reporter 4×PRF_λ1.1_BLCAT2 was constructed by inserting a 1.1-kb fragment of the BamHI/BglII digestion product of the λ phage DNA (Promega) into the BamHI site in the 4×PRF_BLCAT vector.
For the construction of most PRDI-BF1 mammalian expression vectors, DNA fragments were obtained by PCR amplification using appropriate primers, and inserted between the EcoRI and XbaI sites in pcDNA3 vector (Invitrogen) that had been modified by inserting a Flag tag sequence (5′GGTACCTAGCCGCCACATGGACTACAAGGACGACGATGACAAGGAATTC-3′) between the KpnI and EcoRI sites. To obtain the Gal4/PRDI-BF1, Gal4/hGrg, and Gal4/TLE1 fusion vectors, DNA fragments were made by PCR using appropriate primers and cloned into the EcoRI and XbaI sites in the pBXG vector. GST/hGrg, GST/TLE1, and GST/TLE2 constructs were made by inserting the corresponding DNA fragments into pGEX-5x-1 (Pharmacia) or pGEX-A (a gift from M. Tian, Children’s Hospital, Harvard Medical School, Cambridge, MA) vector. All constructs were confirmed by either sequencing and/or restriction digestion.
Yeast two-hybrid screen
The two-hybrid screen was performed essentially as outlined in the Clontech Laboratories protocol using the strain HF7c. The repression domain of PRDI-BF1, amino acids 331–429, was cloned into the EcoRI and SmaI sites in the DNA-binding domain vector pGBT9 (Clontech), and cDNA libraries (a gift from L. Gaudreau, Memorial Sloan-Kettering Cancer Institute, New York, NY) were built into the Gal4 activation domain vector pGAD10. Among 5 × 105 library plasmids screened, five interacting clones were identified that support growth on His−Leu−Trp−Glu medium, and show blue color on His−Leu−Trp−Glu plates coated with X-gal substrate. The plasmids that tested positive were reintroduced into yeast (HF7c) to test their interaction with two baits, one containing the repression domain and another lacking amino acids 331–397, on His−Leu−Trp−Glu medium. The five positive clones were analyzed by ABI sequencing and characterized using the BLASTN program on the NCBI database. All were identified as hGrg (GenBank accession no. U04241).
In vitro transcription/translation and binding
PRDI-BF1 proteins were synthesized and labeled in vitro using rabbit reticulocyte lysate and [35S]methionine, according to the instructions of the manufacturer (Promega). For the analysis of direct protein–protein interactions between Groucho family proteins and PRDI-BF1 in vitro, hGrg, TLE1, and TLE2 were subcloned into the pGEX-5X-1 vector (Pharmacia) to create fusion proteins of GST. These plasmids were transformed into the bacterial strain DH5α. Transformed bacteria were grown at 37°C to OD595 = 0.5, induced with 1 mm IPTG, and allowed to grow for an additional 2 hr. Cells were pelleted by centrifugation and resuspended in buffer A (10% glycerol, 20 mm HEPES at pH 7.9, 100 mm KCl, 1 mm DTT, 0.5 mm PMSF, 1% NP-40, 1 μg/ml leupeptin, 1 μg/ml pepstatin A). Cells were lysed by sonication, and the crude extract was centrifuged at 8000 for 30 min. Glutathione–agarose beads were swollen overnight in buffer A and washed three times before use. The washed beads were added to the bacterial lysate and incubated with rocking for 2 hr at 4°C. After incubation, the beads were washed five times with buffer A, and most of the supernatant removed after the last wash. GST–hGrg, GST–TLE1, and GST–TLE2 fusion proteins were analyzed on 10% SDS-PAGE. The 35S-labeled, PRDI-BF1 full-length and truncated proteins (5 μl of each) were incubated with equivalent amounts (2 μg) of the GST fusion proteins in 0.25 ml of +BSA buffer (50 mm KCl, 0.5% NP-40, 20 mm HEPES at pH 7.9, 0.1 mm DTT, 5% glycerol, 50 μm ZnSO4, 0.2 mm PMSF, 1% BSA) for 1 hr at 25°C, washed twice with +BSA buffer and three times with −BSA buffer, and analyzed on an 11% SDS-polyacrylamide gel and autoradiography.
Mammalian cell transfections
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM)/10% fetal bovine serum (FBS) and were seeded 1 day prior to transfection at a concentration of 1 × 105 cells/well. Cells were transfected by the calcium phosphate precipitation method with 0.125 m CaCl2 and 1× BBS (N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid-buffered solution). Typical transfections included 2 μg of pCMV–lacZ, 3 μg of reporter construct, 1 μg of effector, and 6 μg of the expression plasmid pXM, which includes the adenovirus major-late promoter, or 6 μg of a 1:2 mixture of pXM:sp72 plasmid vectors. At 48 hr post-transfection, the cells were harvested by scraping, and the extracts were prepared for CAT assays. The chloramphenicol products were resolved on TLC, and CAT activity was quantified. Transfection efficiencies were normalized using a cotransfected β-galactosidase plasmid. All transfections were performed in triplicate. For SEAP assays in Figure 8C, 100 μl of cell medium was collected at the indicated time points, and SEAP activity was determined according to Cullen and Malim (1992).
Protein secondary structure prediction
The PHD server can be found at http://www.embl-heidelberg.de/predictprotein/predictprotein.html. The PairCoil and MultiCoil servers can be found at http://www.wi.mit.edu/kim/computing.html.
Acknowledgments
We are grateful to Luc Gaudreau for providing the yeast two-hybrid library, and to Xun Zhao for help on the secondary structure prediction of hGrg. We thank Stefano Stifani for making TLE1 and TLE2 plasmids available. We also thank Ming Tian for providing the pGEX-A vector. Many thanks go to Bhavin Parekh, Tae Kook Kim, Niranjan Pandey, Neal Silverman, and other members of the Maniatis group for valuable advice and comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL maniatis@biohp.harvard.edu; FAX (617) 495-3537.
References
- Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, West RW, Jr, Johnson SL, Gans H, Kruger B, Ma J. TSF3, a global regulatory protein that silences transcription of yeast GAL genes, also mediates repression by alpha 2 repressor and is identical to SIN4. Mol Cell Biol. 1993;13:831–840. doi: 10.1128/mcb.13.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Roth SY, Simpson RT. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes & Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- Courey AJ, Huang JD. The establishment and interpretation of transcription factor gradients in the Drosophila embryo. Biochim Biophys Acta. 1995;1261:1–18. doi: 10.1016/0167-4781(94)00234-t. [DOI] [PubMed] [Google Scholar]
- Cullen BR, Malim MH. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 1992;216:362–368. doi: 10.1016/0076-6879(92)16033-g. [DOI] [PubMed] [Google Scholar]
- Dehni G, Liu Y, Husain J, Stifani S. TLE expression correlates with mouse embryonic segmentation, neurogenesis, and epithelial determination.Mech. Dev. 1995;53:369–381. doi: 10.1016/0925-4773(95)00452-1. [DOI] [PubMed] [Google Scholar]
- DeMaeyer E, DeMaeyer-Guignard J. Interferons and other regulatory cytokines. New York, NY: John Wiley and Sons; 1988. [Google Scholar]
- DePinho RA. Transcriptional repression. The cancer-chromatin connection. Nature. 1998;391:533–536. doi: 10.1038/35257. [DOI] [PubMed] [Google Scholar]
- Downes M, Burke LJ, Bailey PJ, Muscat GE. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with Rev-erbA alpha and RVR: Physical association is dependent on the E region of the orphan receptors. Nucleic Acids Res. 1996;24:4379–4386. doi: 10.1093/nar/24.22.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes & Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes & Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- Fisher A L, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Levine M. Transcriptional repression in development. Curr Opin Cell Biol. 1996;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- Grbavec D, Stifani S. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem Biophys Res Commun. 1996;223:701–705. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- Hartley DA, Preiss A, Artavanis-Tsakonas S. A deduced gene product from the Drosophila neurogenic locus, enhancer of split, shows homology to mammalian G-protein beta subunit. Cell. 1988;55:785–795. doi: 10.1016/0092-8674(88)90134-1. [DOI] [PubMed] [Google Scholar]
- Herschbach BM, Arnaud MB, Johnson AD. Transcriptional repression directed by the yeast alpha 2 protein in vitro. Nature. 1994;370:309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- Hoffman-Liebermann B, Liebermann DA. Suppression of c-myc and c-myb is tightly linked to terminal differentiation induced by IL6 or LIF and not growth inhibition in myeloid leukemia cells. Oncogene. 1991;6:903–909. [PubMed] [Google Scholar]
- Huang L, Zhang W, Roth SY. Amino termini of histones H3 and H4 are required for a1-alpha2 repression in yeast. Mol Cell Biol. 1997;17:6555–6562. doi: 10.1128/mcb.17.11.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes & Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- Kakkis E, Calame K. A plasmacytoma-specific factor binds the c-myc promoter region. Proc Natl Acad Sci. 1987;84:7031–7035. doi: 10.1073/pnas.84.20.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkis E, Riggs KJ, Gillespie W, Calame K. A transcriptional repressor of c-myc. Nature. 1989;339:718–721. doi: 10.1038/339718a0. [DOI] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Keller AD, Maniatis T. Identification and characterization of a novel repressor of β-interferon gene expression. Genes & Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- Komachi K, Redd MJ, Johnson AD. The WD repeats of Tup1 interact with the homeodomain protein α2. Genes & Dev. 1994;8:2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- Kuchin S, Carlson M. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol. 1998;18:1163–1171. doi: 10.1128/mcb.18.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B-cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- Madden SL, Cook DM, Morris JF, Gashler A, Sukhatme VP, Rauscher FJ., III Transcriptional repression mediated by the WT1 Wilms tumor gene product. Science. 1991;253:1550–1553. doi: 10.1126/science.1654597. [DOI] [PubMed] [Google Scholar]
- Mallo M, Franco del Amo F, Gridley T. Cloning and developmental expression of Grg, a mouse gene related to the Groucho transcript of the Drosophila Enhancer of split complex. Mech Dev. 1993;42:67–76. doi: 10.1016/0925-4773(93)90099-j. [DOI] [PubMed] [Google Scholar]
- Mallo M, Gendron-Maguire M, Harbison ML, Gridley T. Protein characterization and targeted disruption of Grg, a mouse gene related to the Groucho transcript of the Drosophila Enhancer of split complex. Dev Dyn. 1995;204:338–347. doi: 10.1002/aja.1002040311. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Whittemore L, Du W, Fan C, Keller AD, Palombella VJ, Thanos DN. Positive and negative control of human interferon-β gene expression. In: McKnight SL, Yamamoto KP, editors. Transcriptional regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 1193–1220. [Google Scholar]
- Melchers F. B-lymphocyte-lineage cells from early precursors to Ig-secreting plasma cells: Targets of regulation by the myc/mad/max families of genes? Curr Top Microbiol Immunol. 1997;224:19–30. doi: 10.1007/978-3-642-60801-8_2. [DOI] [PubMed] [Google Scholar]
- Messika EJ, Lu PS, Sung Y, Yao T, Chi J, Chien Y, Davis MM. Differential effect of B lymphocyte-induced maturation protein (Blimp-1) expression on cell fate during B cell development. J Exp Med. 1998;188:515–525. doi: 10.1084/jem.188.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- Miyasaka H, Choudhury BK, Hou EW, Li SS. Molecular cloning and expression of mouse and human cDNA encoding AES and ESG proteins with strong similarity to Drosophila enhancer of split Groucho protein. Eur J Biochem. 1993;216:343–352. doi: 10.1111/j.1432-1033.1993.tb18151.x. [DOI] [PubMed] [Google Scholar]
- Mock BA, Liu L, LePaslier D, Huang S. The B-lymphocyte maturation promoting transcription factor BLIMP1/PRDI-BF1 maps to D6S447 on human chromosome 6q21-q22.1 and the syntenic region of mouse chromosome 10. Genomics. 1996;37:24–28. doi: 10.1006/geno.1996.0516. [DOI] [PubMed] [Google Scholar]
- Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pinto M, Lobe CG. Products of the grg (Groucho-related gene) family can dimerize through the amino-terminal Q domain. J Biol Chem. 1996;271:33026–33031. doi: 10.1074/jbc.271.51.33026. [DOI] [PubMed] [Google Scholar]
- Potter M, Marcu KB. The c-myc story: Where we’ve been, where we seem to be going. Curr Top Microbiol Immunol. 1997;224:1–17. doi: 10.1007/978-3-642-60801-8_1. [DOI] [PubMed] [Google Scholar]
- Redd MJ, Arnaud MB, Johnson AD. A complex composed of tup1 and ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- Riggs KJ, Saleque S, Wong KK, Merrell KT, Lee JS, Shi Y, Calame K. Yin-yang 1 activates the c-myc promoter. Mol Cell Biol. 1993;13:7487–7495. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- Stifani S, Blaumueller CM, Redhead NJ, Hill RE, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- Treitel MA, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8–Tup1 corepressor complex to differentially regulated promoters. Genes & Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- Varanasi US, Klis M, Mikesell PB, Trumbly RJ. The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol. 1996;16:6707–6714. doi: 10.1128/mcb.16.12.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahi M, Johnson AD. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Qiu QQ, Gurrieri M, Huang J, Deuel TF. WT1, the Wilms’ tumor suppressor gene product, represses transcription through an interactive nuclear protein. Oncogene. 1995;10:1243–1247. [PubMed] [Google Scholar]
- Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinatedly activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Weintraub SJ, Chow KN, Luo RX, Zhang SH, He S, Dean DC. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- Weintraub SJ, Prater CA, Dean DC. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- Whittemore LA, Maniatis T. Postinduction repression of the beta-interferon gene is mediated through two positive regulatory domains. Proc Natl Acad Sci. 1990a;87:7799–7803. doi: 10.1073/pnas.87.20.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Postinduction turnoff of beta-interferon gene expression. Mol Cell Biol. 1990b;10:1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams FE, Trumbly RJ. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf E, Kim PS, Berger B. MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- Zamir I, Harding HP, Atkins GB, Horlein A, Glass CK, Rosenfeld MG, Lazar MA. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]