Abstract
The TGF-β superfamily of growth and differentiation factors, including TGF-β, Activins and bone morphogenetic proteins (BMPs) play critical roles in regulating the development of many organisms. These factors signal through a heteromeric complex of type I and II serine/threonine kinase receptors that phosphorylate members of the Smad family of transcription factors, thereby promoting their nuclear localization. Although components of TGF-β/Activin signaling pathways are well defined in vertebrates, no such pathway has been clearly defined in invertebrates. In this study we describe the role of Baboon (Babo), a type I Activin receptor previously called Atr-I, in Drosophila development and characterize aspects of the Babo intracellular signal-transduction pathway. Genetic analysis of babo loss-of-function mutants and ectopic activation studies indicate that Babo signaling plays a role in regulating cell proliferation. In mammalian cells, activated Babo specifically stimulates Smad2-dependent pathways to induce TGF-β/Activin-responsive promoters but not BMP-responsive elements. Furthermore, we identify a new Drosophila Smad, termed dSmad2, that is most closely related to vertebrate Smads 2 and 3. Activated Babo associates with dSmad2 but not Mad, phosphorylates the carboxy-terminal SSXS motif and induces heteromeric complex formation with Medea, the Drosophila Smad4 homolog. Our results define a novel Drosophila Activin/TGF-β pathway that is analogous to its vertebrate counterpart and show that this pathway functions to promote cellular growth with minimal effects on patterning.
Keywords: Activin, Baboon, dSmad2, optic proliferation centers, imaginal disc, TGF-β
Cytokines of the transforming growth factor-β (TGF-β) superfamily are potent regulators of cell growth, differentiation, and morphogenesis (for review, see Derynck and Feng 1997; Kingsley 1994). On the basis of sequence similarities, TGF-β molecules can be subdivided into TGF-βs, bone morphogenetic proteins (BMPs), and Activins. All members of the TGF-β superfamily studied to date appear to share common features in their downstream signaling mechanisms (for review, see Heldin et al. 1997). At the cell surface, these ligands associate with two classes of transmembrane serine/threonine kinases known as type I and type II receptors. The type II receptor possesses a constitutively active intracellular kinase and, in the case of Activins and TGF-βs, is the primary determinant of ligand recruitment. On binding ligand, the type II receptor associates with an appropriate type I partner whose intracellular domain serves as a substrate for type II-dependent phosphorylation (Wrana et al. 1994a). Phosphorylation of the type I receptor results in the activation of its kinase domain and in phosphorylation of downstream components such as the Smad transcriptional activators.
The genetic identification of a Smad protein required for BMP signaling in Drosophila (Raftery et al. 1995; Sekelsky et al. 1995) led to the isolation of related molecules in numerous other organisms (for review, see Heldin et al. 1997; Attisano and Wrana 1998). Smad family members can be grouped into subfamilies according to functional and phylogenetic relationships. The receptor-regulated class of Smads (R-Smads) are direct substrates of the type I receptor kinase and are phosphorylated on serine residues within a carboxy-terminal SSXS motif (Macías-Silva et al. 1996; Abdollah et al. 1997; Kretzschmar et al. 1997; Liu et al. 1997b). Activation of R-Smads is pathway specific. Thus, Smad1, Smad5, and Smad8 are preferentially targeted by type I BMP receptors (Hoodless et al. 1996; Kretzschmar et al. 1997), whereas TGF-β and Activin regulate Smad2 and Smad3 (Macías-Silva et al. 1996; Liu et al. 1997b). On phosphorylation, the R-Smads can associate with the common-mediator class of Smads represented by Smad4. After nuclear translocation of the complex (Lagna et al. 1996; Liu et al. 1997a; Zhang et al. 1997), some Smads can bind directly to DNA (Kim et al. 1997). Tissue-specific DNA binding partners, such as FAST1 or FAST2 (Chen et al. 1996; Labbé et al. 1998), are likely to help recruit the complexes to specific promoters. This recruitment facilitates the low-affinity interaction of Smads with adjacent DNA elements, which in turn enhances transcriptional responses to TGF-β (Labbé et al. 1998), perhaps by recruiting CBP/p300 to the promoter (Feng et al. 1998; Janknecht et al. 1998). A third, more distant subfamily of Smad proteins, the inhibitory Smads, function to antagonize the activity of TGF-β superfamily proteins (Hayashi et al. 1997; Tsuneizumi et al. 1997).
In Drosophila, representative molecules from all the different classes of TGF-β-like signaling components have been described. At the ligand level, three BMP-type factors, Decapentaplegic (Dpp), Screw (Scw), and Glass bottom boat (Gbb also known as 60A), have been identified and characterized by loss-of-function mutations (Padgett et al. 1987; Arora et al. 1994; Chen et al. 1998; Khalsa et al. 1998). Dpp is a functional ortholog of vertebrate BMPs 2 and 4 (Padgett et al. 1993) and controls many aspects of Drosophila development including the specification of dorsal tissue in the early blastoderm embryo, induction of dorsal mesoderm at germ-band extension, midgut differentiation during late embryogenesis, regulation of imaginal disc cell fate and proliferation, and regulation of follicular cell fate specification during oogenesis (Gelbart 1989; Twombly et al. 1996). The Scw and Gbb-60A ligands are more closely related to the BMP-5/BMP-6/BMP-7 subfamily, and in certain tissues, both appear to act by augmenting Dpp activity (Arora et al. 1994; Chen et al. 1998; Haerry et al. 1998; Khalsa et al. 1998). All three factors appear to share a common set of receptors that include the type II receptor Punt, and the type I receptors Thickveins (Tkv) and Saxophone (Sax) (Haerry et al. 1998 and references therein). At the intracellular level, loss-of-function mutations in Mad and the Drosophila Smad4/DPC4 homolog, Medea, suggest that both these downstream factors transduce BMP-type signals (Raftery et al. 1995; Sekelsky et al. 1995; Das et al. 1998; Hudson et al. 1998; Wisotzkey et al. 1998). In addition, an inhibitory Smad known as Dad has also been identified that appears to antagonize the Dpp signal during imaginal disc growth (Tsuneizumi et al. 1997).
Although these examples provide compelling proof for the existence of invertebrate BMP-like signaling pathways, the evidence for invertebrate TGF-β or Activin-like signaling pathways has been scant. In this paper, we present a genetic and biochemical characterization of the Activin type I receptor Babo (Atr-I). Null mutations and germ-line clonal analysis demonstrate that baboon (babo) is not required during embryogenesis but is essential for proper pupation and adult viablity. Loss of babo function results in late larval or early pupal lethality. The major defect in these mutants is a reduction of cell proliferation within the primordia for adult structures. Clonal analysis of babo mutants as well as overexpression of constitutively activated Babo receptors in imaginal discs further suggest that Babo primarily regulates cell proliferation and has only minimal affects on patterning. Mutations in babo do not affect the expression patterns of Dpp target genes in embryos or imaginal discs, implying that the Babo pathway is distinct from Drosophila BMP signaling. Consistent with this view, we find that activated Babo can signal to vertebrate TGF-β/Activin, but not to BMP-responsive promoters in cell culture. Activated Babo cannot bind to or interact with Drosophila Mad in tissue culture but can utilize a new Drosophila Smad homolog, dSmad2, which is most related to the vertebrate Smads 2 and 3. Drosophila dSmad2 is highly expressed in tissues that require babo function and can be phosphorylated by either overexpression of activated Babo or by overexpression of wild-type Punt and Babo together. On the basis of these results, we propose that an Activin-like signaling pathway exists in Drosophila, which is required for proper cell proliferation in many primordial adult tissues. The identification and characterization of this pathway in Drosophila may provide additional insights into conserved features of Activin-like signaling pathways in vertebrates.
Results
babo genomic structure and identification of mutants
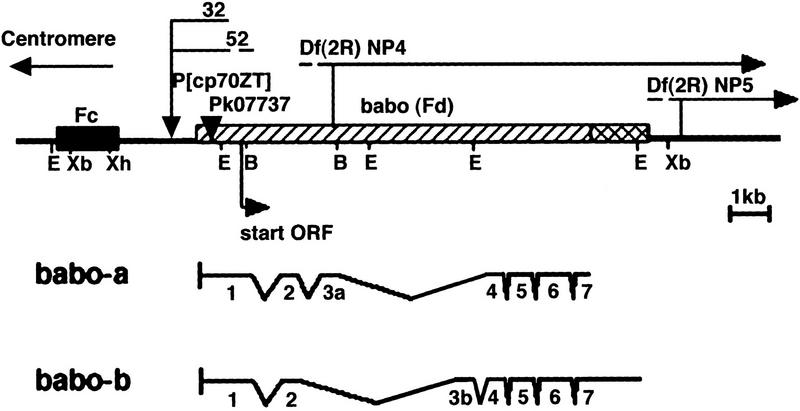
The babo locus maps to the 44F–45A region on the right arm of chromosome two and encodes two transcripts that differ in the sequence of their extracellular domains (Wrana et al. 1994b). The boundaries of these transcripts and their exon–intron structures were determined by comparison of different cDNA clones to genomic DNA by PCR, sequencing, and Southern blot analysis. Both babo transcripts are encoded within a 9-kb genomic fragment and contain seven exons with exon 3a/b being differentially spliced to produce the two isoforms (Fig. 1).
Figure 1.
Schematic representation of the babo genomic region. The region is displayed with the centromere at left. The Fc and Fd loci are indicated by boxed areas. The babob transcript contains an alternatively spliced exon 3 and an alternative polyadenylation site producing a longer transcript. (▾) The two P-element insertions. The uncertainties in the endpoints of deficiencies Np4 and Np5 as well as babo32 and babo52 are indicated by broken lines. The start site of translation is indicated by an arrow below the map. (E) EcoRI; (B) BalI; (Xb) XbaI; (Xh) XhoI.
For initial genetic characterization of the babo locus, we obtained all available mutations, deficiencies, and P-element transposons that mapped to this region. Using RFLP analysis, we found that two P elements, P[lac w+]k07737 and P[cp70ZT] were inserted near the 5′ end of the babo transcript. The P[lac w+]k07737 line is homozygous lethal, whereas the P[cp70ZT] line is homozygous viable. The exact insertion point for P[lac w+]k07737 was determined, by plasmid rescue and DNA sequencing, to be in the 5′-untranslated region of the babo transcript at base 25. Southern blot analysis of P[cp70ZT] demonstrated that it is inserted ∼300 bp upstream of the babo transcription start site.
To demonstrate that the insertion of P[lac w+]k07737 was responsible for the lethality of these flies, excisions were induced by hybrid dysgenesis. Seventy percent (n = 25) of the w− excision events resulted in a reversion to viability over the original P[lac w+]k07737. This demonstrates that the insertion of P[lac w+]k07737 into the 5′-untranslated region of babo is responsible for the lethality. Complementation testing showed that P[lac w+]k07737 is allelic to the Fd complementation group of Konev et al. (1994). Other babo alleles produced by the mobilization of the P[cp70ZT] were identified by testing P-element lines for RFLPs in the babo region. Three of these lines, babo52, babo50, and babo32, displayed polymorphisms and failed to complement P[lac w+]k07737 and Fd mutants. One line, babo50, did not complement either Fd or Fc alleles and was found to be a small deletion that removed portions of both the Fd and Fc transcription units. Both the babo52 and babo32 mutations were the result of imprecise excision events that produced deletions eliminating the translational start site of babo (Fig. 1). Specifically, babo32 deletion removed a portion of P[cp70ZT] and the first 850 bp of the babo transcript, which includes the translational start codon and the first 53 amino acids. Southern blot analysis indicated that babo52 contained a similar deletion. Therefore, babo32 and babo52 likely represent null alleles. In addition, we generated transgenic fly lines carrying a genomic rescue construct, P[Gen babo], which contained only the babo transcription unit and ∼2.5 kb of upstream and 300 bp of downstream DNA. The P[Gen babo] transgene fully rescued various heterozygous combinations of Fd alleles and homozygotes of P[lac w+]k07737, babo52, and babo32.
The babo loss-of-function phenotype suggests a role for Babo in cell proliferation
The lethal phase of babo loss-of-function mutants was determined by use of embryos derived from either the null babo32/InGl, Bc, or the hypomorphic P[lac w+]k07737/InGl, Bc stocks. After the second instar stage, babo homozygotes could be distinguished by the absence of pigmented granules caused by the dominant Bc marker. No significant embryonic lethality was found for either line. The homozygous mutant larvae (Fig. 2B) from both lines, however, developed more slowly than balancer-containing larvae (Fig. 2A), were smaller in length, and exhibited enlarged anal pads. The penetrance for all these phenotypes approaches 100%. A significant fraction of the P[lac w+]k07737 homozygotes pupate and survive to pharate adults (Table 1). On dissection, they were found to have normal thoracic cuticle and adult appendages, but portions of the abdominal cuticle were missing, suggesting a possible defect in the proliferation of the abdominal histoblasts (Fig. 2C). In contrast, very few of the babo32 or babo52 homozygotes progressed to the pharate stage, implying that the P[lac w+]k07737 allele is likely to be hypomorphic. All babo alleles exhibited two additional defects: One is the failure to leave food and the second is a failure to evert the anterior spiracles upon pupation (data not shown). Because babo is maternally expressed (Wrana et al. 1994b), we also generated germ-line clones with both the null and hypomorphic alleles. Maternally depleted animals derived from both alleles showed a lethal phase similar to the zygotic mutants.
Figure 2.
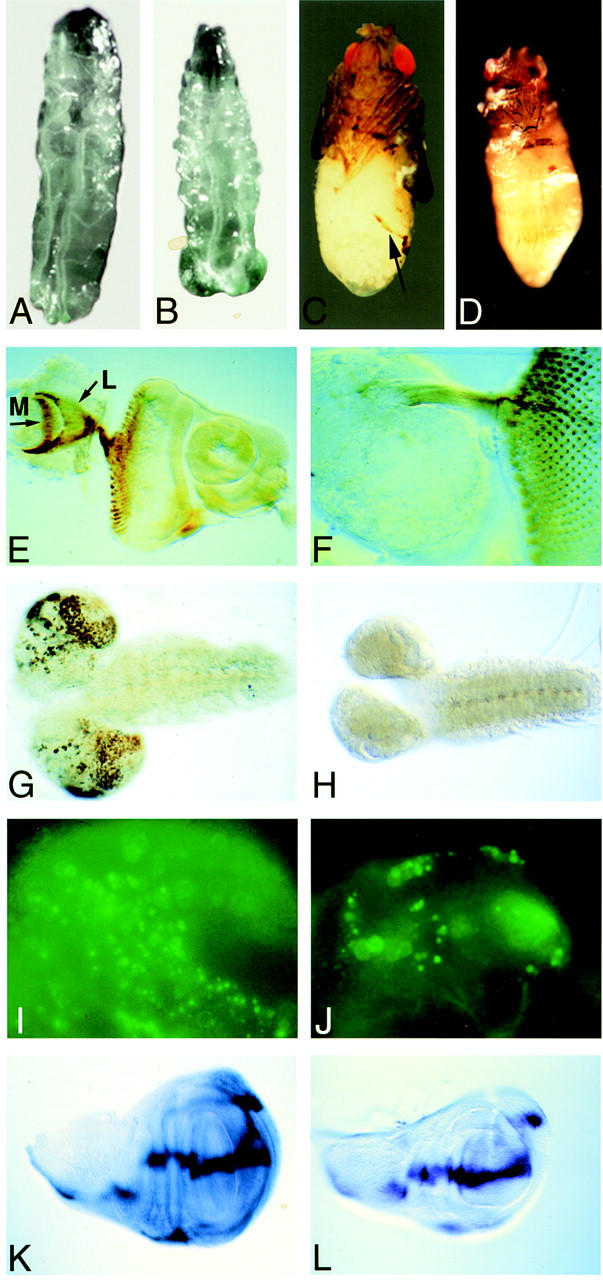
babo zygotic loss-of-function phenotpyes. (A) Wild-type early third instar larvae. (B) babo32 null mutant early third instar larvae exhibit large anal pads and a short body. (C) babok07737 pharate adult. (Arrow) The edge of the incomplete abdominal cuticle. (D) Adult structures of a babo52/babo32 mutant derived from germ-line clones are reduced in size. (E) Wild-type eye–antennal disc and portion of a brain lobe probed with mAb 24B10, which stains the R1–8 photoreceptor axons. The R7 and R8 axons reach targets in the medulla, whereas R1–6 project into the lamella (arrows). (F) babo32 null mutant brain and eye antennal disc stained with mAb 24B10. Note that the axons project to the brain but do not find their proper targets. (G) BrdU labeling of a wild-type brain and ventral nerve cord. Note heavy labeling of cells in the optic proliferation centers. (H) BrdU incorporation into a babo32 mutant brain and ventral nerve cord. Note the small size of the brain hemispheres and the lack of actively dividing cells compared to wild type. (I) Acridine orange incorporation into a portion of a wild-type brain lobe marks dying cells. (J) babo32 mutant brain lobe stained with acridine orange shows no increase in cell death. (K) dpp in situ hybridization to a wild-type late third instar wing disc. (L) dpp staining is unaltered in a late third instar babo32 null mutant wing disc. Note the smaller size of the mutant disc.
Table 1.
Survival rates
|
|
babok07737(%)
|
babo32 (%)
|
Wild type (%)
|
|---|---|---|---|
| Embryo | 90–100 | 90–100 | 90–100 |
| Second | 86 | 80 | 90 |
| Third | 80 | 50 | 86 |
| Early pupa | 70 | 35 | 83 |
| Pharate | 10–30 | <1 | 80 |
| Adult | 0 | 0 | 75 |
Lethality staging of wild-type, babok07737 and babo32 mutants at 25°C. Both babok07737 and babo32 mutants were found to grow slowly, taking an average of 2–3 days longer than wild type to pupate. We present the average from three experiments using 200 embryos each. Percent survival was determined by dividing the number of animals counted at each stage, by the number of fertilized embryos.
To investigate the cause of the lethality, we examined the internal organs from mutant third instar larvae. No significant abnormalites were noted in the morphology of the gut, fat body, or malphigian tubules. However, we did observe a severe reduction in the size of the brain hemispheres as well as a partial reduction in the size of the imaginal discs (Fig. 2). The small discs give rise to small adult structures in the dead pharates (Fig. 2D). Because the brain lobes appeared to be small, we examined the development of the visual center in babo mutants. As the eye disc matures, axons from photoreceptor cells posterior to the morphogenetic furrow fasciculate and send projections into the brain in a specific pattern (Fig. 2E). The axon bundles of photoreceptors one through six extend into the lamina, whereas those of photoreceptors seven and eight extend deeper into the medulla (Bausenwein et al. 1992). In babo mutants, photoreceptor differentiation and axon bundling proceed normally, but the axons fail to extend into the brain (Fig. 2, cf. E and F). This phenotype is consistent with a number of explanations, the simplest of which is that the medulla and lamina targets in the optic lobe fail to form.
The small size of the brains and discs might result either from a decrease in cell proliferation or an increase in cell death. To test for a reduction in cell division, larval tissue was labeled with BrdU, which is incorporated into S-phase cells. Wild-type brains displayed a characteristic mitotic replication pattern with high levels of cell division in the optic proliferation center, whereas the mutant brains showed little or no DNA replication during the time interval of BrdU incorporation (Fig. 2G,H). No striking difference in BrdU incorporation into imaginal discs was observed consistent with the observation that the discs are only ∼30% smaller. This slight reduction in size could be due to a more subtle change in cell proliferation or cell death. To test whether cell death might also be contributing to the reduced size of the mutant brains and discs, dying cells were stained with acridine orange. No increase in the number of dying cells was observed in either mutant brain (Fig. 2I,J) or discs (not shown). We also analyzed whether the observed reduction in cell proliferation is the result of secondary changes in the expression levels or patterns of several important growth factors. When we examined the expression of dpp, wg, and hh in brains and discs by in situ hybridization, no significant alterations in expression levels or patterns of these genes were observed (Fig. 2K,L; data not shown). Taken together, these results suggest that loss of babo strongly reduces cell proliferation within the brain, whereas imaginal discs cell proliferation is affected in a more subtle way. This is accomplished without an increased cell death or noticeable alterations in the expression of several patterning genes within these tissues.
The babo loss-of-function phenotype is distinct from that exhibited by mutations in components of Drosophila BMP signaling pathways
Reduced size of imaginal discs and brain tissue is not unique to babo mutations. It is also a characteristic feature of mutations in Drosophila BMP signaling pathways (Gelbart 1989; Chen et al. 1998; Khalsa et al. 1998). Accordingly, we wished to determine whether babo could be functioning as a receptor for BMP-type ligands. In the embryo, all three characterized BMP ligands, Dpp, Scw, and Gbb, participate in specific developmental processes. Because Scw and Dpp are both critically important for formation of the amnioserosa tissue at the blastoderm stage (Gelbart 1989; Arora et al. 1994), we used a Kr–lacZ line to follow the fate of amnioserosa tissue in babo mutants and found it to be unaffected (data not shown). Next, we analyzed the role of Babo in midgut development. Between stages 14 and 16, three constrictions form in the midgut producing a four-chambered vessel. Dpp is required for formation of the second midgut constriction, whereas gbb mutants lack the first constriction (Chen et al. 1998; K. Wharton, pers. comm.). Mutant embryos derived from babo germ-line clones were found to exhibit a normal four-chambered midgut at stage 16 (Fig. 3A). Furthermore, visceral mesoderm expression of the Gbb target gene Antennapedia, and the Dpp target gene Ubx, were both normal in babo mutants (data not shown). In addition to these activities, both Dpp and Gbb are required for growth and patterning of imaginal discs and larval brains (Gelbart 1989; Chen et al. 1998; Khalsa et al. 1998). Therefore, we also analyzed the expression of several target genes in babo mutant discs. In wild-type discs, the genes optormotor blind (omb) and spalt (sal) respond to Dpp in a dose-dependent fashion (Lecuit et al. 1996; Nellen et al. 1996). In addition, proper omb expression also requires input from the Gbb ligand (Haerry et al. 1998). In leg discs, Dpp represses wg in the ventral portion of the disc (Theisen et al. 1996). We found no alterations in the expression patterns of omb, sal, and wg in any babo mutant disc (Fig. 3E,F; data not shown). These results indicate that babo does not likely function as a receptor for Dpp, Scw, or Gbb during Drosophila development.
Figure 3.
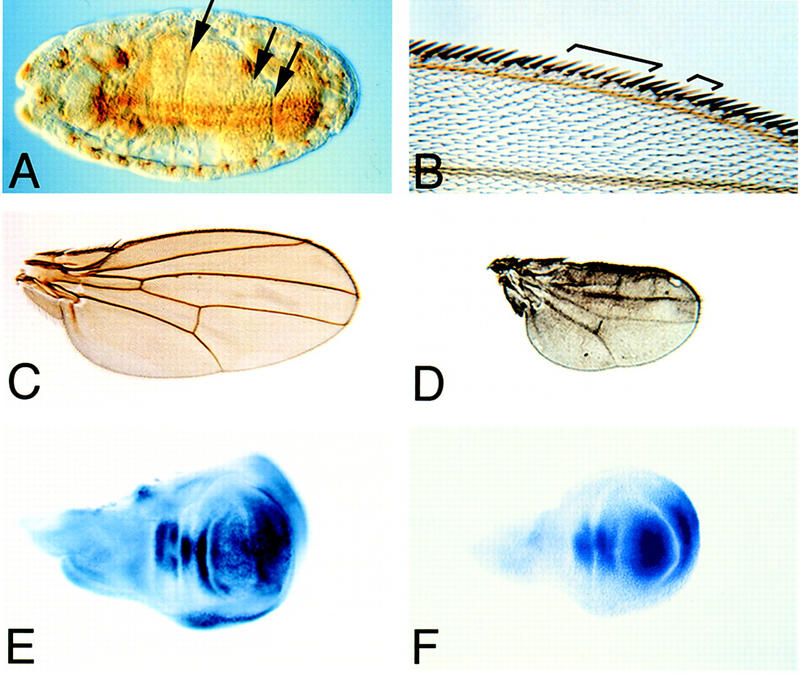
Clonal analysis of babo loss-of-function mutants. (A) Stage 16 babo52 germ-line null mutant embryo showing normal midgut constrictions (arrows). (B) Wing margin clones (marked with y−) of babo52 mutants cells show no pattern defects. (C) Wild-type wing. (D) Wings of a babo52/babo32 germ-line clone derived from pharates are dramatically reduced in size and exhibit only minor venation defects. (E) Omb staining of a wild-type wing disc. (F) Omb staining is unaltered in a babo32 mutant disc.
Babo affects growth but not patterning processes
To further determine whether babo may have an effect on imaginal disc patterning, we performed mosaic analysis using the FRT/FLP system (Xu and Rubin 1993). Using a Myc marker to monitor clone production in the imaginal discs, we found that clones were induced at high frequency in the discs. As shown in Table 2 however, no adult defects were found to be associated with clones in either the wing margins or legs in which we could relibably score for clones that were marked with yellow. An example of wing margin clones with no patterning defect are shown in Figure 3B. In addition to these FRT/FLP mediated adult clones, we could also occasionally obtain a pharate adult from a germ-line clone. These animals exhibited smaller appendages than wild type (Figs. 2D and 3D) but lacked major patterning defects.
Table 2.
Clonal analysis
|
|
babok07737
|
babo52
|
Wild type
|
|||
|---|---|---|---|---|---|---|
| Total flies examined for defects after heat shock | 300 | 350 | 500 | |||
| Flies with pattern defects | 8 | 10 | 8 | |||
| Flies in which defects associated with clones | 0 | 0 | 0 | |||
| Body parts examined | wings | legs | wings | legs | wings | legs |
| for clones | 90 | 85 | 80 | 90 | 30 | 35 |
| Size of clonesa | ||||||
| large | 4 | 6 | 1 | 4 | 0 | 0 |
| medium | 9 | 10 | 10 | 4 | 0 | 0 |
| small | 7 | 14 | 5 | 17 | 0 | 0 |
(Large) more than 5 leg or wing margin bristles; (medium) 3–5 bristles; (small) 1–2 bristles.
We also examined the consequences of expressing a constitutively active form of Babo (Babo*, Q302D) in imaginal discs via the UAS–Gal4 system (Brand and Perrimon 1993). Using the A9–Gal4 line (Haerry et al. 1998), we found that ubiquitous expression of the constitutively active receptor leads to tissue overgrowth in the wing, but only limited pattern abnormalities (Fig. 4A). Control experiments in which the wild-type receptor was expressed by use of the same driver showed no overgrowth. Surface area measurements indicate an ∼30% increase in wing size for a particular UAS–Babo* line grown at 25°C (Fig. 4A). To determine whether this increase in wing surface area results from an increase in cell size or number, we counted wing hairs, single apical extensions found on the surface of each wing blade cell, within a fixed area at two locations on the dorsal surface of the wing (Fig. 4B). We found an ∼20% increase in the density of cells at both positions compared with control flies expressing only the A9–Gal4 driver (Fig. 4B). Assuming that there is no decrease in cell death, and taking into account the ∼30% increase in wing size, these results suggest that for this particular transgene line, one of every two cells undergoes an extra round of division during wing formation as a result of ectopic activated Babo expression.
Figure 4.
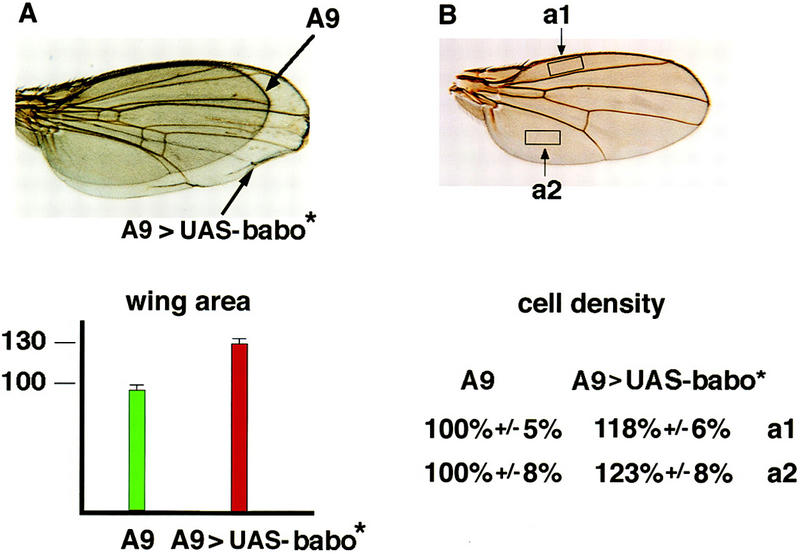
Overexpression of activated Babo in the wing leads to overgrowth. (A) Wings that overexpress a constitutively active Babo receptor are ∼30% larger than wild type. The area was determined for 12 wings of each genotype. Error bars are indicated. (B) The boxes indicate the regions in which wing hairs were counted for six wings of each genotype. The average number of hairs found within these areas on A9 wings equal 100%.
Babo activates TGF-β/Activin type response elements
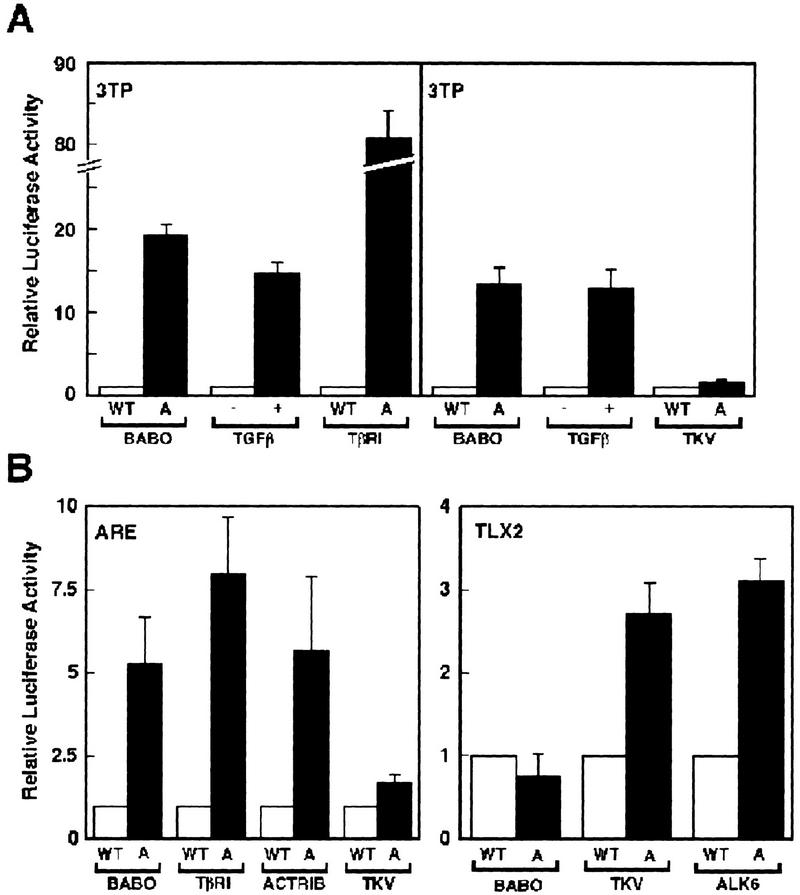
Because our genetic data indicate that in Drosophila Babo is not a component of BMP signaling, we examined the specificity of Babo activity in mammalian cell culture assays using several different pathway specific promoters. Using the TGF-β/Activin responsive promoter p3TP fused to luciferase (p3TP–Lux; Wrana et al. 1992), we cotransfected HepG2 cells with p3TP–Lux alone or with wild-type forms of Babo or the TGF-β type-I receptor, TβRI (Fig. 5A). In the absence of TGF-β, only basal levels of luciferase activity were observed. As described previously (Hayashi et al. 1997), addition of TGF-β or transfection with an activated TβRI results in a strong induction of the p3TP–Lux promoter. A similar induction was observed when the constitutively active form of Babo was cotransfected with p3TP–Lux. In contrast, cotransfection with the constitutively active Dpp receptor Tkv, did not stimulate the 3TP reporter. Similar results were obtained with a second Activin/TGF-β responsive element known as ARE (Hayashi et al. 1997). Cotransfection of P19 embryonic carcinoma cells with pARE–Lux and activated Babo resulted in a fivefold induction in luciferase activity (Fig. 5B). This stimulation was similar to levels of induction produced by the activated forms of TβRI or ActRIB. Consistent with our analysis of 3TP–Lux, Tkv did not modulate induction of the ARE promoter.
Figure 5.
Activation of TGF-β/Activin and BMP transcriptional responses in mammalian cells by Drosophila type I receptors. (A) HepG2 cells were transiently transfected with p3TP–Lux alone or with wild-type (WT, open bar) or activated (A, solid bar) forms of the indicated type I receptors. Luciferase activity was measured in cell lysates 2 days post-transfection and for each type I receptor the activity is expressed relative to luciferase activity in cells transfected with the reporter construct and the respective wild type type I receptor. To determine TGF-β-dependent induction, cells that were transfected with the reporter construct alone were treated with (solid bar) or without (open bar) 100 pm TGF-β for 16–24 hr prior to lysis. TGF-β-dependent induction is expressed relative to untreated cells. (B,C) Signalling by Drosophila type I receptors in P19 cells. Cultures of the embryonic carcinoma cell line P19 were transfected with reporter constructs that were driven either by the ARE or the BMP-responsive Tlx2 promoter (TLX2) together with wild-type or activated forms of the indicated type I receptors. Luciferase activity in cell lysates was determined 2 days after transfection and luciferase activity is expressed as in A.
Finally, we examined whether activated Babo can stimulate the BMP-responsive promoter Tlx2 in P19 cells (Tang et al. 1998). Consistent with their role in BMP/Dpp signaling, both activated Tkv and ALK6 induced the Tlx2 promoter (Fig. 5B), whereas activated Babo had no effect on this reporter. Together these data indicate that Babo specifically activates a TGFβ/Activin-like pathway in mammalian cells and suggest that this receptor may regulate a Smad2/Smad3-like pathway in Drosophila.
Cloning of dSmad2
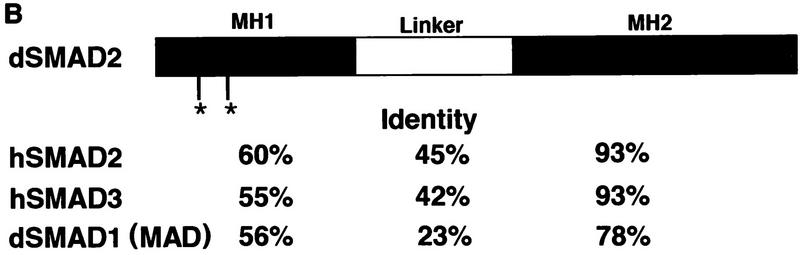
In Drosophila, Mad and Medea, both of which mediate Dpp signaling (Newfeld et al. 1996; Das et al. 1998; Hudson et al. 1998; Wisotzkey et al. 1998), are the only activating Smads that have been identified so far. Because Babo only appears to induce Smad2/3 responsive promoters in mammalian cell culture, we searched the Drosophila ESTs (Berkely Genome Project) database for new Smad-like genes. One clone with significant homology to vertebrate Smad2/3 was identified. Using this clone as a probe, we screened a Drosophila ovarian cDNA library and obtained the full-length cDNA, which was sequenced and named dSmad2. This cDNA encodes a protein of 486 amino acids and contains a carboxy-terminal SSXS motif, which is found in other receptor-regulated SMADs (Macías-Silva et al. 1996; Abdollah et al. 1997; Kretzschmar et al. 1997; Liu et al. 1997; Souchelnytskyi et al. 1997). dSmad2 sequence alignment with other known Smads revealed ∼50% overall identity to Mad or human Smad1 and ∼70% identity to human Smad2 or Smad3 (Fig. 6A). Furthermore, the MH2 domain represented the region of highest homology with >90% identity between dSmad2 and either Smad2 or Smad3. In the MH1 domain, dSmad2 lacks the two inserts that are found in the MH1 domain of Smad2. In the linker, dSmad2 contains a PY motif that is present in other Smads, but in addition has a glycine, serine, and glutamine-rich insert (amino acid residues 177–251) that is absent in either hSmad2 or hSmad3.
Figure 6.
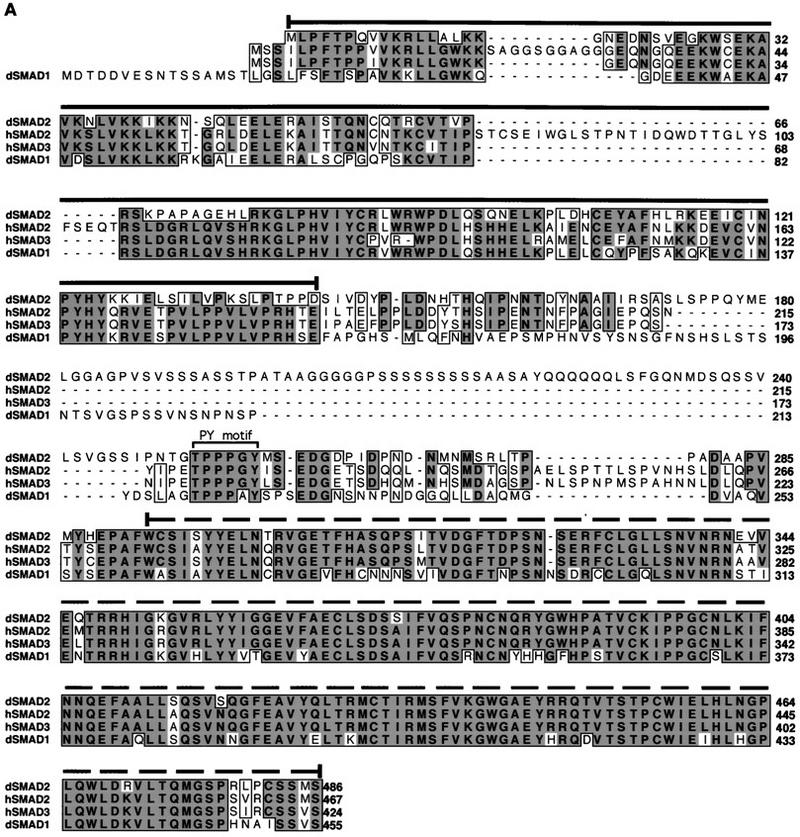
(A) Alignment of the predicted amino acid sequences of dSmad2, hSmad2, hSmad3, and Mad. Conserved residues (bold black type) are shaded in dark gray; letters representing similar residues are shaded light gray. (Dashes) Gaps that are introduced to maximize the alignment. The MH1 and MH2 regions of dSmad2, hSmad2, hSmad3, and Mad are indicated with solid or broken overline, respectively. The PY motif in the linker region is marked with the square bracket. (B) Schematic representation of dSmad2. Percent identity of dSmad2 relative to hSmad2, hSmad3, or Mad is listed for each of the MH1, linker, or MH2 domains. (Asterisks) The absence of the two inserts in MH1 domain of dSmad2, which are found only in hSmad2.
Expression of dSmad2
The expression pattern of dSmad2 in embryonic and larval tissue was determined by in situ hybridization (Fig. 7). We observed high expression of dSmad2 in preblastoderm stage embryos indicating that dSmad2 is a maternally supplied product. The maternal message is rapidly turned over during the blastoderm stage, and the first zygotic expression of dSmad2 is detected during early gastrulation in the ventrally invaginating mesoderm (Fig. 7B). Enriched mesodermal expression remains throughout embryogenesis, particularly in the visceral mesoderm surrounding the midgut. In third instar larvae, we see expression in all the imaginal discs. Most notable, however, is the enriched expression in the brain lobes, specifically in the optic proliferation centers (Fig. 7E), in which the most pronounced effects on cell proliferation are observed in babo mutants.
Figure 7.
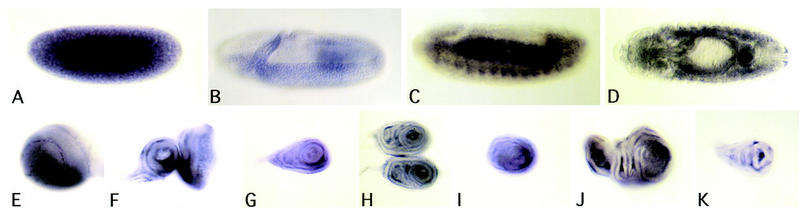
dSmad2 expression pattern. Embryos, imaginal discs, and brains from a yw stock where hybridized with digoxigenin-labled antisense dSmad2 mRNA. (A) Precellularized embryo showing a strong maternal contribution of dSmad2 mRNA. (B) First zygotic expression of dSmad2 appears in the invaginating mesoderm. (C,D) high level dSmad2 expression in mesodermal derivatives at stage 13. (E) Brain hemisphere showing high-level dSmad2 expression in the optic proliferation center. (F) Eye-antennal disc. (G) First leg. (H) Second leg. (I) Third leg disc. (J) Wing disc. (K) Haltere disc.
Babo phosphorylates dSmad2 on the last two carboxy serines and induces dSmad2–Medea interactions
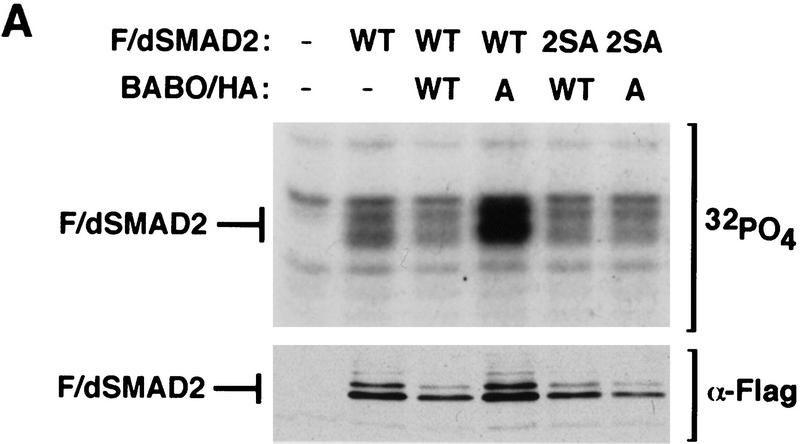
The specificity of Smad interactions with receptors and nuclear targets is dictated by the MH2 domain. This suggests that dSmad2 might function in a Drosophila TGFβ/Activin-like signaling pathway that involves Babo. To investigate this possibility, we tested whether Babo could mediate the phosphorylation of dSmad2. COS-1 cells were transfected with an epitope-tagged dSmad2 either alone or together with wild-type or constitutively active versions of Babo. Analysis of dSmad2 in the absence of signaling showed some basal phosphorylation of the protein (Fig. 8A). However, on coexpression of activated Babo, we observed a strong increase in phosphorylation of the protein (Fig. 8A). Previous work has shown that receptor-dependent phosphorylation of Smads occurs on the last two serines of the protein (Macías-Silva et al. 1996; Abdollah et al. 1997; Kretzschmar et al. 1997; Souchelnytski et al. 1997). To confirm that Babo induced phosphorylation of dSmad2 on these residues, we mutated the last two serines to alanines (dSmad2-2SA). Unlike wild-type dSmad2, this mutant was not phosphorylated by activated Babo (Fig. 8). These data suggest that dSmad2 is a downstream target of Babo and is phosphorylated on the last two serine residues in the carboxyl terminus.
Figure 8.
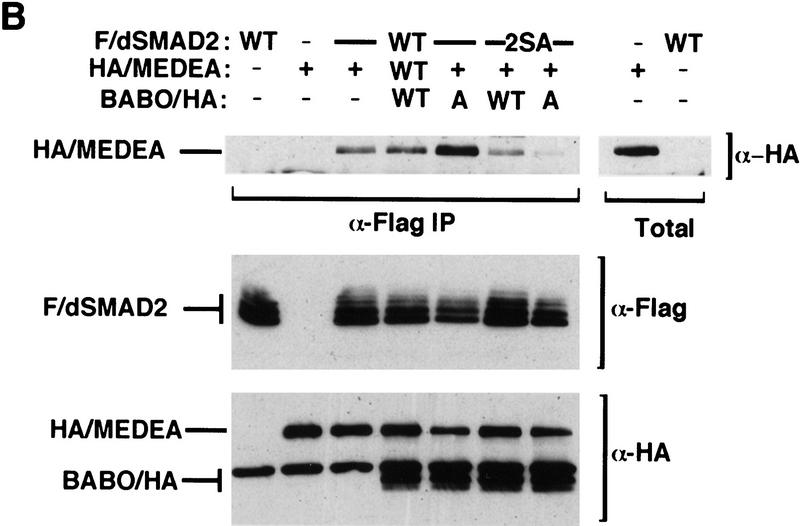
Activated Babo induces the phosphorylation of dSmad2 and formation of dSmad2–Medea heteromeric complexes. (A) COS-1 cells were transiently transfected with wild-type Flag-tagged dSmad2 (F/dSmad2) or a mutant in which the last two serines in the carboxyl terminus were mutated into alanines (2SA) together with either empty vector, wild-type (WT) or constitutively active (A) forms of Babo as indicated. Two days following transfection, cells were labeled with [32P]phosphate and Flag-tagged dSmad2 was purified by subjecting cell lysates to anti-Flag immunoprecipitation followed by SDS-PAGE and autoradiography. The migration of dSmad2 is marked. To determine F/dSmad2 protein levels in each transfection, aliquots of cell lysates run in parallel were resolved by immunoblotting with antibodies against the anti-Flag epitope. (B) To investigate the interaction of dSmad2 with Medea, COS-1 cells were transfected with the indicated expression constructs and cell lysates subjected to anti-Flag immunoprecipitation. The immunoprecipitates were then immunoblotted with anti-HA antibodies to detect HA/MEDEA coprecipitating with Flag/dSmad2. Aliquots of total cell lysates were probed with anti-Flag and anti-HA antibodies to confirm equivalent expression of dSmad2, Medea, and Babo.
One functional consequence of phosphorylating receptor-regulated Smads is the induction of heteromeric complex formation with the common partner Smad4 (Lagna et al. 1996; Kretzschmar et al. 1997). In Drosophila, the Smad4 homolog Medea similarly associates with Mad and is required for a subset of Dpp signaling (Wisotzkey et al. 1998). Thus, we investigated whether Babo-dependent phosphorylation of dSmad2 might induce association with Medea. In the absence of signaling, we observed that dSmad2 and Medea form some heteromeric complexes, however, the level of complex formation was substantially increased on cotransfection with a constitutively active form of Babo (Fig. 8B). Furthermore, use of dSmad2-2SA abolished this receptor-dependent increase in heteromeric complex formation. These data indicate that phosphorylation of dSmad2 on the last two serines is necessary for receptor-dependent induction of heteromeric complexes of dSmad2 and Medea.
dSmad2 interacts transiently with Babo–Punt receptor complexes
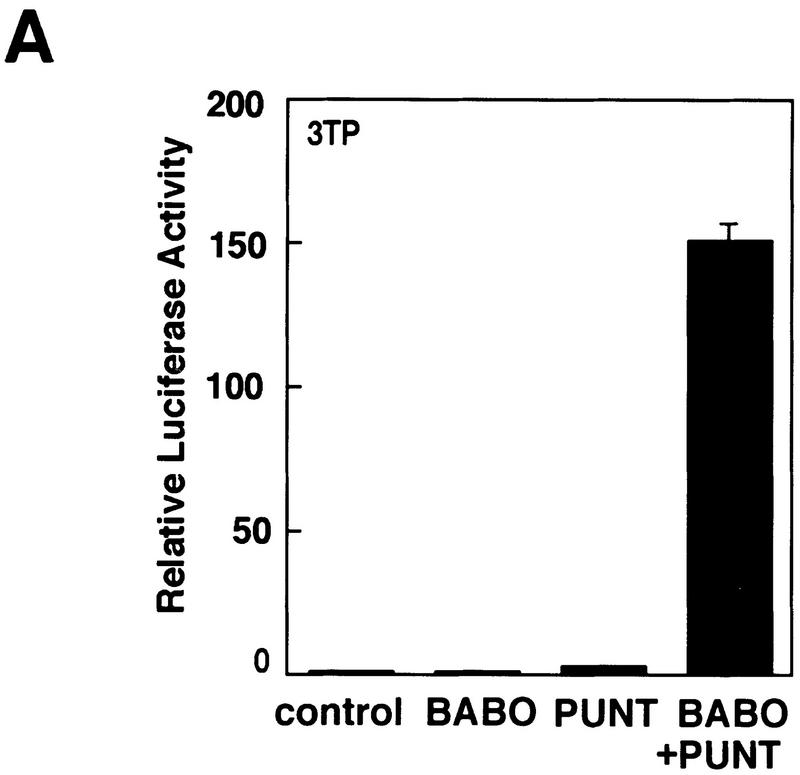
To further characterize the activation of dSmad2, we sought to determine whether it might interact with a Babo receptor complex. Previously, we demonstrated that Punt and Babo form a heteromeric Activin receptor complex in the presence of human Activin (Wrana et al. 1994b). Because recognition of type I receptors by Smads requires activation by the type II receptor, we first confirmed that Punt can function to activate Babo by testing whether Punt/Babo complexes can induce the 3TP promoter. In HepG2 cells, transient transfection of either Punt or Babo alone had no effect on the 3TP promoter (Fig. 9). However, overexpression of Punt together with Babo led to a strong induction of the promoter. This is consistent with previous observations that type II and type I ser/thr kinase receptors have intrinsic affinity for each other and on overexpression can associate and signal in the absence of ligand (Vivien et al. 1995).
Figure 9.
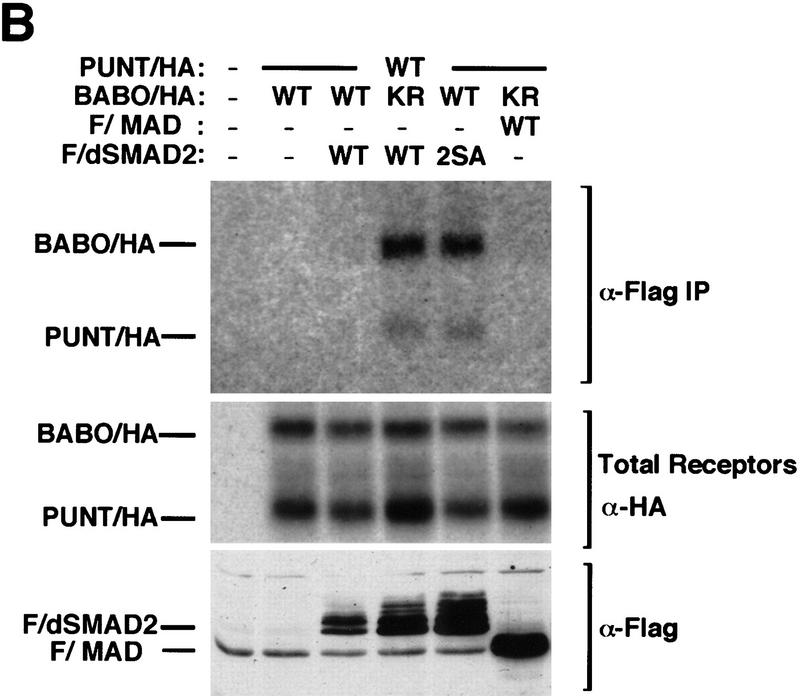
dSmad2 associates specifically and transiently with Babo/Punt receptor complexes. (A) Punt activates BABO in mammalian cells. HepG2 cells were transiently transfected with p3TP–Lux alone or together with the indicated combination of Babo and Punt. Luciferase activity was measured 2 days post-transfection and expressed relative to cells transfected with the reporter construct alone. (B) dSmad2 associates with heteromeric complexes of Babo and Punt. COS-1 cells were transfected with the indicated expression vectors and receptor complexes were affinity labeled and cross-linked with 125I-labeled Activin. Cell lysates were subjected to anti-Flag immunoprecipitation followed by SDS-PAGE and autoradiography to visualize receptor complexes associated with Flag/dSmad2 (anti-Flag IP). The migration of Punt/HA and Babo/HA is indicated. We confirmed equivalent receptor expression in these transfectants by analyzing aliquots of total cell lysates by SDS-PAGE followed by autoradiography (total receptors), whereas expression of Flag/dSmad2 or Flag/Mad was confirmed by immunoblotting total cell lysates with anti-Flag antibodies (anti-Flag blot).
Next, we tested whether dSmad2 interacts with Punt/Babo complexes. For this experiment, we used a kinase-deficient Babo, which functions to stabilizes Smad-receptor interactions (Macías-Silva et al. 1996). When dSmad2 was immunoprecipitated in the presence of wild-type receptors, no complexes could be detected (Fig. 9B). However, when the kinase-deficient form of Babo was utilized, receptor complexes coprecipitating with dSmad2 were readily detected. In addition, when the phosphorylation site mutant of dSmad2 (2SA) was tested, we detected stable complex formation between the mutant protein and wild-type receptor complexes. We also tested for the association of Mad with Punt–Babo receptor complexes. In contrast to dSmad2, we failed to detect any receptor complexes coprecipitating with Mad even when kinase deficient Babo was used in the transfection. Thus, dSmad2 interacts transiently and specifically with Punt–Babo receptor complexes. Taken together, our functional and biochemical analyses strongly suggest that dSmad2 is a Drosophila homolog of Smad2/Smad3 and functions as a downstream signaling component that directly interacts with Babo.
Discussion
Components of an Activin signaling pathway in Drosophila
Early embryonic patterning events in both vertebrates and invertebrates require signals from members of the TGF-β superfamily. One example is the conservation in function of BMP-type molecules in organizing early axial patterning events in both amphibian and Drosophila embryos (Ferguson 1996). Whether other TGF-β pathways also exist in lower eukaryotes and what their functions might be has been unclear. Previously, we reported that the type II receptor Punt and the type I receptor Atr-I (Babo) exhibited close sequence homology to the vertebrate Activin receptors and that they could form a complex in vitro with human Activin (Childs et al. 1993; Wrana et al. 1994b). In this paper we provide three lines of evidence for the existence of a non-BMP-type signaling pathway in Drosophila. First, we demonstrate that Babo mutants do not disrupt any of the known processes that are affected by mutations in BMP-type ligands. These include the establishment of amnioserosa cell fate, midgut morphogenesis in the embryo, and the regulation of the Dpp response genes omb, sal, and wg in imaginal discs. Second, we show that in tissue culture, Babo can stimulate Activin/TGF-β, but not BMP, downstream target elements. Third, we demonstrate that Babo interacts with dSmad2, a Drosophila homolog of the vertebrate Activin/TGF-β pathway-specific Smads, but not Mad, a BMP-specific transcriptional activator.
Although these data provide compelling evidence in favor of an Activin-like signaling pathway in Drosophila, many questions remain unresolved. One important issue is the identity of endogenous ligand(s) for Babo. Recently, Kutty et al. (1998) reported the identification of a Drosophila Activin βB-like subunit. In addition, a ligand, which by phylogenetic tests appears to be more closely related to the Activin/TGF-β subfamily than to BMPs (S. Newfeld pers. comm.), can be found in the Drosophila genomic database (DS07149). Either one or both of these proteins could be the ligands for the two Babo receptor isoforms. Likewise, in vivo proof that Punt, dSmad2, and Medea all participate in an Activin signaling system will have to await the identification of a downstream target gene that is specifically activated by this pathway. Once such a gene is in hand, an analysis of its expression in punt, dsmad2, and medea mutant backgrounds should answer the question of whether these genes provide endogenous components for an Activin signaling pathway.
Babo influences cell growth and proliferation but not patterning
The simplicity of the babo loss-of-function phenotype in Drosophila is striking in light of the number of speculated roles for Activin signaling in vertebrates. In particular, the fact that babo loss-of-function mutations primarily affects cell growth and proliferation in late development, but not cell fate specification or patterning processes, stands in marked contrast to the situation in vertebrates in which Activin or Activin-like signaling pathways have been implicated in a wide range of early developmental events, as well as adult functions such as reproductive potential. Similarly, in Drosophila the BMP-like factor Dpp has been implicated in the control of cell fate, cell proliferation, and patterning processes. Thus, the Babo signaling pathway appears to be unique in that it seems to act primarily in only one of these interconnected processes. It should be recognized, however, that Babo signaling may contribute to some patterning processes and/or cell fate decisions. For example, in the brain, we have yet to determine whether all of the morphological defects can be accounted for by reduced proliferation rates of certain neuroblast populations or whether there are also changes in cell fate specification. Likewise, the cause of the enlarged anal pads during larval development has not been investigated. As the anal organ is involved in regulating salt homeostasis in insects (Brugger 1974), the enlarged anal pads might simply represent swelling due to a salt imbalance or the inability to regulate water content. Larvae missing babo function also fail to exhibit a typical behavioral trait, which is to move away from moist food before pupation. Thus, the inability to assess hydration as the result of abnormal brain development may underlie both the enlarged anal pads and the failure to seek a drier climate for pupation.
At least one other signal-transducing component has been implicated in the control of imaginal disc cell growth but not patterning. Ectopic expression of the Drosophila phosphoinositide 3-kinase (PI3K), Dp110, in the wing disc results in an increase in cell proliferation but no patterning defects (Leevers et al. 1996). Likewise, reduction in the levels of PI3K via expression of dominant-negative forms reduces cell proliferation, resulting in small wings (Leevers et al. 1996) as do loss-of-function clones of babo. Whether Babo signaling could be connected to a PI3K is unclear. The PI3K pathway is generally thought to be activated via coupling to receptor tyrosine kinases. In one recent report however, the PI3K regulatory subunit p85 was found to bind directly to Alk5 and to stimulate PIK-3 activity under certain conditions (Krymaskay et al. 1997). This observation is intriguing because Alk5 and Alk4 are the closest vertebrate homologs of Babo receptor.
The issue of how patterning and growth are coupled in a developing tissue to regulate both size and form is not well understood. Imaginal disc development is a case in point. Despite the fact that many growth-inducing signals such as Dpp are highly localized, cell proliferation, when viewed at a global level, appears to be nearly uniform (Milán et al. 1996). At higher resolution, however, local clusters of nonclonally related cells are seen to divide in synchrony, which has led to the suggestion that some as yet undefined cell–cell communication pathway is responsible for coordinating division within small subpopulations of disc cells (Milán et al. 1996). It will be interesting to learn whether Babo might contribute to this process and how it might be tied to other growth-promoting signals like Dpp.
How might Babo signaling couple to cell proliferation?
In vertebrates, TGF-β and Activins are known to influence the cell cycle. In the best documented cases, the effects appear to be negative and reflect alterations in the levels or activities in one or more cell cycle control proteins (for review, see Derynck and Feng 1997; Hocevar and Howe 1998). However, in certain human tumor cell lines, TGF-β can either stimulate or inhibit cell growth via up or down regulation of a common target, p21/Waf1/Cip1 (Miyazaki et al. 1998). Therefore, the type of response mediated by either Activin or TGF-β likely depends on the biochemical makeup of the cell. Thus, despite the fact that most characterized examples of Activin signaling appear to be negative with respect to their effects on cell proliferation, it is not surprising that the Babo pathway appears to act in a positive manner to stimulate cell proliferation.
Recent analysis of cell growth and division in the Drosophila wing imaginal discs has led to the conclusion that the cell division rate is normally coupled to increases in cell mass such that a relatively constant ratio between the two processes is maintained (Neufeld et al. 1998). Because mutations that affect protein synthesis, such as Minutes, retard cell proliferation without changing cell size, Neufeld et al. (1998) favored a model in which tissue growth is upstream and dominant to cell cycle control. Consistent with this view, they found that if the cell cycle is artificially stimulated to give a higher division rate, then the size of the cells decreases. Conversely, if mitosis in the wing is blocked by inactivation of the Drosophila cdc2 kinase, then wings of normal size are produced but they contain smaller numbers of larger cells (Weigmann et al. 1997). We have found that overexpression of activated Babo in the wing appears to alter the process that couples growth and proliferation rates with the determination of tissue size. The simplest explanation for our results is that Babo signaling stimulates some aspect of the cell cycle resulting in an increased proliferation rate and reduced cell size similar to that described by Neufeld et al. (1998). However, the fact that we see larger wings despite the smaller cell size suggests that Babo may independently affect tissue growth parameters as well as the cell cycle. Therefore, additional studies on the mechanism of Babo signaling could help reveal how cell growth and proliferation are linked to the determination of final tissue size.
Materials and methods
babo mutant strains
The babo mutant P[lac w+]k07737 is from (Török et al. 1993). P[cp70ZT] is from Simon et al. (1985). The deficiencies Df(2R) Np4, Np5, and G75 as well as the 44F Fd and Fc alleles are described in Konev et al. (1994). The babo32, babo52, and babo50 lines were generated by the mobilization of P[cp70ZT]. Mobilization was carried out by crossing P[cp70ZT]/CyO; Ki delta 2–3 males to CyO/Sco; ry506 females in mass matings. P[cp70ZT]*/CyO Ki+ progeny were tested by crossing each fly individually to Df(2R)NP1/Gla flies. Lines in which P[cp70ZT]* failed to complement the NP1 deficiency were saved.
Mutant mapping and identification
A genomic map of babo was generated by restriction mapping and Southern blot analysis of λ fixII genomic phage (Stratagene) and P1 clone DS07727 from the Berkeley Drosophila genome project. Precise exon–intron boundaries were determined by PCR amplification of genomic DNA followed by cycle sequencing (Promega) and comparision to the babo cDNA sequence (Wrana et al. 1994b). The deletion endpoint of the babo52 allele was determined by sequencing a 1.8-kb BalI fragment that contained the junction between babo genomic DNA and the imprecisely excised P[cp70ZT]*.
Germ-line transformation
All germ-line transformations were carried out by standard protocols. A babo rescue construct was generated by cloning the 12-kb XhoI–XbaI genomic fragment into P[Casper Hs70]. An activated form of Babo (isoform a) was generated by changing glutamine 302 to aspartic acid (Wieser et al. 1995) via PCR mutagenesis of a full-length babo cDNA (Wrana et al. 1994b). A 3.2-kb EcoRI fragment containing the activated or wild-type form of babo was then subcloned into pUAST (Brand and Perrimon 1993).
Clonal analysis
The P[lac w+]k07737 and babo52 mutants were recombined onto the FRT-G13 chromosome (Chou et al. 1993). Females from these lines were crossed to males of either yw; Hs Flp; FRT-G13, y+, or yw; Hs Flp π-Myc (Singer et al. 1997) or to yw; Hs Flp; FRT-G13 ovoD. Clones were generated by heat shocking the early first instar larvae. Adult clones were scored by the presence of y− tissue. In discs, clones were detected by the absence of Myc staining by use of confocal microscopy. Germ-line clonal analysis was performed by crossing babo32/Cyo-wg lacZ males to heatshocked females of genotype yw; Hs Flp; FRT-G13 babo52/FRT G13 ovoD. Cuticle preparations and embryo staining were performed as described in Brummel et al. (1994).
Lethality staging and mutant characterization
The lethal phase of babo mutants was determined by monitoring the development of 500 freshly laid eggs from babo/Gla,Bc stocks. Developmental timing and lethality phase were determined by counting the number of animals at each stage of both B+ and Bc− genotypes. Labeling with BrdU and acridine orange were performed as described in Tix et al. (1997) and Abrams et al. (1993), respectively. In situ hybridizations were carried out as described previously (Brummel et al. 1994). The monoclonal antibody 24B10 (gift of L. Zipursky) was used at a dilution of 1:100 and visualized with an ABC Vectastain HRP kit (Vecta Labs). The Omb antibody was a gift from G. Pflugfelder and was used at a 1:100 dilution.
Cloning of dSmad2
The clone CK 10802 was obtained from the EST database (BDGP) and used to probe an ovarian cDNA library (a gift from P. Tolias, Public Health Research Institute, New York). Of six cDNA isolates examined, all had similar-sized insert and both strands of one clone were sequenced by primer walking.
Construction of expression vectors
The construction of Babo-HA (Atr-1-HA) and Punt-HA in pCMV5 have been described previously (Child et al. 1993; Wrana et al. 1994b). pCMV5/HA-Medea is described in Wisotzkey et al. (1998). A kinase-deficient Babo was generated by mutating the lysine residue at position 330 to arginine. The introduction of each mutation was confirmed by sequencing. By use of convenient restriction sites, the amplified regions were subcloned into the appropriate plasmids containing Babo/HA. For expression in mammalian cells, full-length dSmad2 was subcloned from pBluescript into pCMV5-Flag. Serine residues 484 and 486 were changed to alanine by a PCR-based strategy to generate dSmad2 (2SA) in pCMV5. The PCR-amplified region was subcloned into the full-length Flag-dSmad2 and the mutation was confirmed by sequencing.
Mammalian cell lines and transfections
HepG2 cells were cultured in minimal essential media (MEM) containing 10% fetal calf serum and nonessential amino acids (NEAA). P19 cells were grown in MEM containing 7.5% calf serum, 2.5% fetal calf serum, and NEAA. COS-1 cells were maintained in DMEM containing high glucose and 10% fetal calf serum. The COS-1 cells were transiently transfected by the DEAE-dextran method as described previously (Attisano et al. 1996) or by lipofectamine (GIBCO-BRL), whereas HepG2 and P19 cells were transiently transfected with the indicated vectors by the calcium phosphate–DNA precipitation method.
Transcriptional response assay
For TGF-β/Activin-inducible luciferase reporter assays, cells were transiently transfected with the reporter plasmid p3TP-lux (HepG2 cells) or pARE-Lux (P19 cells), pCMV–β-gal and the indicated constructs or with vector alone. For BMP-inducible luciferase reporter assays, P19 cells were transfected with pTlx2-Lux reporter construct, pCMV–β-gal and the indicated type I receptor constructs. Cells were seeded at 25%–50% confluency in 24-well plates and grown overnight. The medium was changed 2–3 hr prior to transfection, and cells were transfected by incubating overnight with 1.0–0.5 μg of DNA per well by the calcium phosphate–DNA precipitation method as described previously (Hoodless et al. 1996). To induce the 3TP-luciferase reporter, cells were incubated overnight in 100 pm TGF-β. Luciferase activity was measured by use of the luciferase assay system (Promega) in a Berthold Lumat LB 9501 luminometer. Transfection efficiency was determined by measuring the β-galactosidase activity (Abdollah et al. 1997). All luciferase data were normalized with respect to β-galactosidase activity.
Immunoprecipitations and immunoblotting
For α-Flag immunoprecipitations, cell lysates were subjected to immunoprecipitation with anti-Flag M2 monoclonal antibody (IBI, Eastman Kodak), followed by adsorption to protein G-sepharose (Pharmacia) as described previously (Hoodless et al. 1996) Precipitates were washed with TNTE buffer (50 mm Tris-HCl at pH 7.4, 150 mm NaCl, 0.1% Triton X-100 and 1 mm EDTA), separated by SDS-PAGE, and visualized by autoradiography. For determination of Flag- or HA-tagged transfected proteins, aliquots of total cell lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes. Flag-tagged and HA-tagged proteins were probed with anti-Flag M2 antibody (Kodak, 1:3000 dilution) or anti-HA 12CA5 antibody (Boehringer, 1:1000 dilution), respectively, and were detected with horseradish peroxidase-conjugated anti-mouse antibody and chemiluminesence as recommended by the manufacturer (ECL kit, Amersham).
Affinity labeling
COS-1 cells transfected with DEAE–dextran were affinity labeled using 5 nm 125I-labeled Activin in medium containing 0.2% fetal calf serum at 37°C for 30 min and the receptors were cross-linked to the ligand as described previously. Cells were lysed in lysis buffer containing 10% glycerol and then subjected to immunoprecipitation with anti-Flag M2 monoclonal antibody as described previously (Macías-Silva et al. 1996).
Acknowledgments
We thank Joumana Jamal for help with the Southern blot analysis of babo alleles and Alexander Konev for suppling us with various deficiencies and mutants from the 45A region. We are grateful to Lil Attisano for providing a Medea expression construct, Larry Zipursky for the gift of monoclonal antibody 24B10, Jamana Jamal for help with localizing babo mutants, and Markus Friedrich for his assistance in performing the phylogenetic anlysis. This manuscript was impoved by the thoughtful comments of Micheline Laurent, Robinette Aley, and Leslie Bell. This work was supported by grants from the March of Dimes and U.S. Public Health Service (GM47462) to M.B.O. and by a grant from the American Cancer Society (DB147) to L.R. T.H. was supported by fellowships from the Ciba-Geigy Jubiläumsstiftung and the American Cancer Society-California Division. Work in J.L.W.’s laboratory was supported by grants from the National Cancer Institute of Canada with funds from the Terry Fox Run and from the Medical Research Council (MRC). J.L.W. is an MRC Scholar. M.B.O. is an Associate Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
The accession no. for the dSmad2 sequence is AF101386.
Footnotes
E-MAIL moconnor@gene.med.umn.edu; FAX (612) 626-7031.
References
- Abdollah S, Macias SM, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Abrams JM, White K, Fessler LI, Steller H. Programmed cell death during Drosophila embryogenesis. Development. 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- Arora K, Levine M, O’Connor M. The screw gene encodes a ubiquitously expressed member of the TGF-β family required for specification of dorsal cell fates in the Drosophila embryo. Genes & Dev. 1994;8:2588–2601. doi: 10.1101/gad.8.21.2588. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL, Montalvo E, Massagué J. Activation of signalling by the activin receptor complex. Mol Cell Biol. 1996;16:1066–1073. doi: 10.1128/mcb.16.3.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Mads and Smads in TGFβ signalling. Curr Opin Cell Biol. 1998;10:188–194. doi: 10.1016/s0955-0674(98)80141-5. [DOI] [PubMed] [Google Scholar]
- Bausenwein B, Dittrich AP, Fischbach KF. The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 1992;267:17–28. doi: 10.1007/BF00318687. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brugger C. Research on the ultrastructure and function of the anal organ of the normal and lethal (l(3)tr) larvae of the Drosophila melanogaster. J Insect Physiol. 1974;20:121–41. doi: 10.1016/0022-1910(74)90128-0. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Brummel TJ, Twonbly V, Marques G, Wrana JL, Newfeld SJ, Attisano L, Massagué J, O’Connor MB, Gelbart WM. Characterization and relationship of Dpp receptors encoded by the saxophone and thick veins gene in Drosophila. Cell. 1994;78:251–261. doi: 10.1016/0092-8674(94)90295-x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Riese MJ, Killinger MA, Hoffmann FM. A genetic screen for modifiers of Drosophila decapentaplegic signaling identifies mutations in punt, Mothers against dpp and the BMP-7 homologue, 60A. Development. 1998;125:1759–1768. doi: 10.1242/dev.125.9.1759. [DOI] [PubMed] [Google Scholar]
- Childs SR, Wrana JL, Arora K, Attisano L, O’Connor MB, Massagué J. Identification of a Drosophila activin receptor. Proc Natl Acad Sci. 1993;90:9475–9479. doi: 10.1073/pnas.90.20.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Das P, Maduzia L, Wang H, Finelli A, Cho SH, Smith M, Padgett R. The Drosophila gene Medea demonstrates the requirement for different classes of Smads in dpp signaling. Development. 1998;125:1519–1528. doi: 10.1242/dev.125.8.1519. [DOI] [PubMed] [Google Scholar]
- Derynck R, Feng XH. TGF-beta receptor signaling. Biochim Biophys Acta. 1997;1333:F105–150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- Feng XH, Zhang Y, Wu RY, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-β-induced transcriptional activation. Genes & Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL. Conservation of dorsal-ventral patterning in arthropods and chordates. Curr Opin Genet Dev. 1996;6:424–431. doi: 10.1016/s0959-437x(96)80063-3. [DOI] [PubMed] [Google Scholar]
- Gelbart, W.M. 1989. The decapentaplegic gene: A TGF-β homolog controlling pattern formation in Drosophila. Development (Suppl.) 107: 65–74. [DOI] [PubMed]
- Haerry T, Khalsa O, O’Connor K, O’Connor M. Synergistic signaling by two BMP ligands through the Sax and TKV receptors controls wing growth and patterning in Drosophila. Development. 1998;125:3977–3987. doi: 10.1242/dev.125.20.3977. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MJ, Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGF beta receptor and functions as an antagonist of TGF-β signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten DP. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hocevar BA, Howe PH. Mechanisms of TGF-beta-induced cell cycle arrest. Miner Electrolyte Metab. 1998;24:131–135. doi: 10.1159/000057360. [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Haerry T, Abdollah S, Stapleton M, O’Connor MB, Attisano L, Wrana JL. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- Hudson J, Podos S, Keith K, Simpson S, Ferguson E. The Drosophila Medea gene is required downstream of dpp and encodes a functional homolog of human Smad4. Development. 1998;125:1407–1420. doi: 10.1242/dev.125.8.1407. [DOI] [PubMed] [Google Scholar]
- Janknecht R, Wells NJ, Hunter T. TGF-β-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes & Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalsa O, Yoon J, Torres-Schumann S, Wharton KA. TGF-β/BMP superfamily members, Gbb-60A and Dpp, cooperate to provide pattern information and establish cell identity in the Drosophila wing. Development. 1998;125:2723–2734. doi: 10.1242/dev.125.14.2723. [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kingsley DM. The TGF-β superfamily: New members, new receptors, and new genetic tests of function in different organisms. Genes & Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- Konev A, Varentsova ER, Khromykh I. Cytogenetic analysis of the chromosome region containing the Drosophila radiosensitivity gene. II. The vitally important loci of the 44F-45C region of chromosome 2. Genetika. 1994;30:201–211. [PubMed] [Google Scholar]
- Kretzschmar M, Liu F, Hata A, Doody J, Massagué J. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes & Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- Krymaskay VP, Hoffman R, Eszterhas A, Ciocca V, Panettieri RA. TGF-β 1 modulates EGF-stimulated phosphatidylinositol 3-kinase activity in human airway smooth muscle cells. Am J Physiol. 1997;273:12220–12227. doi: 10.1152/ajplung.1997.273.6.L1220. [DOI] [PubMed] [Google Scholar]
- Kutty G, Kutty RK, Samuel W, Duncan T, Jaworski C, Wiggert B. Identification of a new member of transforming growth factor-beta superfamily Drosophila: The first invertebrate activin gene. Biochem Biophys Res Comm. 1998;246:644–649. doi: 10.1006/bbrc.1998.8678. [DOI] [PubMed] [Google Scholar]
- Labbé E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA binding protein, FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Partnership between DPC4 and SMAD proteins in TGFβ signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD. The Drosophila phophoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massagué J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes & Dev. 1997a;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci. 1997b;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macías-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana J. MADR2 is a substrate of the TGFβ receptor and its phosphorylation is required for nuclear accumulation and signalling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Milán M, Campuzano S, García-Bellido A. Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc Natl Acad Sci. 1996;93:640–645. doi: 10.1073/pnas.93.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Ohashi R, Tsuji T, Mihara K, Gohda E, Namba M. Transforming growth factor-beta1 stimulates or inhibits cell growth via down-or up-regulation of p21/Waf1. Biochem Biophys Res Commun. 1998;246:873–880. doi: 10.1006/bbrc.1998.8712. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Flor A, della Cruz A, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Newfeld SJ, Chartoff EH, Graff JM, Melton DA, Gelbart WM. Mothers against dpp encodes a conserved cytoplasmic protein required in DPP/TGF-beta responsive cells. Development. 1996;122:2099–2108. doi: 10.1242/dev.122.7.2099. [DOI] [PubMed] [Google Scholar]
- Padgett RW, Johnston RDS, Gelbart WM. A transcript from a Drosophila pattern gene predicts a protein homologous to the transforming growth factor-(beta) family. Nature. 1987;325:81–84. doi: 10.1038/325081a0. [DOI] [PubMed] [Google Scholar]
- Padgett RW, Wozney JM, Gelbart WM. Human BMP sequences can confer normal dorsal-ventral patterning in the Drosophila embryo. Proc Natl Acad Sci. 1993;90:2905–2909. doi: 10.1073/pnas.90.7.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery L, Twonbly V, Wharton K, Gelbart WM. Genetic screen to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics. 1995;139:241–254. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Sutton CA, Lobell RB, Glaser RL, Lis JT. Determinants of heat shock-induced chromosome puffing. Cell. 1985;40:805–817. doi: 10.1016/0092-8674(85)90340-x. [DOI] [PubMed] [Google Scholar]
- Singer MA, Penton A, Twombly V, Hoffmann FM, Gelbart WM. Signaling through both type I DPP receptors is required for anterior- posterior patterning of the entire Drosophila wing. Development. 1997;124:79–89. doi: 10.1242/dev.124.1.79. [DOI] [PubMed] [Google Scholar]
- Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten DP, Heldin CH. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Hoodless PA, Lu Z, Breitman ML, McInnes RR, Wrana JL, Buchwald M. The Tlx-2 homeobox gene is a downstream target of BMP signalling and is required for mouse mesoderm development. Development. 1998;125:1877–1887. doi: 10.1242/dev.125.10.1877. [DOI] [PubMed] [Google Scholar]
- Theisen H, Haerry TE, O’Connor MB, Marsh JL. Developmental territories created by mutual antagonism between Wingless and Decapentaplegic. Development. 1996;122:3939–3948. doi: 10.1242/dev.122.12.3939. [DOI] [PubMed] [Google Scholar]
- Tix S, Eule E, Fischbach KF, Benzer S. Glia in the chiasms and medulla of the Drosophila melanogaster optic lobes. Cell Tissue Res. 1997;289:397–409. doi: 10.1007/s004410050886. [DOI] [PubMed] [Google Scholar]
- Török T, Tick G, Alvarado M, Kiss I. P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: Isolation of lethals with different overgrowth phenotypes. Genetics. 1993;135:71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart WM. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development. 1996;122:1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- Vivien D, Attisano L, Wrana JL, Massagué J. Signaling activity of homologous and heterologous transforming growth factor-beta receptor kinase complexes. J Biol Chem. 1995;270:7134–7141. doi: 10.1074/jbc.270.13.7134. [DOI] [PubMed] [Google Scholar]
- Weigmann K, Cohen SM, Lehner CF. Cell cycle progression, growth and patterning in imaginal discs despite inhibition of cell division after inactivation of Drosophila Cdc2 kinase. Development. 1997;124:3555–3563. doi: 10.1242/dev.124.18.3555. [DOI] [PubMed] [Google Scholar]
- Wieser R, Wrana JL, Massagué J. GS domain mutations that constitutively activate T beta R-I, the downstream signaling component in the TGF-beta receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisotzkey R, Mehra A, Sutherland D, Dobens L, Liu X, Dohrmann C, Attisano L, Raftery L. Medea is a Drosophila Smad4 homolog that is differentially required to potentiate DPP responses. Development. 1998;125:1433–1445. doi: 10.1242/dev.125.8.1433. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massagué J. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-beta receptor. Nature. 1994a;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Tran H, Attisano L, Arora K, Childs SR, Massagué J, O’Connor MB. Two distinct transmembrane serine/threonine kinases from Drosophila melanogaster form an activin receptor complex. Mol Cell Biol. 1994b;14:944–950. doi: 10.1128/mcb.14.2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]