We used whole exome sequencing of a single patient with combined malonic and methylmalonic aciduria (CMAMMA) to identify mutations in ACSF3, a putative malonyl-CoA and methylmalonyl-CoA synthetase (MCS). Follow-up sequencing of eight additional patients, including an individual who was diagnosed after mining an exome database as well as an affected canine, showed pathogenic mutations. ACSF3 mutant alleles occur with a minor allele frequency (MAF) of 0.0058 in ~1,000 control individuals predicting a CMAMMA population incidence ~ 1:30,000. CMAMMA is the first human disorder caused by mutations in a member of the acyl-CoA synthetase family, a diverse group of evolutionarily conserved proteins, and may emerge as one of the more common human metabolic disorders.
Methylmalonic acidemias (MMAemias) are heterogeneous disorders that exhibit elevated methylmalonic acid (MMA) in body fluids. Deficiency of methylmalonyl-CoA mutase (MUT) or the enzymes (MMAA, MMAB, MMADHC) that synthesize 5′-adenosylcobalamin comprise most disease subtypes. Some patients have atypical forms of MMAemia, e.g., combined malonic and methylmalonic aciduria (CMAMMA) that lack enzymatic and molecular definition. CMAMMA was first reported in a child with immunodeficiency, failure to thrive, seizures, increased urinary MMA compared to malonic acid (MA) and normal malonyl-CoA decarboxylase activity1. A Labrador retriever with similar biochemical features and neurodegeneration has also been described2.
To determine the cause of CMAMMA, we took a multifaceted approach that included exome and candidate gene sequencing in nine patients, identification of the canine orthologue and mutation analysis in an affected dog, and a novel strategy of hypothesis-generating clinical research in an exome cohort3. We establish ACSF3 mutations as the cause of CMAMMA and describe the first disease association with a member of the acyl-CoA synthetase (ACS) family, enzymes that activate fatty acids for intermediary metabolism4.
Nine subjects with CMAMMA participated and six were evaluated at the NIH. The age of diagnosis and symptoms were variable (Table 1). After uneventful early decades, four patients were diagnosed in adulthood with neurological manifestations (seizures, memory problems, psychiatric disease, and/or cognitive decline) without vitamin B12 deficiency. Five subjects presented during childhood with symptoms suggestive of an intermediary metabolic disorder (coma, ketoacidosis, hypoglycemia, failure to thrive, elevated transaminases, microcephaly, dystonia, axial hypotonia, and/or developmental delay).
Table 1.
Clinical and biochemical features of subjects with combined malonic and methylmalonic aciduria (CMAMMA)
| Patient Subjects | Age (years) & Sex | Age and clinical symptoms at diagnosis | MLYCD enzyme assay/mutation testing | Propionyl-CoA pathway studies | Propionyl-carnitine μM | Plasma MMA μM (nl <0.4) | Plasma MA μM (nl <0.89) | Urine MMA mmol/mol Cr (nl <3) | Urine MA mmol/mol Cr (nl undetectable) | Urine MMA/MA ratio | ACSF3 Variant 1 | ACSF3 Variant 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 46, F | 43, ocular migraine, memory problems, T2 hyperintensities on brain MRI | n.d./normal | Normal 1-14C-propionate incorporation, MUT, MMAA, MMAB, MMADHC, MCEE negative | 0.44 nl <0.88 |
15.4 | 4.5 | 104.5 | 17.9 | 5.8 | c.1385A>C p.Lys462Thr c.del1394_1411, p.Gln465_Gly470 |
c.1672C>T p.Arg558Trp MAF=0.004 |
| 2 | 51, M | 51, complex partial seizures and memory problems with onset at 43 | normal/n.d. | Normal 1-14C-propionate incorporation | 0.47 nl <0.88 |
11.8 | 2.1 | 89.7 | 14.1 | 6.4 | c.1567C>T p.Arg523X |
c.1672C>T p.Arg558Trp MAF=0.004 |
| 3 | 56, F | 55, psychiatric symptoms, T2 hyperintensities on brain MRI, died at age 60 | n.d./n.d. | Normal 1-14C-propionate incorporation | 0.19 nl <0.88 |
10.0 | 5.5 | 29.2 | 2.9 | 10.1 | c.1075G>A p.Glu359Lys MAF=0.001 |
c.1672C>T p.Arg558Trp MAF=0.004 |
| 4 | 16, F | 22 mo, seizure, encephalopathy; recurrent ketoacidosis | normal/n.d. | n.d. | 0.70 nl <0.88 |
7.7 | 1.7 | 101.2 | 20.0 | 5.2 | c.1672C>T p.Arg558Trp MAF=0.004 |
c.1672C>Ta p.Arg558Trp MAF=0.004 |
| 5 | 5, F | 4, hypoglycemia, acidosis, poor weight gain, diarrhea episodes | n.d./normal | n.d. | 0.34 nl <1.78 |
8.3 | 3.3 | 230.4 | 72.0 | 3.2 | c.1073C>T p.Thr358Ile |
c.1412G>A p.Arg471Gln |
| 6 | 66, F | 66, exome sequencing, incontinence, mild memory problems | n.d./normal | MUT, MMAA, MMAB, MMADHC, MCEE negative | 0.77 nl <0.88 |
48.0 | 11.3 | 206.0 | 26.3 | 7.8 | c.1411C>T p.Arg471Trp MAF=0.001 |
c.1411C>Tb p.Arg471Trp MAF=0.001 |
| 7 | 16 mo, M | 6 mo, failure to thrive, elevated transaminases | n.d./Heterozygous c.642-5C>T |
n.d. | 0.67 nl <1.78 |
5.6 | n.d. | 196.0 | 37.0 | 5.3 | c.728C>T p.Pro243Leu |
c.728C>Tc p.Pro243Leu |
| 8 | 17 mo, M | 17 mo, psychomotor delay without regression at 5 mo; microcephly, dystonia, axial hypotonia, speech delay | n.d./n.d. | n.d. | normal | n.d. | n.d. | 583.0 | 91.0 | 6.4 | c.593T>G p.Met198Arg |
c.593T>Gd p.Met198Arg |
| 9 | 26 mo, M | 26 mo, psychomotor delay, axial hypotonia, loss of speech | normal/normal | n.d. | normal | n.d. | n.d. | 112.0 | 47.5 | 2.4 | none in coding exons or splice sites | |
| Dog Subject | Age | Clinical symptoms at diagnosis | MLYCD enzyme assay/mutation testing | Propionyl-CoA pathway studies | Propionyl-carnitine | Plasma MMA uM | Plasma MA μM | Urine MMA mmol/mol Cr | Urine MA mmol/mol Cr | Urine MMA/MA ratio | ACSF3 Variant 1 | ACSF3 Variant 2 |
| Podell et al 1996 | 12 weeks | Neurodegeneration of brain and spinal cord | normal/n.d. | n.d. | n.d. | n.d. | n.d. | 449 | 101 | 4.4 | c.1741G>A, p.Gly581Ser | c.1741G>A, p.Gly581Ser |
parents unavailable for testing,
analysis of read depth with whole exome sequencing not suggestive of a deletion,
both parents heterozygous,
parents second cousins and mother heterozygous, n.d.=not determined, MAF= minor allele frequency in 1000 Genomes and ClinSeq™ datasets. If the MAF is not specified, that variant was not detected in those datasets.
Methylmalonic and malonic aciduria with urinary MMA/MA >5 was present in seven of nine affecteds (Table 1). Serum MMA was elevated but serum B12 levels, acylcarnitines, and total homocysteine were normal, as were malonyl-CoA decarboxylase activity, 1-C14-propionate incorporation, malonyl-CoA decarboxylase (MLYCD) genetic testing, and sequencing of known MMAemia genes (Table 1). Plasma MA was measured by GC/MS in six patients and was also markedly elevated (Table 1). We conclude that these subjects all have CMAMMA, which is distinct from other forms of MMAemia.
For Subject 1, we sequenced target-selected libraries in the paired-end 101 bp configuration, yielding 114,467 variant genotypes. We used genetic filters for homozygosity or compound heterozygosity. We included nonsynonymous, splice, frameshifting, and nonsense variants as potential mutations but excluded dbSNP variants. We used control exome data3 to exclude homozygous variants or variants with >10% frequency (Supplementary Table 1 and Online Methods).
The filtering strategy yielded 12 genes, from which we selected for further evaluation ACSF3, an orphan member of the acyl-CoA synthetase family, based on its putative function and predicted mitochondrial localization. We found three ACSF3 exome variants in Subject 1 (c.1385A>C p.Lys462Thr, c.del1394_1411, p.Gln465_Gly470del, and c.1627C>T, p.Arg558Trp) and confirmed them by Sanger sequencing (Table 1, Supplementary Figure 1, Supplementary Table 2). The variants p.Lys462Thr and p.Gln465_Gly470del were in trans with p.Arg558Trp based on parental genotypes and segregated in two unaffected siblings who, like their parents, had normal serum MMA levels. We sequenced ACSF3 exons in seven additional patients with CMAMMA; six had ACSF3 variations (Table 1, Supplementary Figure 1). One patient had no damaging mutations detected.
Next, we identified a putative canine ACSF3 orthologue and sequenced DNA from the CMAMMA Labrador retriever, this showed a homozygous alteration (c.1288G>A, p.Gly430Ser; orthologous to human p.Gly480) in a conserved residue (Figure 1, Table 1, Supplementary Figure 1). This variant was absent in 40 control Labrador DNAs selected for maximum diversity based on American Kennel Club numbers. Finally, we took a novel approach to patient discovery by analyzing exome data of 401 individuals ascertained for cardiovascular phenotypes3. We identified a 66 year-old female, apparently homozygous for a c.1411C>T, p.Arg471Trp ACSF3 variant. She had no known metabolic disease symptoms but reported incontinence and mild memory problems. Her laboratory evaluation showed 48 μM MMA and 11.3 μM MA in plasma and 206 mmol/mol Cr MMA and 26.3 mmol/mol Cr MA in urine, and normal serum B12 levels and acylcarnitines. We did not find mutations in other known MMAemia genes in her exome (Table 1).
Figure 1.
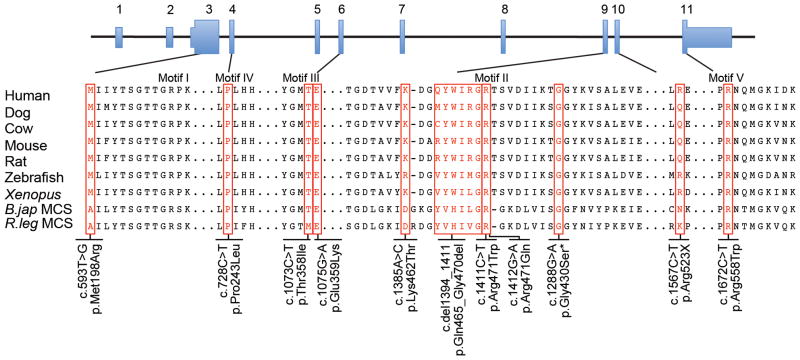
Alignment of the motif regions in ACSF3 orthologues and the malonyl-CoA synthase enzymes in bacteria. The sequences (see methods) were aligned using MegAlign via the Clustal W method. An additional three amino acids amino-terminal to Motif I are shown. Motif II was aligned independent of the full-length protein to improve the alignment of the ACSF3 and MCS proteins. The ACSF3 alterations identified in the eight subjects and affected dog are indicated. The asterisk (*) indicates the dog variant p.Gly430Ser, which is orthologous to position p.Gly480 in human ACSF3.
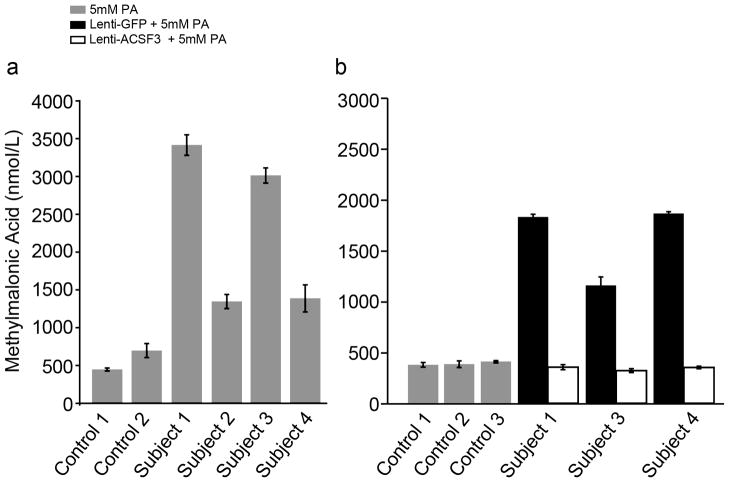
We identified nine missense, one in-frame deletion and one nonsense mutation (Figure 1). Four subjects were apparently homozygous for ACSF3 variants. Although it is possible that unidentified deletions might play a role in this disorder, this is unlikely for these individuals (Table 1). Most of the variants resided in the C-terminal half of ACSF3. Eight out of nine missense mutations and the in-frame deletion were located in conserved ACS motifs predicted to be involved in AMP binding (Motif I), conformational change and catalytic function (Motif II), substrate binding (Motifs III, IV), or catalysis (Motif V)5 (Figure 1). Western analyses using fibroblasts from Subjects 1–4 and 7 showed the presence of cross-reactive ACSF3 (Supplementary Figure 2). Fibroblasts from Subjects 1–4 produced 2.4- to 6-fold more MMA than control cells (Figure 2A) after chemical stimulation. Viral expression of ACSF3, but not GFP (Figure 2B) restored metabolism, and provided validation of ACSF3 function in a cell culture biochemical assay.
Figure 2.
MMA production by CMAMMA fibroblasts and lentiviral complementation with ACSF3. A. Control fibroblasts and fibroblasts from Subjects 1–4 were incubated in medium containing 5 mM sodium propionate at 37°C for 72 hours and the media was removed for GC/MS analysis of MMA. The CMAMMA patient cells showed increased accumulation of MMA in the media, which were 6, 2.4, 5.3 and 2.4 fold elevated compared to the control cell lines. Error bars are +/− 1 standard deviation (n=3 measurements per cell line). B. Fibroblasts from Subjects 1, 3 and 4 were transduced with lentivirus designed to express ACSF3 or GFP and then incubated in medium containing 5 mM sodium propionate as described. Fibroblasts transduced with ACSF3, but not GFP, exhibit MMA production similar to control fibroblasts treated in a similar fashion. Error bars are +/− 1 standard deviation (n=3 replicates per cell line).
These data establish a candidate gene for CMAMMA using exome sequencing in a single affected with validation using four approaches. First, seven additional probands harbored two mutations in ACSF3. Second, an affected dog had a single, unique sequence variant in the canine ACSF3 orthologue in a conserved residue that was absent in 40 diverse controls. Third, one patient with two ACSF3 mutations was identified in a cohort of subjects not ascertained for metabolic disease and had biochemical features of CMAMMA. Finally, viral complementation of ACSF3 in patient fibroblasts corrected the cellular metabolic defect. Based on these observations, we conclude that mutations in ACSF3 cause CMAMMA.
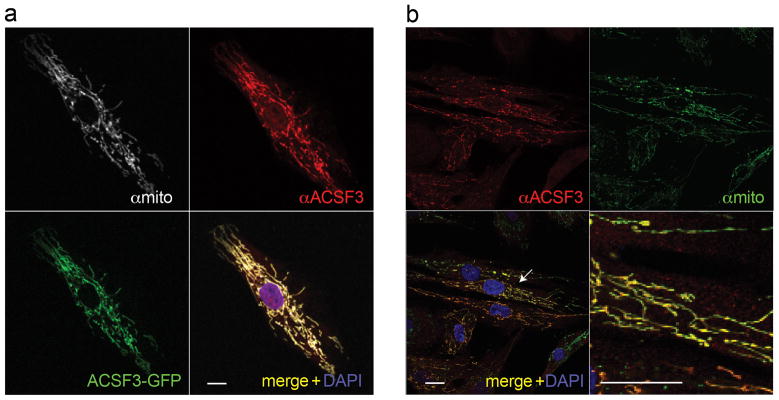
The ACSF3 gene is an orphan member of the acyl-coenzyme A synthetase gene family, enzymes that thioesterify substrates into CoA derivatives, and weakly activated C24:0 fatty acid4. The biochemical abnormalities in the patients led us to reassess the possible function of ACSF3. When we compared human ACSF3 to Bradyrhizobium japonicum malonyl-CoA synthetase (MCS), a well-characterized enzyme, the proteins were more identical (32%) and similar (50%) to each other than ACSF3 was to the next closest human ACS family member (ACSM3v1, 28% identity). Phylogenetic analyses rooted human ACSF3 with the MCS enzymes versus other ACSs (Supplementary Figure 3). To provide preliminary experimental evidence for predicted function, we examined purified, GST-tagged ACSF3 under MCS assay conditions and found that the enzyme activated malonate and methylmalonate, but not acetate, into the respective coenzyme thioesters (Supplementary Table 3). The specific activity of GST-tagged ACSF3 was higher with malonate as a substrate compared to methylmalonate, similar to its prokaryotic homologues. Because the first 58 amino acids of ACSF3 are predicted to encode a mitochondrial leader sequence (Supplementary Figure 4), we performed immunostaining with fibroblasts overexpressing ACSF3 and a C-terminal GFP-ACSF3 fusion protein. ACSF3 staining showed a distinct mitochondrial distribution and co-localized with a mitochondrial antibody (Figure 3). The comparative sequence analysis, enzymatic data, and subcellular localization lead us to propose that ACSF3 is a mitochondrial malonyl-CoA and methylmalonyl-CoA synthetase (MCS), an enzyme postulated to catalyze the first step of intramitochondrial fatty acid synthesis5.
Figure 3.
ACSF3 mitochondrial localization. A) Control fibroblasts transfected with a plasmid expressing C-terminal GFP-tagged ACSF3 were co-stained with anti-ACSF3 (red) and a mitochondrial antibody (white). Scale bar=10 μm B) Fibroblasts from Subject 4 that express ACSF3 after lentiviral transduction were co-stained with anti-ACSF3 (red) and a mitochondrial antibody (green). The bottom right depicts an enlargement of the area surrounding the arrow. Scale bar=20 μm. All images were collected using a confocal microscope using a 63X objective with 0.7 zoom.
The assignment of ACSF3 as an MCS provides a framework to understand the consequences of the ACSF3 mutations and the metabolic perturbations of CMAMMA. MCS from R. trifolii and B. japonicum activate malonate and methylmalonate as substrates in vitro6,7 as does ACSF3 (Supplementary Table 3), suggesting that malfunction of this enzyme causes accretion of the proximal substrates that manifests as methylmalonic and malonic aciduria. Site-directed mutagenesis experiments with B. japonicum MCS showed that p.Glu308Gln abolishes malonate binding7. The corresponding human ACSF3 position is the residue mutated in Subject 3, p.Glu359Lys in Motif III, and predicts that this mutation is likely to effect the Km for malonate. Arg471 in motif II is nearly invariant in the ACS family4 and essential for acyl-CoA synthetase activity8–10. Therefore, an Arg471 alteration in ACSF3, as in Subjects 5 and 6, likely affects enzymatic function. Other missense alterations (p.Pro243Leu, p.Thr358Ile, p.Gly430Ser (dog), p.Arg558Trp) map to conserved residues in B. japonicum and R. leguminosarum MCS (Figure 1) or to conserved residues in other ACSF3 family members (p.Met198Arg, p.Lys462Thr), and are likely to be deleterious.
In the ClinSeq™ cohort, there were an additional four participants heterozygous for ACSF3 variants (p.Glu359Lys n=1, p.Arg558Trp n=3) also found in patients with CMAMMA. The 1000 genomes dataset (estimated coverage of 629 genomes) there were six individuals with ACSF3 mutations (p.Glu359Lys n=1, p.Arg558Trp n=5). Combining these data yields an overall MAF of 0.0058 (95% CI, .0033–.0106) for an estimated disease incidence of ~1/30,000 (95% CI, 1/9,000 – 1/92,000). We predict that CMAMMA is one of the most common forms of MMAemia11, and perhaps, one of the most common inborn errors of metabolism. Clearly, the spectrum of symptoms and natural history of this disorder are highly variable and require further delineation. The identification of an affected using exome sequencing highlights an interesting and alterative diagnostic approach because CMAMMA is not identified through routine newborn screening (via elevated propionylcarnitine [C3]). We speculate that CMAMMA and other metabolic disorders that have escaped early diagnosis could be identified using genomic techniques.
Online Methods
Subjects
Patient studies were approved by the National Human Genome Research Institute Institutional Review Board as part of NIH studies: 04-HG-0127 “Clinical and Basic Investigations of Methylmalonic Acidemia and Related Disorders”(ClinicalTrials identifier: NCT00078078), 10-HG-0065 “Whole Genome Medical Sequencing for Genome Discovery” (ClinicalTrials identifier: NCT01087320), 07-HG-0002 “ClinSeq™: A Large-Scale Medical Sequencing Clinical Research Pilot Study” (ClinicalTrials identifier: NCT00410241), and/or 94-HG-0193 “Genetic and Clinical Studies of Congenital Anomaly Syndromes” (ClinicalTrials identifier: NCT00001404) and were performed in compliance with US 45.CFR.46. Adult participants and parents of the younger subjects signed written informed consent for their participation. Patients with CMAMMA were evaluated at the NIH Clinical Center and/or outside records from referring centers were reviewed. Plasma MMA was determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS) stable isotope dilution analysis and urine organic acids were measured by gas chromatography-mass spectrometry (GC/MS) (Mayo Medical Laboratories). A GC/MS assay was developed to measure MA in patient samples (Gavrilov et al, unpublished). In brief, D3 methylmalonic and 13C2-malonic acid were added to plasma, serum or urine, adjusted with NaCl, and acidified. An ethyl acetate extraction was performed and the organic layer was concentrated under N2 flow. The resulting residue was silylated with BSTFA + 1% TMCS. The samples were then analyzed by GC/MS in the selected ion monitoring mode. Plasma samples from 19 anonymous controls with normal MMA levels were used to develop the reference ranges for MA (mean 0.67 μM ± 0.14, range 0.38 to 0.89 μM).
DNA Isolation
DNA was isolated from all human subjects and the canine controls from whole blood using the salting out method (Qiagen) following the manufacturer’s instructions. For the CMAMMA canine sample, DNA was isolated by similar methods from a fibroblast cell line.
Next-Generation sequencing and variant analysis
Solution hybridization exome capture was carried out using the SureSelect Human All Exon System (Agilent Technologies). Manufacturer’s protocol version 1.0 compatible with Illumina paired end sequencing was used, with the exception that DNA fragment size and quality was measured using a 2% agarose gel. Flow cell preparation and 101 bp paired-end read sequencing were carried out as per protocol for the GAIIx sequencer (2) (Illumina Inc). A single 101 base pair paired-end lane on a GAIIx flow cell was used per exome sample to generate sufficient reads to generate the aligned sequence. Image analyses and base calling on all lanes of data were performed using Illumina Genome Analyzer Pipeline software (GAPipeline versions 1.4.0 or greater) with default parameters.
Read mapping, variant calling and annotation
Reads were aligned to a human reference sequence (UCSC assembly hg18, NCBI build 36) using the package called “efficient large-scale alignment of nucleotide databases” (ELAND). Reads that align uniquely were grouped into genomic sequence intervals of about 100 kb, and reads that fail to align were binned with their paired-end mates. Reads in each bin were subjected to a Smith-Waterman-based local alignment algorithm, cross_match using the parameters – min score 21 and – mask level 0 to their respective 100 kb genomic sequence. Genotypes were called at all positions where there were high-quality sequence bases (Phred-like Q20 or greater) using a Bayesian algorithm (Most Probable Genotype – MPG)12.
Filtering Strategy
The filters used in this study included a number of criteria that were implemented using the VarSifter software program for exome and whole genome data management (Teer et al., unpublished). The filters for homozygosity or compound heterozygosity in the proband were used because most metabolic diseases are autosomal recessive and those for mutation type (nonsynonymous, splice, frameshift, and nonsense) were selected because they encompass the majority of disease-causing variants. However, these filters would not detect large deletions, regulatory mutations, or non-canonical splice mutations, which can account for several percent of causative mutations. We also used an exclusion of alleles present in dbSNP, reasoning that the causative variants were uncommon and unlikely to be cataloged there. We used a MAF filter of <10% from a cohort of 258 subjects who were sequenced in our center with similar methodology. We recognized that this frequency was quite liberal, but used it as a starting point, anticipating that it would be adjusted should the initial screen not have yielded a plausible candidate. We reasoned, incorrectly in retrospect, that it was appropriate to filter out variants that were homozygous in controls, as we assumed that no member of the control cohort could have this disease.
Sanger Sequence Analysis
Sequence analysis of ACSF3 was performed using standard methods. Sequencing was performed with v3.1 BigDye terminator cycle sequencing kit (Applied Biosystems) and the ABI 3130 (Applied Biosystems) per the manufacturer’s protocol. Sequence data were compared with the published ACSF3 sequence (GenBank reference number NM_174917.2) using Sequencher 4.10.1 (Gene Codes Corp.). Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1.
Canine ACSF3
As no canine orthologue for ACSF3 was known, the Dog Genome (UCSC browser, May 2005 build) was used to predict the sequence for canine ACSF3 and primers were designed to amplify the exonic regions of the gene. Liver cDNA was obtained (Zyagen) and primers for the predicted dog cDNA were used to amplify the transcript. The dog ACSF3 partial cDNA sequence has been submitted to GenBank, Accession number JF907588.1. DNA from forty unrelated Labrador retrievers was obtained (E. Ostrander, NHGRI) and tested for c.1288G>A, p.Gly430Ser.
Sequence alignment and Bioinformatics
ACSF3 orthologues were identified by BLAST search and through Homologene. Sequence alignment of the ACSF3 orthologues: human NP_001120686.1, mouse NP_659181.2, dog JF907588.1, cow NP_001030240.1, rat XP_574249.3, zebrafish XP_690782.2, Xenopus NP_001086314.1, B. japonicum NP_767149.1 and R. leguminosarum AAC83455.1, was performed by the Clustal W method in MacVector version 9.0.2. The phylogenetic tree was created in MegAlign (Lasergene) by the Clustal W method. Similarity was determined by BLAST-P using the BLOSUM 62 matrix. The mitochondrial leader sequence was predicted using MitoProtII.
Expression of human ACSF3 cDNA in human fibroblast cells
Wild-type ACFS3 cDNA was generated by RT-PCR from total RNA extracted from normal human liver tissue and sequence validated. This gene was cloned into a Gateway (Invitrogen) retroviral expression vector, pLenti6/V5-DEST, as recommended by the manufacturer. The viral constructs express ACSF3 or GFP under the control of the CMV promoter; the backbone also has a blasticidin cassette driven by the E7 promoter. Human fibroblast cell lines were transduced with virus containing either the ACSF3 or GFP. The transduced cells were selected and expanded in DMEM with 5% fetal bovine serum containing 10 μg/ml blasticidin, for selection, prior to propionate loading. ACSF3 with a C-terminal GFP fusion was cloned into pCMV6 and sequence verified. Control fibroblasts were electroporated with 3 μg of plasmid DNA using an Amaxa nucleofector electroporator (Amaxa GmbH, Walkersville, MD). Transfected fibroblasts were grown for 48 hours before immunofluorescence experiments.
MMA production by cultured CMAMMA fibroblasts
A modified chemical stimulation study was performed as described13. Six well tissue culture plates were seeded at a density of 2 or 5×105 per well in high glucose (4 g/L) DMEM supplemented with 10% fetal bovine serum, penicillin streptomycin, L-glutamine and sodium pyruvate. The next day, the DMEM growth media was removed and replaced with 1 ml of DMEM growth media containing sodium propionate at a concentration of 5 mM. After 72 hours the media was collected for GC/MS analysis of MMA.
Western blot analyses
Thirty to forty micrograms of clarified fibroblast extract were analyzed by Western blot using a rabbit polyclonal anti-ACSF3 (ab100860; Abcam) or mouse monoclonal anti-PDH-E2 (MSP05; MitoSciences) at a dilution of 1:1,500. Mouse monoclonal anti-β-actin (ab8226, Abcam) was used as a loading control for immunoblotting at a dilution of 1:1,000. Horseradish peroxidase–conjugated anti-rabbit IgG or anti-mouse IgG (NA934 or NA931; GE Healthcare Life Sciences) was used as the secondary antibody and was visualized with chemiluminescence detection (Pierce Biotechnology).
Enzyme Assay
Full-length ACSF3 containing an N-terminal GST fusion, expressed in wheat germ extract, was obtained from Novus Biologicals and used to assay malonyl- and methylmalonyl-CoA synthetase activity with a previously described spectrophotometric method7. The reaction mixture contained the following components in a volume of 500 μL: 100 mM potassium phosphate buffer (pH 7.0), 8 mM malonate, methylmalonate, or acetate, 2 mM MgCl2, 0.4 mM ATP, 0.2 mM CoA, and 1.43 μg of GST-tagged, purified ACSF3. An increase in absorbance at 232 nm was used to measure the formation of the thioester bond (ε232=4.5 × 10−3 M−1 cm−1) and determine enzyme activity, represented as specific activity (nmol/min/mg) toward the three substrates assayed.
Immunofluorescence
Control fibroblasts transfected with pCMV-ACSF3-GFP and fibroblasts from Subject 4 stably expressing ACSF3 as described above were grown on chamber slides, fixed with 3% paraformaldehyde in 1X PBS, permeabilized with 0.5% Triton X 100 in 1X PBS and blocked in 1% donkey serum, 0.1% saponin and 100 μM glycine in PBS. Fibroblast slides were incubated with rabbit polyclonal ACSF3 antibody (ab100860; Abcam) and mouse monoclonal mitochondrial MTC02 antibody (ab3298; Abcam) in a solution containing 1X PBS, 0.1% BSA and 0.1% saponin overnight at 4 °C. The cells were washed and incubated with donkey anti-rabbit IgG conjugated to Alexa Fluor 555 and donkey anti-mouse IgG conjugated to Alexa Fluor 488 or Alexa Fluor 633 (Invitrogen, Carlsbad, CA) for 1 hour at room temperature. Slides were washed with 1X PBS and mounted with VectaShield containing DAPI. Slides were imaged using a Zeiss LSM 510 META confocal laser-scanning microscope using 488 nm Argon, a 543 nm HeNe and 405 nm lasers (Carl Zeiss, Microimaging Inc., Thornwood, NY) equipped with a Plan-Apochromat 63x/1.4 Oil DIC objective.
Supplementary Material
Acknowledgments
We thank the patients and families for participating; E. Ostrander for the gift of unrelated Labrador DNA samples, K.M. Gibson and R. Wander for performing the malonyl-CoA decarboxylase assay on fibroblasts from Subjects 2 and 9, J. Teer for bioinformatics assistance, S. Suchy for patient referral, M. Podell and W. Gahl for helpful discussions, R. Fisher, I. Bernardini, A. Gropman, L. Hecht, F. Facio, C. Gitiaux, C. Ottolenghi, D. Rabier, G. Touati for laboratory assistance and clinical care, M. Oglesbee and A. Genders searched for dogs related to the affected Labrador retriever.
J.L.S., J.J.J, I.M., R.J.C., N.C.C., S.D.C., J.R.S., H.M.D, M.H., K.O., N.S.H., J.C.S., C.K., L.G.B., and C.P.V. were supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. M.D.G. was supported by NIA K23 AG021989 and R01-AG031189
Footnotes
Author contributions
J.C.S. and L.G.B. developed the exome sequencing protocol, J.L.S., I.M., J.C.S., L.G.B., and C.P.V. designed clinical research studies, NISC performed exome sequencing, J.J.J., J.L.S., I.M., R.J.C., L.G.B. and C.P.V. performed bioinformatic analyses, J.J.J. and C.K. performed mutational analysis and genotyping, R.J.C., N.C.C., S.D.C. and J.R.S. performed cell culture studies and Western blot analyses, R.J.C. created viral vectors and performed cellular complementation studies, J.L.S., M.H., and H.M.D. performed immunofluorecense experiments, J.L.S., J.R.S. and I.M. performed enzymatic assays, J.L.S., I.M., N.S.H., K.O., J.C.S., B.A.B., S.A.B., P.M.J., N.L.C., P.D., V.V., M.D.G., W.L.N., L.G.B. and C.P.V. contributed to clinical evaluations and the delineation of the patient phenotypes, D.K.G. performed organic acid measurements, J.L.S., J.J.J., L.G.B. and C.P.V. prepared manuscript, L.G.B. and C.P.V. conceived and supervised the study.
Competing financial interest
The authors declare no competing financial interests. LGB is an uncompensated advisor to the Illumina Corporation.
URLs
GeneReviews, http://www.ncbi.nlm.nih.gov/books/NBK1231/
ClinicalTrials, http://clinicaltrials.gov
Phrap assembler, http://www.phrap.org
Homologene, http://www.ncbi.nlm.nih.gov/homologene
MitoProtII, http://ihg.gsf.de/ihg/mitoprot.html
Accession numbers
Canine ACSF3, JF907588.1; Human ACSF3, NM_174917.2. The accession numbers for ACSF3 orthologues and ACS homologues are listed in Online Methods.
References
- 1.Gregg AR, Warman AW, Thorburn DR, O’Brien WE. Combined malonic and methylmalonic aciduria with normal malonyl-coenzyme A decarboxylase activity: a case supporting multiple aetiologies. J Inherit Metab Dis. 1998;21:382–90. doi: 10.1023/a:1005302607897. [DOI] [PubMed] [Google Scholar]
- 2.Podell M, et al. Methylmalonic and malonic aciduria in a dog with progressive encephalomyelopathy. Metab Brain Dis. 1996;11:239–47. doi: 10.1007/BF02237961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biesecker LG, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–74. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J Lipid Res. 2007;48:2736–50. doi: 10.1194/jlr.M700378-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Hiltunen JK, Chen Z, Haapalainen AM, Wierenga RK, Kastaniotis AJ. Mitochondrial fatty acid synthesis--an adopted set of enzymes making a pathway of major importance for the cellular metabolism. Prog Lipid Res. 2010;49:27–45. doi: 10.1016/j.plipres.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 6.An JH, Kim YS. A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium trifolii--cloning, sequencing, and expression of the enzymes in Escherichia coli. Eur J Biochem. 1998;257:395–402. doi: 10.1046/j.1432-1327.1998.2570395.x. [DOI] [PubMed] [Google Scholar]
- 7.Koo HM, Kim YS. Identification of active-site residues in Bradyrhizobium japonicum malonyl-coenzyme A synthetase. Arch Biochem Biophys. 2000;378:167–74. doi: 10.1006/abbi.2000.1813. [DOI] [PubMed] [Google Scholar]
- 8.Black PN, Zhang Q, Weimar JD, DiRusso CC. Mutational analysis of a fatty acyl-coenzyme A synthetase signature motif identifies seven amino acid residues that modulate fatty acid substrate specificity. J Biol Chem. 1997;272:4896–903. doi: 10.1074/jbc.272.8.4896. [DOI] [PubMed] [Google Scholar]
- 9.Stuible H, Buttner D, Ehlting J, Hahlbrock K, Kombrink E. Mutational analysis of 4-coumarate:CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Lett. 2000;467:117–22. doi: 10.1016/s0014-5793(00)01133-9. [DOI] [PubMed] [Google Scholar]
- 10.Zou Z, DiRusso CC, Ctrnacta V, Black PN. Fatty acid transport in Saccharomyces cerevisiae. Directed mutagenesis of FAT1 distinguishes the biochemical activities associated with Fat1p. J Biol Chem. 2002;277:31062–71. doi: 10.1074/jbc.M205034200. [DOI] [PubMed] [Google Scholar]
- 11.Sniderman LC, et al. Outcome of individuals with low-moderate methylmalonic aciduria detected through a neonatal screening program. J Pediatr. 1999;134:675–80. doi: 10.1016/s0022-3476(99)70280-5. [DOI] [PubMed] [Google Scholar]
- 12.Teer JK, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–31. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandler RJ, et al. Propionyl-CoA and adenosylcobalamin metabolism in Caenorhabditis elegans: evidence for a role of methylmalonyl-CoA epimerase in intermediary metabolism. Mol Genet Metab. 2006;89:64–73. doi: 10.1016/j.ymgme.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.