Abstract
To elucidate how the deficiency of a major corneal proteoglycan lumican affects corneal homeostasis, we used mass spectrometry to derive the proteome profile of the lumican-deficient and the heterozygous mouse corneas and compared these to the wild type corneal proteome. 2,108 proteins were quantified in the mouse cornea. Selected proteins and transcripts were investigated by western blot and quantitative RT-PCR, respectively. We observed major changes in the composition of the stromal extracellular matrix (ECM) proteins in the lumican-deficient mice. Lumican deficiency altered cellular proteins in the stroma and the corneal epithelium. The ECM changes included increases in fibril forming collagen type I and VI, fibromodulin, perlecan, laminin β2, collagen type IV, nidogen/entactin and anchoring collagen type VII in the Lum+/− and the Lum−/− mouse corneas, while the stromal proteoglycans decorin, biglycan and keratocan were decreased in the Lum−/− corneas. Cellular protein changes included increases in alcohol dehydrogenase, superoxide dismutase and decreases in epithelial cytokeratins 8 and 14. We also detected proteins that are novel to the cornea. The proteomes will provide an insight into the lumican-deficient corneal phenotype of stromal thinning and loss of transparency and a better understanding of pathogenic changes in corneal and ocular dystrophies.
Keywords: Proteomics, Lumican, Mass spectrometry, iTRAQ, Cornea, Collagen
1. Introduction
The healthy cornea is an avascular connective tissue-rich barrier of the eye that is both transparent and refractive for normal vision. The stroma, underlying the stratified epithelium, is a key regulator of corneal transparency and refraction, and it is comprised of specialized mesenchymal cells, the keratocytes and a collagen-rich extracellular matrix (ECM) they produce [1]. Injury, infection and corneal diseases can alter the exquisitely balanced cellular and the ECM content of the cornea and compromise its transparency, refractive power and barrier properties. The primary ECM constituents of the corneal stroma are fibrillar collagen types I, III and V and the small leucine-rich repeat proteoglycans (SLRPs), lumican, decorin, biglycan, keratocan and osteoglycin/mimecan [1]. Another SLRP, fibromodulin, is present in the developing cornea but restricted to the corneal periphery at maturity [2]. While the major protein components of the cornea have been identified, little is known of the corneal proteome, or how it is regulated by the major ECM components. The proteomics field has made significant advances in quantitative protein profiling by using isobaric tags for relative and absolute quantification (iTRAQ) that allow simultaneous analysis of four (iTRAQ 4 plex) or eight (iTRAQ 8 plex) different samples [3]. Thus, recent studies of the ocular surface in health and disease are beginning to incorporate these proteomic approaches [4, 5].
Lumican is present in the interstitial ECM of the cornea, skin, intestinal submucosa, cartilage and bone [6]. Lumican is normally expressed by the mesenchymal fibroblasts, and transiently by the injured epithelia [7, 8]. In vitro collagen fibrillogenesis assays and subsequent in vivo studies show that lumican and the other SLRP members of the cornea bind collagen and regulate collagen fibril growth [9–12]. The lumican-deficient (Lum−/−) mice have cloudy corneas that are 40% thinner than those of the wild type mouse [9, 13]. Our previous studies showed that the loss of corneal transparency in the Lum−/− mice was linked to abnormal collagen fibril architecture and increased light scattering [11, 14, 15]. We further found that wound healing in the Lum−/− corneas was delayed, and the stromal cells showed reduced apoptosis and increased proliferation, suggesting lumican-deficiency to have a broad pleiotropic effect in the cornea [16, 17]. By contrast, deficiencies of the other corneal SLRPs, decorin {Danielson, 1997 #254}, biglycan {Zhang, 2009 #8761}, fibromodulin {Chakravarti, 2003 #2177; Chen, 2010 #6268} and keratocan {Meek, 2003 #9218} present a milder corneal phenotype. Therefore, we selected the lumican-null corneas for an in-depth proteomic analysis, and compared the corneal proteomes of lumican-expressing (Lum+/+, Lum+/−) and lumican-deficient (Lum−/−) mice. We performed a multiplexed relative quantification of proteins by LC-MS/MS mass spectrometry using three different iTRAQ tags for Lum+/+, Lum+/− and Lum−/− corneal protein extracts. About 2,108 proteins were identified and quantified in the corneal extracts. These included several known cellular proteins, extracellular matrix collagens and proteoglycans, as well as proteins not previously linked to the cornea. Absence of one or both Lum alleles was associated with increases in several oxidative stress-related proteins, increased collagen type I, VI and decreases in the lumican-related SLRPs, decorin, biglycan and keratocan.
2. Materials and Methods
2.1. Materials
The enzyme chondroitinase ABC was purchased from Associates of CAPE Cod incorporated. TCEP (Tris (2-carboxyethyl) phosphine), cysteine blocking agent (methyl methanethiosulfonate) and SYBR Green master mix were purchased from Applied Biosystems. Sequencing grade trypsin was from Promega. The oligonucleotide primers for PCR analyses were purchased from Eurofins Mwg Operon. KH2PO4, acetonitrile, KCl, formic acid, guanidine-HCl, sodium acetate, Tris-HCl, trifluoroacetic acid (TFA) and NaCl were from Sigma. TRIZOL and SuperScript III first-strand synthesis system were from Invitrogen.
Rabbit anti-decorin (LF113) and anti-biglycan (LF159) were kindly provided by Dr. L. Fisher (NIH-NICDR). The following antibodies and reagents were purchased from Santa Cruz Biotech: goat anti-rabbit and anti-goat IgG-perixodase, and antibodies against β-Actin, Krt8, IGFBP2. The anti-Adh1 was obtained from Cell Signaling and Aldh1a1 from Abcam. The chemiluminescent HRP antibody detection reagent was obtained from Denville Scientific Inc. The source of the anti-collagen antibodies were as follows: Col6a1 from Dr. Monli Chu (Thomas Jefferson University), Col12 and Col14 were kindly provided by Dr. Manuel Koch (University of Cologne).
2.2. Protein sample preparation for mass spectrometry
Lum+/+, Lum+/− and Lum−/− mice were generated as we described before [9]. All animals were housed in a specific pathogen-free mouse facility at Johns Hopkins University, according to protocols approved by the Animal Care and Use Committee. For the proteomics study, eight corneas per genotype were harvested from 8 week-old mice, rinsed in ice-cold PBS with protease inhibitor, and then frozen in liquid nitrogen (2.0 ml Biomasher, USA Scientific). After adding 150 μl of 0.5% SDS, the samples were centrifuged at 15,000 g for 30 sec, sonicated for 1 min and centrifuged again at 15,000 g for 10 min. Since many of the commercially available cocktail protease inhibitors contain free amino groups, which compete for labeling with iTRAQ reagents, we did not include any protease inhibitors in the lysis buffer. Protease activity was minimized by lysing the samples directly in 0.5% SDS and subjecting the samples to ultrasonication, which is expected to cause denaturation and solubilization of proteins. Moreover, the samples were maintained on ice until the addition of trypsin to minimize any residual endogenous protease activity. The protein concentration in the supernatant was measured using the Bradford assay kit (Bio-Rad) and further confirmed by SDS-PAGE.
2.3. iTRAQ labeling
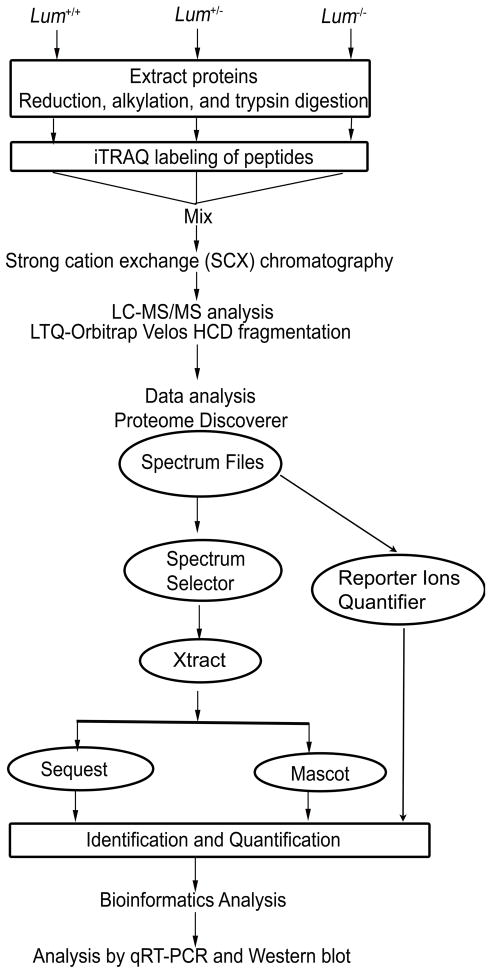
The Lum+/+, Lum+/− and Lum−/− mouse corneal protein samples were differentially labeled using the iTRAQ reagent from Applied Biosystems (Fig. 1). Briefly, 70 μg of protein was treated with 2 μl of reducing agent TCEP at 60°C for 30 min; and alkylated with 1 μl of cysteine blocking agent at room temperature for 10 min. Subsequently, the samples were diluted in iTRAQ dissolution buffer to reach a final SDS concentration of 0.025%. The protein samples were digested with 5 μg of sequencing grade trypsin for 12 h at 37 °C. The Lum+/+, Lum+/− and Lum−/− digested samples, in a final volume of 35 μl were labeled with the iTRAQ reagents 114, 115 and 116 respectively, in 70 μl of ethanol for 2 h at room temperature, and the reactions were terminated by adding 100 μl water. The dried samples were reconstituted in a strong cation exchange (SCX) solvent (10 mM potassium phosphate buffer, pH 2.85, in 25% acetonitrile), combined and adjusted to the final volume of 1ml by solvent A (5 mM KH2PO4, 30% acetonitrile, pH 2.7). The combined mixture was fractionated by SCX chromatography on a Polysulfoethyl A column (PolyLC, USA), using Agilent 1100 HPLC system. The peptides were fractionated by a linear gradient between solvent A and solvent B (5 mM KH2PO4, 30% acetonitrile, 350 mM KCl, pH 2.7) to obtain 24 fractions. Each fraction was desalted by a C18 column, eluting with 0.1% TFA in 60% acetonitrile. Finally the fractions were dried, resuspended in fresh 0.1% formic acid and replicate aliquots of each fraction were analyzed by mass spectrometry.
Fig. 1.
An outline of sample preparation and iTRAQ labeling for LC-MS/MS.
2.4. Nanoflow electrospray ionization tandem mass spectrometry (MS) analysis
Mass spectrometry of the peptide fractions was carried out on an LTQ Orbitrap Velos (Thermo Fisher Scientific), interfaced with an Agilent 1200 nanoflow liquid chromatography system. The fractions were enriched (5 μl/min) on a trap column (5 μm, 100 Å, 75 μm × 2 cm, Magic C18 AQ Michrom Bioresources), and then separated on an analytical column (5 μm, 100 Å, 75 μm × 10 cm, Magic C18 AQ Michrom Bioresources) with the delivery of nanoflow solvent. The resolutions of precursor and product ion scans were 60,000 and 7,500 at m/z 400, respectively. The twenty most abundant peptides were selected for data dependent MS/MS analysis. Higher-energy collision dissociation mode (HCD) was used for MS/MS scans.
2.5. MS data analysis
The MS data was analyzed using the Proteome Discoverer 1.2 software workflow (Thermo Fisher Scientific). This software workflow consists of a spectrum selector and a reporter ion quantifier which include Mascot and Sequest search nodes. The data was also processed using Xtract feature of the Proteome Discoverer under the Mascot search component of the workflow. For both nodes, the same search parameters were selected, which include iTRAQ label at tyrosine, oxidation of methionine, deamidation at aspargine and glutamine as variable modifications. iTRAQ label at the N-terminus and lysine, methylthio label at cysteine were used as fixed modifications. The data was searched against the NCBI Refseq mouse protein database containing 31,183 proteins for analysis. Using the Proteome Discoverer workflow, the data from Mascot and Sequest search nodes were merged to obtain average values from replicates. The reporter ion window tolerance and target false discovery rate (FDR) were fixed at 10 ppm and 0.01, respectively. The precursor range was set at 600 to 8,000 Da. The results were analyzed by Proteome Discoverer software, and unique peptide(s) was used to calculate accurate relative protein content. The peptide and protein data were extracted using high peptide confidence and top one peptide rank filters. The FDR was calculated by enabling the peptide sequence analysis using decoy database. Peptide levels with ≤ 1% FDR were included in our analysis. The average ratio and percentage variability were used for protein quantification wherever multiple peptides were identified for a protein. The NCBI (www.ncbi.nlm.nih.gov/) and the GeneCards data base (www.genecards.org/ Crown Human Genome Center, Department of Molecular Genetics, the Weizmann Institute of Science) were used for gene annotation and functional categorization of the proteins detected by mass spectrometry.
2.6. Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using the TRIZOL reagent from eight corneas of Lum+/+, Lum+/− and Lum−/− mice. First-strand cDNA was prepared using the SuperScript III first-strand synthesis system, with 5 ng/μl random hexamers. The gene transcripts were measured by qRT-PCR using the SYBR Green master mix. The sense and antisense primers were mixed to obtain a final concentration of 2 pg/μl for each reaction. All primer information is listed in Table 1. Amplification was performed using the following PCR program: 50 °C for 2 min, 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 sec, 60 °C for 1 min. Control reaction mixtures without reverse transcriptase were included in all experiments. All reactions were normalized against the mouse Gapdh gene qRT-PCR. The threshold cycle (Ct) for each reaction was determined using the Applied Biosystems software. At least two independent RT-PCR reactions were carried out for each gene. The results are presented as expression relative to Gapdh. Relative expression is defined as 2−ΔCT, where the threshold cycle difference is (Ct of gene - Ct of Gapdh).
Table 1.
Sequence of primers
| Name of Primers | Sequence of primers (From 5′ to 3′) |
|---|---|
| Dpt-F | ACGCGGCGAGCCTAAACGTT |
| Dpt-R | AGGGCCGGCATGGAATGGTACC |
| CalML3-F | TCGTCAGTGCAGCCGAGCTG |
| CalML3-R | GCACCTGGCTGTGCCAGTCT |
| Efemp1-F | GCCCGGGCAGGAAAGTTGCG |
| Efemp1-R | ATCCATCGGTGCATTGCGTGTATG |
| Rab8A-F | GTCCAGCGACCAGTGGCGAGG |
| Rab8A-R | GGCCGTGTCCCATATCTGCAGTTTA |
| Bgn-F | GCACCTCTACGCCCTGGTCTTG |
| Bgn-R | TCCGCAGAGGGCTAAAGGCCT |
| Kera-F | GCTCTTCTTGGTGACCAGTCATCC |
| Kera-R | GGTTGCCATTACAGCACCTTGCT |
| Fmod-F | TCCAGGGCAACAGGATCAATGAGTT |
| Fmod-R | TGCGCTGCGCTTGATCTCGT |
| Col1a1-F | TGCTGGCCCCAAGGGTCCTT |
| Col1a1-R | GGCTGCCAGGACTGCCAGTG |
| Dcn-F | TCTTGGGCTGGACCATTTGAA |
| Dcn-R | CATCGGTAGGGGCACATAGA |
| Fbn1-F | CCGTGGCGAATGCATCGACGT |
| Fbn1-R | CGCGTGACATGAAAGCCCGC |
| Krt14-F | AGATCGCCACCTACCGCCGT |
| Krt14-F | TGGTGGAGGTCACATCTCTGGATG |
| Krt 8-F | AGATTGAAGCCCTCAAAGGCCAGA |
| Krt 8-R | CATCCCAGACTCCAGCCTGCTCTC |
| Gapdh-F | TGGTCCCAGGATCTTACAGAA |
| Gapdh-R | TTGTCTCCTGCGACTTCA |
2.7. Proteoglycan and collagen extraction and western blot
To extract proteoglycans, the mouse corneas were homogenized in 20 fold weight/volume of freshly prepared 4 M guanidine-HCl, 50 mM sodium acetate, pH 5.8, containing the HALT proteinase inhibitor cocktail (Sigma), and incubated with shaking at 4 °C for 48 h. After centrifuging at 14,000 g for 20 min at 4 °C, the samples were dialyzed by the Slide-A-Lyzer Dialysis Cassette (Thermo Scientific, MW cut off 10 KD) against 150 mM Tris-HCl, 150 mM NaCl, pH 7.3. The BCA Protein Assay Kit (Thermo Scientific) was used to determine protein concentration. The extracts were digested with chondroitinase ABC at 0.001 units/10–100 μg of tissue for 24 h at 37 °C to remove glycosaminogycan side chains. Samples (10 μg each) were resolved in 10% bis acrylamide gels electro blotted using a Bio-Rad system at 30 V overnight at 4 °C. For the collagens, 12 corneas were extracted in 1% SDS in 62.5 mM Tris-HCl, pH 6.8 overnight at 4 °C. The extracted material was centrifuged at 14,000 g and the supernatant boiled in reducing SDS sample loading buffer for electrophoresis in a 4–12% Bis-Tris gel at 200 V for 1 h. To extract cellular proteins, 8 corneas were homogenized in T-PER detergent containing HALT proteinase inhibitor. The gels were electro blotted as described above. Primary antibodies were used at the following concentrations: anti-Dcn and Bgn at 1:100, Aldh1a1-1:1,000, Krt8- 1:500, Krt14-1:500, Igfbp2-1:500, Adh1-1:1,000, β-Actin-1:500, Col12-1:3,000, Col14-1:3,000 or Col6a1-1:500. Goat anti-rabbit and anti-goat IgG-perixodase were used as secondary antibodies at 1:5,000 with the HygloQuick spray for 1 min.
3. Results
3.1. MS analysis of iTRAQ-labeled peptides
Analysis of the 24 iTRAQ labeled peptide fractions from each replicate on the LTQ-Orbitrap Velos generated ~1.2 million MS/MS spectral search inputs resulting in ~130,000 peptide spectral matches. A total of 2,173 unique proteins were identified from two replicate analyses at the 1% FDR level. The proteins with shared peptides were ranked based on the score and the peptides were grouped to remove redundancy. Further, the proteins with equal scores were ranked based on their sequence coverage. Differentially regulated proteins were preferentially selected based on two or more unique peptides and presence in both replicates.
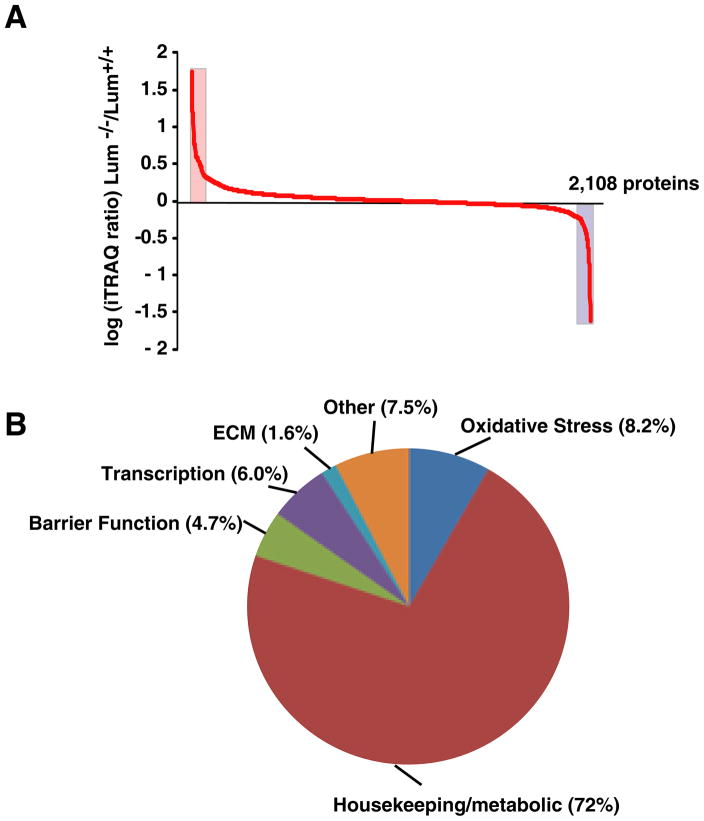
A total of 2,173 proteins were identified. Among these 2,108 proteins could be quantified, the remaining had low reporter ions and were not investigated further (Supplemental Table 1 and Fig. 2A). Following a functional classification that is relevant to the cornea [18, 19], we grouped all the proteins detected into five broad functional categories: housekeeping/metabolic, barrier integrity, redox/oxidative stress, extracellular matrix, transcription/gene regulation and unknown. Housekeeping proteins, including metabolic and soluble enzymes, comprise the largest fraction (72%) of all the proteins detected, while the remaining functional groups contain 2 to 8% of the proteins (Fig. 2B). The overall status of these 2,108 proteins in the Lum+/− and Lum−/− compared to the wild type corneas is shown in Table 2. In the Lum+/− and Lum−/− compared to the wild type corneas, not surprisingly the largest group of 1640 proteins were unchanged, while 113 and 47 were increased and decreased, respectively (Table 2). Without any change in Lum+/−, another 100 and 86 were decreased and increased, respectively in the Lum−/− corneas. With no change in Lum−/−, 41 were decreased and another 70 increased in Lum+/−. Only 11 proteins showed opposite trends in the Lum+/− and Lum−/− extracts compared to the controls. However, the fold changes were too small to consider these to be of significance. The complete set of raw mass spectrometry data (.raw files) generated from this study is available through the Tranche server (http://proteomecommons.org/tranche). The raw data files used for genome annotation may be retrieved using the stable URL https://proteomecommons.org/dataset.jsp?i=75719 and 75726.
Fig. 2.
Overall protein changes in Lumican-deficient (Lum+/− and Lum−/−) compared to wild type mouse corneas. (A) Log ratio of fold increase and decrease in Lum−/− corneas compared to Lum+/+ corneas. Highlighted areas of the graph show significant changes in proteins; (B) The 2,108 identified proteins were classified into 5 broad functional categories. The largest “housekeeping” category includes, housekeeping cellular proteins, metabolic and catabolic enzymes and many soluble proteins that are often described as corneal crystallins.
Table 2.
Protein changes in lumican-deficient compared to wild type mouse corneas.
| Lum−/−/Lum+/+ | Lum+/−/Lum+/+ | ||
|---|---|---|---|
| No change (0.8 – 1.4 fold) | Decreased (≤ 0.8 fold) | Increased (≥1.4 fold) | |
| No change (0.8 – 1.4 fold) | 1640 | 41 | 70 |
| Decreased (≤ 0.8 fold) | 100 | 47 | 6 |
| Increased (≥1.4 fold) | 86 | 5 | 113 |
3.2. Protein changes in Lum+/− and Lum−/− mouse corneas
Considering an increase of ≥ 1.5 fold in any one sample compared to the wild type we identified 78 proteins (Table 3). Approximately 38 of these showed increases in both Lum+/− and Lum−/− corneas. These included metabolic enzymes such as alcohol dehydrogenase 1, detoxification and antioxidant related S-formyl glutathione hydrolase and growth-inhibitory metallothionein-2. Using a cutoff of ≤ 0.6 fold change in any one (Lum+/− or Lum−/−) sample compared to the wild type cornea, we obtained 43 proteins that were decreased (Table 4). Unlike the up regulated proteins, most of the decreases occurred primarily in the absence of both Lum alleles. These included proteins related to oxidative stress, peroxiredoxin 3 and flavin reductase, immune-related complement C3 and the cytokeratins 8, 14 and 19.
Table 3.
Increased proteins at ≥ 1.5 fold level
| Gene Symbol | Description | Experiment 1* | Experiment 2* | ||
|---|---|---|---|---|---|
| Lum+/− | Lum−/− | Lum+/− | Lum−/− | ||
| 1810074P20Rik | Hypothetical protein LOC67490 | 1.4 | 5.5 | ND | ND |
| Acsm1 | Acyl-coenzyme A synthetase ACSM1, mitochondrial | 15.8 | 7.2 | ND | ND |
| Adh1 | Alcohol dehydrogenase 1 | 5.0 | 5.2 | 6.6 | 6.3 |
| Aebp1 | adipocyte enhancer-binding protein 1 | 1.3 | 3.5 | 1.4 | 3.2 |
| Baiap2l2 | Brain-specific angiogenesis inhibitor 1-associated | 5.6 | 1.9 | 9.2 | 3.8 |
| Banf1 | Barrier-to-autointegration factor | 4.6 | 4.0 | 9.0 | 7.7 |
| BC016423 | Peripherial benzodiazepine receptor associated | 2.3 | 15.7 | 2.2 | 15.0 |
| Capn2 | Calpain-2 catalytic subunit | 1.6 | 1.3 | 1.2 | 1.4 |
| Col1a1 | Collagen alpha-1(I) chain | 4.5 | 2.6 | 4.5 | 2.7 |
| Col1a2 | Collagen alpha-2(I) chain | 4.0 | 2.4 | 4.1 | 2.4 |
| Col2a1 | Collagen alpha-1(II) chain isoform 2 | 4.2 | 2.6 | 4.0 | 2.4 |
| Col4a2 | Collagen alpha-2(IV) chain | 1.5 | 3.0 | 1.4 | 3.2 |
| Col5a3 | Collagen, type V, alpha 3 | 2.5 | 1.8 | 1.4 | 1.4 |
| Col6a1 | Collagen alpha-1(VI) chain | 2.5 | 2.0 | 2.4 | 1.9 |
| Col6a2 | Collagen alpha-2(VI) chain | 2.3 | 1.9 | 2.0 | 1.7 |
| Col7a1 | Collagen alpha-1(VII) chain | 2.3 | 2.1 | 2.3 | 2.0 |
| Cox7b | Cytochrome c oxidase subunit 7B, mitochondrial | 1.1 | 1.9 | 1.1 | 2.0 |
| Cyp2f2 | Cytochrome P450 2F2 | 2.4 | 1.6 | 1.9 | 1.3 |
| Dak | ATP-dependent dihydroxyacetone kinase | 1.8 | 1.5 | 2.2 | 2.9 |
| Dpt | Dermatopontin | 2.2 | 1.6 | 2.5 | 2.0 |
| Efemp1 | EGF-containing fibulin-like ECM protein 1 | 2.2 | 1.7 | 2.5 | 2.3 |
| Emilin1 | EMILIN-1 | 1.1 | 1.9 | 1.2 | 2.3 |
| Esd | S-formylglutathione hydrolase | 1.7 | 1.4 | 3.1 | 2.6 |
| Fancf | Fanconi anemia, complementation group F | 1.7 | 10.1 | 1.2 | 5.9 |
| Fbln5 | Fibulin-5 | 2.2 | 1.8 | 2.2 | 1.8 |
| Fbn1 | Fibrillin-1 | 2.9 | 2.5 | 2.5 | 2.4 |
| Fmod | Fibromodulin | 1.6 | 3.0 | 1.6 | 3.1 |
| Gm10131 | Similar to human protein homologous to DROER | 2.8 | 2.8 | 2.5 | 2.0 |
| Gm11575 | PREDICTED: similar to SEC61 gamma | 1.9 | 1.6 | 2.4 | 2.1 |
| Gm9769 | PREDICTED: similar to Sid3177p | 19.3 | 16.2 | 13.3 | 11.6 |
| Hspg2 | Perlecan | 1.8 | 2.2 | 1.8 | 2.1 |
| Krt31 | Keratin, type I cuticular Ha1 | 0.7 | 3.8 | ND | ND |
| Lama5 | Laminin subunit alpha-5 | 1.6 | 2.2 | 1.0 | 1.6 |
| Lamb1 | Laminin subunit beta-1 | 1.2 | 1.7 | 1.1 | 2.0 |
| Lamb2 | Laminin subunit beta-2 | 1.6 | 2.2 | 1.8 | 2.2 |
| Lamc1 | Laminin subunit gamma-1 | 1.2 | 1.9 | 1.4 | 1.8 |
| Las1l | LAS1-like | 2.2 | 33.1 | 1.5 | 22.9 |
| LOC100041471 | PREDICTED: hypothetical protein | 3.1 | 2.2 | 3.3 | 3.1 |
| LOC100045439 | Similar to testis-specific adriamycin sensitivity protein | 1.7 | 2.8 | 1.1 | 2.3 |
| LOC100046627 | PREDICTED: hypothetical protein | 1.5 | 4.6 | 1.8 | 3.4 |
| LOC100047777 | PREDICTED: hypothetical protein | 3.5 | 3.1 | 3.1 | 2.9 |
| LOC100048339 | PREDICTED: similar to ribosomal protein L36a | 6.9 | 6.5 | 4.4 | 4.0 |
| LOC100048552 | PREDICTED: hypothetical protein | 2.1 | 1.9 | 2.6 | 2.2 |
| LOC639119 | PREDICTED: hypothetical protein | 4.4 | 2.7 | 5.5 | 3.3 |
| Lox | Protein-lysine 6-oxidase | 6.0 | 4.9 | 2.7 | 2.2 |
| Ltbp4 | Latent-transforming growth factor β-binding protein 4 | 1.0 | 4.9 | 1.3 | 2.8 |
| Lypd3 | Ly6/PLAUR domain-containing protein 3 | 4.4 | 4.0 | 4.9 | 4.1 |
| Marcks | Myristoylated alanine-rich C-kinase substrate | 2.0 | 4.2 | ND | ND |
| Mcm3 | DNA replication licensing factor MCM3 | 1.8 | 1.7 | 1.2 | 1.4 |
| Mmp2 | 72 kDa type IV collagenase | 1.7 | 1.7 | 1.2 | 1.4 |
| Mosc2 | MOSC domain-containing protein 2, mitochondrial | 2.3 | 2.2 | 2.3 | 2.3 |
| Mt2 | Metallothionein-2 | 5.9 | 5.9 | 5.7 | 5.7 |
| Mtr | Methionine synthase | 2.4 | 11.8 | 3.1 | 14.9 |
| Ncbp1 | Nuclear cap-binding protein subunit 1 | 1.4 | 1.2 | 2.0 | 1.5 |
| Ndufb6 | NADH dehydrogenase 1 β subcomplex subunit 6 | 1.5 | 4.6 | 1.5 | 3.3 |
| Nes | Nestin | 1.3 | 2.0 | 1.3 | 1.9 |
| Nid1 | Nidogen-1 | 1.4 | 1.8 | 1.4 | 1.9 |
| Nid2 | Nidogen 2 | 1.7 | 2.2 | 1.9 | 2.1 |
| Npnt | Nephronectin isoform b | 2.2 | 5.0 | 1.5 | 3.2 |
| Ppp2cb | Serine/threonine phosphatase 2A catalytic subunit β | 2.3 | 1.2 | ND | ND |
| Prpf8 | Pre-mRNA-processing-splicing factor 8 | 5.9 | 5.8 | ND | ND |
| Psmd13 | 26S proteasome non-ATPase regulatory subunit 13 | 2.2 | 1.9 | ND | ND |
| Rab5a | Ras-related protein Rab-5A | 1.4 | 2.4 | 1.2 | 1.1 |
| Rab6 | Ras-related protein Rab-6A isoform 2 | 1.2 | 2.6 | ND | ND |
| Rhobtb2 | Rho-related BTB domain-containing protein 2 | 2.2 | 2.0 | 2.4 | 2.1 |
| Rpe | Ribulose-phosphate 3-epimerase | 2.4 | 4.5 | 1.6 | 2.2 |
| Rpl18a | 60S ribosomal protein L18a | 2.0 | 1.6 | 1.9 | 1.6 |
| Scfd1 | Sec1 family domain-containing protein 1 | 1.1 | 2.5 | ND | ND |
| Sod1 | Superoxide dismutase | 1.2 | 1.9 | 1.3 | 2.0 |
| Sp1 | Transcription factor Sp1 | 1.4 | 2.2 | ND | ND |
| Tgfbi | Transforming growth factor-beta-induced protein ig-h3 | 2.0 | 1.6 | 2.0 | 1.6 |
| Thbs4 | Thrombospondin-4 | 1.8 | 1.3 | 1.9 | 1.2 |
| Tinagl1 | Tubulointerstitial nephritis antigen-like | 2.3 | 2.3 | 1.4 | 1.3 |
| Tmem165 | Transmembrane protein 165 | 0.7 | 0.8 | ND | 2.6 |
| Tsta3 | GDP-L-fucose synthetase | 1.7 | 1.3 | 1.2 | 1.4 |
| Txn1 | Thioredoxin | 1.8 | 1.9 | 2.1 | 1.8 |
| Wt1 | Wilms tumor protein homolog | 0.9 | 7.6 | 0.9 | 7.3 |
Fold change compared Lum+/+
Table 4.
Decreased proteins at ≤ 0.6 fold level
| Gene Symbol | Description | Experiment 1* | Experiment 2* | ||
|---|---|---|---|---|---|
| Lum+/− | Lum−/− | Lum+/− | Lum−/− | ||
| 2700060E02Rik | Hypothetical protein LOC68045 | 0.6 | 0.6 | 0.8 | 0.7 |
| 9930032O22Rik | Serine protease Desc4 | 0.7 | 0.6 | 0.6 | 0.7 |
| Ahsg | Alpha-2-HS-glycoprotein | 0.9 | 0.5 | 0.9 | 0.6 |
| Alb | Albumin | 0.9 | 0.5 | 0.8 | 0.5 |
| Apod | Apolipoprotein D | 1.2 | 0.6 | 1.1 | 0.6 |
| Aqp3 | Aquaporin-3 | 0.7 | 0.7 | 0.8 | 0.8 |
| Blvrb | Flavin reductase | 1.0 | 0.7 | 1.2 | 0.6 |
| C3 | Complement C3 | 1.0 | 0.6 | 1.0 | 0.6 |
| Farsb | Phenylalanyl-tRNA synthetase beta chain | 1.1 | 0.9 | 1.0 | 0.7 |
| Gnas | Protein ALEX XXLb1 | 0.1 | 0.3 | 0.1 | 0.3 |
| Got2 | Aspartate aminotransferase, mitochondrial | 0.9 | 0.6 | 0.9 | 0.7 |
| Hdlbp | Vigilin | 0.7 | 0.8 | 1.0 | 0.9 |
| Hpx | Hemopexin | 1.0 | 0.6 | 1.2 | 0.9 |
| Hrg | Histidine-rich glycoprotein | 1.1 | 0.6 | 1.1 | 0.7 |
| Itih1 | Inter-alpha-trypsin inhibitor heavy chain H1 | 1.1 | 0.8 | 0.9 | 0.7 |
| Kera | Keratocan | 1.0 | 0.6 | 1.0 | 0.6 |
| Khsrp | Far upstream element-binding protein 2 | 1.2 | 0.6 | 0.9 | 0.9 |
| Klc3 | Kinesin light chain 3 | 1.1 | 0.7 | 0.1 | 0.7 |
| Kng1 | Kininogen-1 isoform 2 | 1.0 | 0.5 | 1.0 | 0.5 |
| Krt14 | Keratin, type I cytoskeletal 14 | 0.9 | 0.6 | 1.0 | 0.9 |
| Krt19 | Keratin, type I cytoskeletal 19 | 0.7 | 0.7 | 0.5 | 0.5 |
| Krt8 | Keratin, type II cytoskeletal 8 | 0.2 | 0.2 | 0.3 | 0.4 |
| Lgals7 | Galectin-7 | 0.9 | 0.7 | 0.9 | 0.6 |
| LOC100044537 | Similar to Group specific component | 0.9 | 0.6 | 0.9 | 0.6 |
| LOC100046668 | Similar to yeast ribosomal protein S28 homologue | 0.9 | 0.7 | 0.9 | 0.7 |
| LOC100047628 | LOC100047628 | 0.9 | 0.5 | 0.8 | 0.6 |
| LOC639606 | Hypothetical protein isoform 1 | 1.0 | 0.4 | 1.0 | 0.4 |
| Lrg1 | Leucine-rich alpha-2-glycoprotein | 0.9 | 0.4 | 1.0 | 0.7 |
| Lum | Lumican | 0.8 | 0.1 | 0.8 | 0.1 |
| Nmi | N-myc-interactor | 0.6 | 0.7 | 0.7 | 0.8 |
| Picalm | Phosphatidylinositol-binding clathrin assembly protein | 0.5 | 0.5 | 0.9 | 0.7 |
| Polr2e | DNA-directed RNA polymerases I, II, and III subunit RPABC1 | 0.9 | 0.8 | 1.1 | 0.5 |
| Ppp4c | Serine/threonine-protein phosphatase 4 catalytic subunit | 1.0 | 0.5 | 1.0 | 0.7 |
| Prdx3 | Thioredoxin-dependent peroxide reductase, mitochondrial | 1.0 | 0.6 | 0.9 | 0.6 |
| Rab8a | Ras-related protein Rab-8A | 1.1 | 0.6 | 1.0 | 0.7 |
| Rbm47 | RNA-binding protein 47 | 2.6 | 0.4 | 0.4 | 0.8 |
| Rps24 | 40S ribosomal protein S24 isoform 1 | 0.8 | 0.6 | 0.9 | 0.8 |
| Serpina1d | Alpha-1-antitrypsin 1–4 | 1.0 | 0.6 | 1.0 | 0.6 |
| Sh3bp1 | SH3 domain-binding protein 1 | 0.6 | 0.4 | 1.0 | 0.7 |
| Spcs2 | Signal peptidase complex subunit 2 | 0.8 | 0.6 | 0.8 | 0.8 |
| Tmx3 | Protein disulfide-isomerase TMX3 | 0.6 | 0.6 | 0.5 | 0.7 |
| Trf | Serotransferrin | 0.9 | 0.6 | 0.9 | 0.7 |
Fold change compared Lum+/+
Several extracellular matrix proteins were altered in response to lumican deficiency (Table 5). Basement membrane proteins perlecan, nidogen/entactin, laminin β2 and Collagen type VII (α1 chain) were elevated in the Lum+/− and the Lum−/− corneas. In the interstitial stromal ECM, the known corneal SLRPs decorin, biglycan and keratocan were decreased in the Lum−/− corneas. Two other SLRPs, podocan and PRELP, not known to be associated with the cornea, were slightly increased in the Lum−/− corneas. Fibromodulin, another SLRP, known to be present in the corneal-scleral junction [2], was markedly increased in the Lum−/− corneas. Certain corneal collagens, fibril-forming collagen type I and V, and collagen type VI (Col6a1 and Col6a2), another widely present form, were increased in both Lum+/− and Lum−/− compared to Lum+/+ mice. Other ECM proteins, fibrillin-1 and TGFβ I, also showed some lumican-dosage effects, and were increased in the Lum+/− and the Lum−/− corneas. The collagen type II, a component of cartilaginous tissues, and not known to be present in mammalian corneas, was detected by MS and increased with lumican deficiency. The associated unique peptide (LTGPIGPPGPAGANGEK) used for its identification clearly relates to Col2a1 and suggests its presence in mammalian corneas.
Table 5.
Corneal extracellular matrix proteins
| Gene Symbol | Description | Experiment 1* | Experiment 2* | ||
|---|---|---|---|---|---|
| Lum+/− | Lum−/− | Lum+/− | Lum−/− | ||
| Bgn | Biglycan | 0.98 | 0.70 | 1.03 | 0.74 |
| Col12a1 | Collagen alpha-1(XII) chain | 1.34 | 1.30 | 1.31 | 1.28 |
| Col14a1 | Collagen alpha-1(XIV) chain | 1.28 | 1.45 | 1.18 | 1.38 |
| Col17a1 | Collagen alpha-1(XVII) chain | 0.92 | 0.80 | 0.81 | 0.74 |
| Col18a1 | Collagen alpha-1(XVIII) chain isoform 2 | 1.21 | 1.14 | ND | ND |
| Col1a1 | Collagen alpha-1(I) chain | 4.45 | 2.56 | 4.51 | 2.66 |
| Col1a2 | Collagen alpha-2(I) chain | 4.02 | 2.39 | 4.11 | 2.41 |
| Col2a1 | Collagen alpha-1(II) chain isoform 2 | 4.17 | 2.63 | 4.00 | 2.41 |
| Col3a1 | Collagen alpha-1(III) chain | 5.06 | 3.95 | ND | ND |
| Col4a2 | Collagen alpha-2(IV) chain | 1.55 | 2.99 | 1.44 | 3.16 |
| Col4a4 | Collagen alpha-4(IV) chain | 1.44 | 3.33 | 1.39 | 3.43 |
| Col5a1 | Collagen alpha-1(V) chain | 1.67 | 1.29 | 1.63 | 1.32 |
| Col5a2 | Collagen alpha-2(V) chain | 1.57 | 0.84 | 1.58 | 0.91 |
| Col5a3 | Collagen, type V, alpha 3 | 2.48 | 1.81 | 1.39 | 1.44 |
| Col6a1 | Collagen alpha-1(VI) chain | 2.50 | 1.95 | 2.39 | 1.89 |
| Col6a2 | Collagen alpha-2(VI) chain | 2.32 | 1.85 | 2.05 | 1.72 |
| Col7a1 | Collagen alpha-1(VII) chain | 2.27 | 2.07 | 2.31 | 2.00 |
| Dcn | Decorin | 0.99 | 0.74 | 0.96 | 0.73 |
| Fmod | Fibromodulin | 1.62 | 3.00 | 1.62 | 3.10 |
| Hspg2 | Basement membrane-specific perlecan | 1.76 | 2.20 | 1.78 | 2.09 |
| Kera | Keratocan | 0.98 | 0.61 | 0.98 | 0.61 |
| Lama2 | Laminin subunit alpha-2 | 1.62 | 1.92 | 1.57 | 1.74 |
| Lama3 | Laminin subunit alpha-3 | 1.17 | 1.17 | 1.07 | 1.10 |
| Lama5 | Laminin subunit alpha-5 | 1.58 | 2.16 | 1.00 | 1.56 |
| Lamb1 | Laminin subunit beta-1 | 1.18 | 1.69 | 1.09 | 2.00 |
| Lamb2 | Laminin subunit beta-2 | 1.65 | 2.25 | 1.77 | 2.25 |
| Lamb3 | Laminin subunit beta-3 | 1.21 | 1.43 | 1.28 | 1.35 |
| Lamc1 | Laminin subunit gamma-1 | 1.22 | 1.92 | 1.40 | 1.84 |
| Lamc2 | Laminin subunit gamma-2 | 1.26 | 1.46 | 1.35 | 1.34 |
| Lum | Lumican | 0.80 | 0.13 | 0.83 | 0.11 |
| Muc4 | Mucin-4 | 0.79 | 0.82 | 0.76 | 0.81 |
| Nid1 | Nidogen-1 | 1.40 | 1.79 | 1.41 | 1.93 |
| Nid2 | Nidogen 2 | 1.71 | 2.16 | 1.88 | 2.11 |
| Podn | Podocan | 1.18 | 1.38 | ND | ND |
| Prelp | Prolargin | 1.21 | 1.21 | 1.23 | 1.28 |
| Tgfbi | Transforming growth factor-beta-induced protein ig-h3 | 2.02 | 1.63 | 2.00 | 1.56 |
| Thbs1 | Thrombospondin-1 | 1.28 | 1.06 | 1.23 | 0.99 |
| Thbs2 | Thrombospondin 2 | 0.98 | 1.29 | 1.00 | 1.16 |
| Thbs3 | Thrombospondin-3 | 1.21 | 0.93 | 1.05 | 0.80 |
| Thbs4 | Thrombospondin-4 | 1.77 | 1.27 | 1.94 | 1.22 |
| Tnmd | Tenomodulin | 3.56 | 1.86 | 2.51 | 1.58 |
Fold change compared Lum+/+
3.3. Quantitative Real-Time PCR (qRT-PCR) of selected genes
For selected proteins we examined their gene expression by qRT-PCR on total RNA from each group (Table 6). In agreement with the increased protein levels detected by mass spectrometry, we detected increases in the expression of Dpt, Igfbp2, Fbn-1, Fmod and Col1a1 (Table 6). On the other hand, Krt8, Bgn, Kera and Dcn gene expression was decreased coinciding with corresponding decreases in their protein levels measured by mass spectrometry. Calm3 and Rab8A, which showed very little change in the encoded proteins, were also relatively unchanged at the transcript level. Altogether, the gene expression patterns coincided reasonably well with the protein profiles.
Table 6.
qRT-PCR measurements of selected genes
| Gene symbol | Gene Name | Proteomics analysis Fold change | qRT-PCR analysis Fold change | ||
|---|---|---|---|---|---|
| Lum+/−/Lum+/+ | Lum−/−/Lum+/+ | Lum+/−/Lum+/+ | Lum−/−/Lum+/+ | ||
| Fbn-1 | Fibrillin-1 | 2.7 | 2.4 | 1.6 | 3.5 |
| Dpt | Dermatopontin | 2.3 | 1.8 | 1.4 | 1.8 |
| Igfbp-2 | insulin-like growth factor-binding protein 2 | 1.9 | 1.5 | 1.1 | 2.2 |
| Fmod | Fibromodulin | 1.6 | 3.1 | 2.4 | 2.7 |
| Col1a1 | Collagen alpha-1(I) | 4.5 | 2.6 | 3.5 | 2.7 |
| Calml3 | Calmodulin-like protein3 | 0.9 | 0.7 | 1.1 | 0.9 |
| Efemp1 | EGF-containing fibulin-like ECM protein | 2.4 | 2.0 | 1.4 | 1.0 |
| Rab8A | Ras-related protein | 1.1 | 0.7 | 0.7 | 0.9 |
| Krt14 | Keratin 14 | 0.9 | 0.7 | 0.9 | 0.8 |
| Krt8 | Keratin 8 | 0.3 | 0.3 | 0.2 | 0.5 |
| Bgn | Biglycan | 1.0 | 0.7 | 1.0 | 0.8 |
| Kera | Keratocan | 1.0 | 0.6 | 0.5 | 0.2 |
| Dcn | Decorin | 1.0 | 0.7 | 0.2 | 0.7 |
3.4. Western blot of selected proteins
We used western blotting techniques to further examine changes in proteins of particular interest to the cornea. The corneal stromal SLRPs decorin and biglycan, detectable after chondroitinase ABC digestion of the glycosamonoglycan side chains, were decreased in the Lum−/− samples confirming the mass spectrometry trends (Fig. 3A). The stromal collagen type VI, type XII and type XIV were considered to have increased slightly with lumican-deficiency by MS and western blotting (Fig. 3B). By MS, cytokeratin 8 appeared to have decreased and the western blot analysis also indicated its decrease in the Lum−/− corneas. Also, in concordance with the mass spectrometry data, the western blots showed very little change in aldehyde dehydrogenase 1 (Aldh1a1), a hall mark of the corneal stroma, and increases in Igfbp2, another known component of the cornea, in the Lum−/− the corneas, confirming the MS data. Alcohol dehydrogenase 1, markedly increased by mass spectrometry, also appeared to have increased by western blotting (Fig. 3C).
Fig. 3.
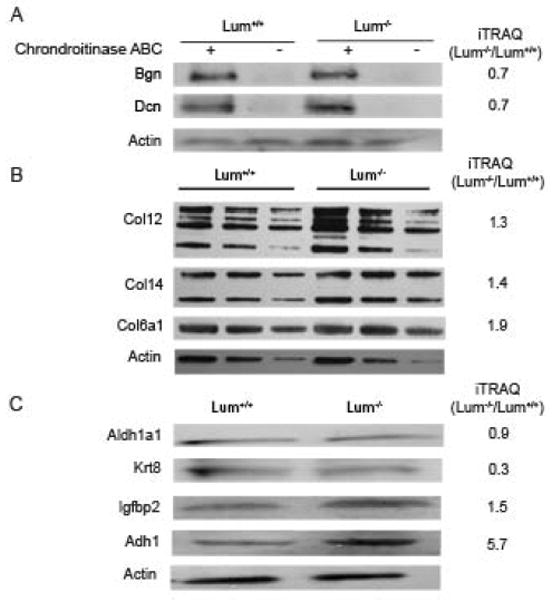
Western blot of selected proteins from extracts of Lum+/+ and Lum−/− corneas with protein-fold change detected by iTRAQ mass spectrometry shown along the right margin. (A) The core proteins of the corneal SLRPs decorin (Dcn) and biglycan (Bgn) are detectable after Chondroitinase ABC digestion to remove the GAG side chains of the proteoglycans; (B) ECM collagens were detected in SDS extracted fractions of the cornea loaded in three different doses. Actin was used as a comparative loading control; (C) The following cellular proteins were detected: Aldh1a1-aldehyde dehydrogenase 1a1; Krt8-cytokeratin 8; Igfbp2–Insulin growth factor binding protein 2; Adh–alcohol dehydrogenase 1.
4. Discussion
The proteomes of the heterozygous and the lumican-deficient mouse corneas compared to the wild type show a broad range of changes in cellular and extracellular matrix proteins. The results suggest that lumican deficiency disrupts normal cellular metabolic functions possibly involving the epithelial and the stromal cells. In addition, significant changes are seen in proteins of the basement membranes and interstitial ECM.
Increases in several basement membrane proteins, perlecan [20], nidogen/entactin [21], several laminin isoforms [22] and the basement membrane anchoring collagen type VII [23], in the Lum+/− and Lum−/− corneas, suggest that lumican deficiency in the cornea perturbs the basement membrane, and the adjacent corneal epithelium somehow. Accordingly, several epithelial proteins were altered. For example mucin-4, a membrane tethered mucin found in the corneal and conjunctival epithelia [24], was decreased with lumican-deficiency. Cytokeratin 8 is considered to be a conjunctival epithelial cytokeratin; its presence in the corneal epithelium is controversial due to antibody cross reactivity [25]. However, by mass spectrometry cytokeratin 8 was detectable in the wild type mouse cornea, and reduced in the Lum+/− and the Lum−/− corneas.
Lumican is a major stromal protein; not surprisingly the corneal stroma of Lum+/− and Lum−/− mice was affected in a number of ways. The ECM collagens seemed to be particularly sensitive to lumican-dosage effects; the fibril forming collagen type I (Col1a1 and Col1a2) was increased. The fibril associated collagen types XII and XIV, were also increased in the Lum+/− and the Lum−/− corneas. Other collagen binding SLRPS of the cornea, keratocan, decorin and bigycan, were decreased, and the limbal/scleral SLRP fibromodulin, was increased in the Lum−/− corneas. These changes provide a molecular understanding of the abnormal collagen fibril structure seen in the Lum−/− mice. In very general terms, it seems that lumican deficiency leads to an immature cornea that may have increased fibrillar collagens, and possibly their turnover, while fibromodulin, which is reduced in the adult cornea remains at higher levels. Others have also reported a lumican-dosage effect on keratocan levels in the cornea [26]. Contrary to our findings in the cornea of the Lum−/− mice, in tendons we reported a decrease in fibromodulin [27]. In the same study, we also found a marked increase in lumican in the tendons of the Fmod+/− and Fmod−/− mice. Thus, the inter-relationship between these SLRPS is tissue specific and likely to depend on the cell types and other components of the ECM. Increases in certain ECM proteins, such as collagen type VI, suggest activities related to ECM repair and remodeling in the lumican-deficient corneas. Interestingly, the TGFβ inducible protein (kerato-epithelin) or BIGH3, known to promote collagen VI aggregation, was also elevated in the Lum+/− and Lum−/− corneas. Mutations in BIGH3 and abnormal deposits of this protein are associated with a number of corneal dystrophies [28]. Other protein changes relating to ECM remodeling and TGFβ signals that we detected in the Lum−/− corneas include increased Ltbp4 (latent TGF binding protein 4), fibulin 5 and fibrillin 1. Some ECM related changes we see in the Lum−/− corneas may also be orchestrated by the IGF signaling pathway, since Igfbp2, a regulator of IGF ligands and signaling via the IGF1 receptor, was elevated in the Lum+/− and the Lum−/− corneas.
The cornea, like the lens contains several crystallins and large amounts of specific soluble enzymes, such as aldehyde dehydrogenase, enolase, glutathione S-transferase, gelsolin and transketolase, that in addition to metabolic functions, are thought to reduce light scattering and increase the transparent and refractive quality of the cornea and its resident cells [29–31]. In the current study, we detected aldehyde dehydrogenase, several glutathione S-transferase subtypes and gelsolin in the mouse cornea, but found no difference in these between the Lum+/− and the Lum−/− corneas, either by mass spectrometry or western blotting. However, alcohol dehydrogenase (Adh1) was markedly increased in the Lum+/− and the Lum−/− corneas. Although alcohol dehydrogenases have not been explicitly discussed as corneal crystallins, they are present in the lens and the cornea at high levels. The alcohol dehydrogenase enzymes convert vitamin A (retinol) to retinoic acid that mediate functions of the retinoic acid and retinoid X receptors in gene transcription relevant to corneal health, ocular development and morphogenesis [32, 33]. The reason for Adh1 increased in the lumican-deficient mouse corneas is unclear at the moment, but stress-related metabolic changes in the stromal keratocytes may be one possibility.
Our mass spectrometry data also detected several proteins that are relatively novel to the cornea. Kininogen 1, novel to the cornea, was reduced in the Lum−/− corneas. Based on its role as a regulator of vascular endothelial cell apoptosis [34], it may have an anti-angiogenesis function in the cornea. Not reported in the cornea before, Lrg1 is a leucine rich glycoprotein that was markedly decreased in the Lum−/− corneas. It has the leucine rich repeat as in the other SLRP members of the cornea, and may be an acute phase response protein [35]. An ECM protein nephronectin, known for its role in kidney development [36] was increased in the Lum−/− corneas. We speculate that its known interactions with α8β1 integrin and role in promoting GDNF (glial cell-derived neurotrophic) expression may be relevant to corneal epithelial migration and health [37, 38].
We speculated if the proteomic changes seen in lumican deficiency resemble pathogenic changes in corneal diseases. Keratoconus is a complex corneal thinning disease involving environmental factors and effects of multiple genes, where excessive ECM remodeling and oxidative stress may be pathogenic responses of the cornea [39]. The lumican-deficient corneal proteome also shows overall changes in the stromal ECM, and in the balance of anti-oxidants and redox sensors. However, there were no clear overlaps in specific proteins between keratoconus and the Lum−/− mouse corneas. For example, Adh1, which was down regulated in keratoconus corneas [40], was increased in the Lum−/− mice; while superoxide dismutase, known to be decreased in keratoconus stromal cells, was increased in the Lum−/− corneas. With respect to other changes, collagen type XII reported as decreased in keratoconus [41], was increased in the Lum−/− corneas. Myopia, another complex multi factorial disease, involves increased axial length, and biochemical and biomechanical alterations in the sclera and the cornea. Specific forms of myopia have been linked to Lum [42] and IGF polymorphisms [43]. Lumican and fibromodulin deficient mice show biochemical and biomechanical connective tissue changes and increased axial growth of the eye [44]. The proteome of the Lum−/− cornea presented in this study shows changes in Igfbp2 and underscores IGF signaling in ocular axial growth and may help to unravel molecular changes in myopia. To the best of our knowledge, this mass spectrometric analysis reports the largest collection of proteins in the corneal proteome. Differences in the corneal proteome of the wild type and lumican-deficient mouse cornea may shed some light on signals controlling ocular growth and biomechanical changes in the sclera and cornea. Our study further demonstrates that disturbances in stromal extracellular matrix impact the health of the stromal and the epithelial cells and overall corneal homeostasis.
Supplementary Material
Acknowledgments
Funding support: EY11654 (National Eye Institute, USA) to SC, EY05129 (National Eye Institute, USA) to DB and the Department of Biotechnology (DBT), Government of India research support to the Institute of Bioinformatics, Bangalore, India.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hassell JR, Birk DE. The molecular basis of corneal transparency. Exp Eye Res. 2010;91:326–35. doi: 10.1016/j.exer.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S, Oldberg A, Chakravarti S, Birk DE. Fibromodulin regulates collagen fibrillogenesis during peripheral corneal development. Dev Dyn. 2010;239:844–54. doi: 10.1002/dvdy.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaerkady R, Pandey A. Quantitative proteomics for identification of cancer biomarkers. Proteomics Clin Appl. 2007;1:1080–9. doi: 10.1002/prca.200700284. [DOI] [PubMed] [Google Scholar]
- 4.Nichols JJ, Green-Church KB. Mass spectrometry-based proteomic analyses in contact lens-related dry eye. Cornea. 2009;28:1109–17. doi: 10.1097/ICO.0b013e3181a2ad81. [DOI] [PubMed] [Google Scholar]
- 5.Pannebaker C, Chandler HL, Nichols JJ. Tear proteomics in keratoconus. Mol Vis. 2010;16:1949–57. [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarti S. Functions of lumican and fibromodulin: lessons from knockout mice. Glycoconj J. 2002;19:287–93. doi: 10.1023/A:1025348417078. [DOI] [PubMed] [Google Scholar]
- 7.Saika S, Shiraishi A, Liu CY, Funderburgh JL, Kao CW, Converse RL, et al. Role of lumican in the corneal epithelium during wound healing. J Biol Chem. 2000;275:2607–12. doi: 10.1074/jbc.275.4.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu F, Chakravarti S. Differential Expression of Inflammatory and Fibrogenic Genes and Their Regulation by NF-{kappa}B Inhibition in a Mouse Model of Chronic Colitis. J Immunol. 2007;179:6988–7000. doi: 10.4049/jimmunol.179.10.6988. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarti S, Magnuson T, Lass JH, Jepsen KJ, LaMantia C, Carroll H. Lumican regulates collagen fibril assembly: skin fragility and corneal opacity in the absence of lumican. J Cell Biol. 1998;141:1277–86. doi: 10.1083/jcb.141.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezura Y, Chakravarti S, Oldberg A, Chervoneva I, Birk DE. Differential expression of lumican and fibromodulin regulate collagen fibrillogenesis in developing mouse tendons. J Cell Biol. 2000;151:779–88. doi: 10.1083/jcb.151.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quantock AJ, Meek KM, Chakravarti S. An x-ray diffraction investigation of corneal structure in lumican-deficient mice. Invest Ophthalmol Vis Sci. 2001;42:1750–6. [PubMed] [Google Scholar]
- 12.Rada JA, Cornuet PK, Hassell JH. Regulation of corneal collagen fibrillogenesis in vitro by corneal keratan sulfate proteoglycan (lumican) and decorin core proteins. Exp Eye Res. 1993;56:635–48. doi: 10.1006/exer.1993.1081. [DOI] [PubMed] [Google Scholar]
- 13.Song J, Lee YG, Houston J, Petroll WM, Chakravarti S, Cavanagh HD, et al. Neonatal corneal stromal development in the normal and lumican-deficient mouse. Invest Ophthalmol Vis Sci. 2003;44:548–57. doi: 10.1167/iovs.02-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beecher N, Chakravarti S, Joyce S, Meek KM, Quantock AJ. Neonatal development of the corneal stroma in wild-type and lumican-null mice. Invest Ophthalmol Vis Sci. 2006;47:146–50. doi: 10.1167/iovs.05-0907. [DOI] [PubMed] [Google Scholar]
- 15.Chakravarti S, Petroll WM, Hassell JR, Jester JV, Lass JH, Paul J, et al. Corneal opacity in lumican-null mice: defects in collagen fibril structure and packing in the posterior stroma. Invest Ophthalmol Vis Sci. 2000;41:3365–73. [PMC free article] [PubMed] [Google Scholar]
- 16.Vij N, Roberts L, Joyce S, Chakravarti S. Lumican suppresses cell proliferation and aids Fas-Fas ligand mediated apoptosis: implications in the cornea. Exp Eye Res. 2004;78:957–71. doi: 10.1016/j.exer.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Vij N, Roberts L, Joyce S, Chakravarti S. Lumican regulates corneal inflammatory responses by modulating Fas-Fas ligand signaling. Invest Ophthalmol Vis Sci. 2005;46:88–95. doi: 10.1167/iovs.04-0833. [DOI] [PubMed] [Google Scholar]
- 18.Wu F, Lee S, Schumacher M, Jun A, Chakravarti S. Differential gene expression patterns of the developing and adult mouse cornea compared to the lens and tendon. Exp Eye Res. 2008;87:214–25. doi: 10.1016/j.exer.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norman B, Davis J, Piatigorsky J. Postnatal gene expression in the normal mouse cornea by SAGE. Invest Ophthalmol Vis Sci. 2004;45:429–40. doi: 10.1167/iovs.03-0449. [DOI] [PubMed] [Google Scholar]
- 20.Noonan D, Hassell J. Perlecan, the large low-density proteoglycan of basement membranes: Structure and variant forms. Kidney International. 1993;43:53–60. doi: 10.1038/ki.1993.10. [DOI] [PubMed] [Google Scholar]
- 21.Chakravarti S, Tam M, Chung A. The basement membrane glycoprotein entactin promotes cell attachment and binds calcium ions. J Biol Chem. 1990;265:10597–603. [PubMed] [Google Scholar]
- 22.Ekblom M, Falk M, Salmivirta K, Durbeej M, Ekblom P. Laminin isoforms and epithelial development. Ann N Y Acad Sci. 1998;23(857):194–211. doi: 10.1111/j.1749-6632.1998.tb10117.x. [DOI] [PubMed] [Google Scholar]
- 23.Knight DP. Unconventional collagens. J Cell Sci. 2000;113 (Pt 23):4141–2. [Google Scholar]
- 24.Govindarajan B, Gipson IK. Membrane-tethered mucins have multiple functions on the ocular surface. Exp Eye Res. 2010;90:655–63. doi: 10.1016/j.exer.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merjava S, Brejchova K, Vernon A, Daniels JT, Jirsova K. Cytokeratin 8 is expressed in human corneoconjunctival epithelium, particularly in limbal epithelial cells. Invest Ophthalmol Vis Sci. 52:787–94. doi: 10.1167/iovs.10-5489. [DOI] [PubMed] [Google Scholar]
- 26.Carlson EC, Liu CY, Chikama T, Hayashi Y, Kao CW, Birk DE, et al. Keratocan, a cornea-specific keratan sulfate proteoglycan, is regulated by lumican. J Biol Chem. 2005;280:25541–7. doi: 10.1074/jbc.M500249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jepsen KJ, Wu F, Peragallo JH, Paul J, Roberts L, Ezura Y, et al. A syndrome of joint laxity and impaired tendon integrity in lumican- and fibromodulin-deficient mice. J Biol Chem. 2002;277:35532–40. doi: 10.1074/jbc.M205398200. [DOI] [PubMed] [Google Scholar]
- 28.Munier F, Korvatska E, Djemai A, Paslier DL, Zografos L, Pescia G, et al. Kerato-epithelin mutations in four 5q31-linked corneal dystrophies. Nature Genetics. 1997;15:247–51. doi: 10.1038/ng0397-247. [DOI] [PubMed] [Google Scholar]
- 29.Jester JV. Corneal crystallins and the development of cellular transparency. Semin Cell Dev Biol. 2008;19:82–93. doi: 10.1016/j.semcdb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lassen N, Black WJ, Estey T, Vasiliou V. The role of corneal crystallins in the cellular defense mechanisms against oxidative stress. Semin Cell Dev Biol. 2008;19:100–12. doi: 10.1016/j.semcdb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Piatigorsky J. Enigma of the abundant water-soluble cytoplasmic proteins of the cornea: the “refracton” hypothesis. Cornea. 2001;20:853–8. doi: 10.1097/00003226-200111000-00015. [DOI] [PubMed] [Google Scholar]
- 32.Nezzar H, Chiambaretta F, Marceau G, Blanchon L, Faye B, Dechelotte P, et al. Molecular and metabolic retinoid pathways in the human ocular surface. Mol Vis. 2007;13:1641–50. [PubMed] [Google Scholar]
- 33.Ubels JL, Iorfino A, O’Brien WJ. Retinoic acid decreases the number of EGF receptors in corneal epithelium and Chang conjunctival cells. Exp Eye Res. 1991;52:763–5. doi: 10.1016/0014-4835(91)90029-e. [DOI] [PubMed] [Google Scholar]
- 34.Zhang JC, Claffey K, Sakthivel R, Darzynkiewicz Z, Shaw DE, Leal J, et al. Two-chain high molecular weight kininogen induces endothelial cell apoptosis and inhibits angiogenesis: partial activity within domain 5. FASEB J. 2000;14:2589–600. doi: 10.1096/fj.99-1025com. [DOI] [PubMed] [Google Scholar]
- 35.Shirai R, Hirano F, Ohkura N, Ikeda K, Inoue S. Up-regulation of the expression of leucine-rich alpha(2)-glycoprotein in hepatocytes by the mediators of acute-phase response. Biochem Biophys Res Commun. 2009;382:776–9. doi: 10.1016/j.bbrc.2009.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linton JM, Martin GR, Reichardt LF. The ECM protein nephronectin promotes kidney development via integrin alpha8beta1-mediated stimulation of Gdnf expression. Development. 2007;134:2501–9. doi: 10.1242/dev.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi H, Shine HD, Li DQ, de Paiva CS, Farley WJ, Jones DB, et al. Glial cell-derived neurotrophic factor gene delivery enhances survival of human corneal epithelium in culture and the overexpression of GDNF in bioengineered constructs. Exp Eye Res. 2008;87:580–6. doi: 10.1016/j.exer.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You L, Ebner S, Kruse FE. Glial cell-derived neurotrophic factor (GDNF)-induced migration and signal transduction in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2001;42:2496–504. [PubMed] [Google Scholar]
- 39.Kenney MC, Brown DJ. The cascade hypothesis of keratoconus. Cont Lens Anterior Eye. 2003;26:139–46. doi: 10.1016/S1367-0484(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 40.Mootha VV, Kanoff JM, Shankardas J, Dimitrijevich S. Marked reduction of alcohol dehydrogenase in keratoconus corneal fibroblasts. Mol Vis. 2009;15:706–12. [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng EL, Maruyama I, SundarRaj N, Sugar J, Feder RS, Yue BY. Expression of type XII collagen and hemidesmosome-associated proteins in keratoconus corneas. Curr Eye Res. 2001;22:333–40. doi: 10.1076/ceyr.22.5.333.5491. [DOI] [PubMed] [Google Scholar]
- 42.Lin HJ, Wan L, Tsai Y, Chen WC, Tsai SW, Tsai FJ. The association between lumican gene polymorphisms and high myopia. Eye (Lond) 2010;24:1093–101. doi: 10.1038/eye.2009.254. [DOI] [PubMed] [Google Scholar]
- 43.Metlapally R, Ki CS, Li YJ, Tran-Viet KN, Abbott D, Malecaze F, et al. Genetic association of insulin-like growth factor-1 polymorphisms with high-grade myopia in an international family cohort. Invest Ophthalmol Vis Sci. 2010;51:4476–9. doi: 10.1167/iovs.09-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakravarti S, Paul J, Roberts L, Chervoneva I, Oldberg A, Birk DE. Ocular and scleral alterations in gene-targeted lumican-fibromodulin double-null mice. Invest Ophthalmol Vis Sci. 2003;44:2422–32. doi: 10.1167/iovs.02-0783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.