Abstract
The zebrafish is an ideal model for elucidating the cellular and molecular mechanisms that underlie development of the peripheral nervous system. A transgenic line that selectively labels all the sensory circuits would be a valuable tool for such investigations. In this study, we describe such a line: the enhancer trap zebrafish line Tg(SKIV2L2:gfp)j1775 which expresses green fluorescent protein (gfp) in the peripheral sensory ganglia. We show that this transgene marks all peripheral ganglia and sensory nerves, beginning at the time when the neurons are first extending their processes, but does not label the efferent nerves. The trapped reporter is inserted just upstream of a previously poorly described gene: lhfpl4 on LG6. The expression pattern of this gene by in situ hybridization reveals a different, but overlapping, pattern of expression compared to that of the transgene. This pattern also does not mimic that of the gene (skiv2l2), which provided the promoter element in the construct. These findings indicate that reporter expression is not dictated by an endogenous enhancer element, but instead arises through an unknown mechanism. Regardless, this reporter line should prove to be a valuable tool in the investigation of peripheral nervous system formation in the zebrafish.
1. Results and Discussion
The sensory peripheral nervous system carries information from the internal organs and the external environment to the CNS. In vertebrates, the nerves that carry such signals originate in ganglia lying outside the CNS (Butler and Hodos, 2005). Although the anatomy of these nerves has been known for over a century, the cellular and molecular mechanisms that underlie the establishment of their connections are poorly understood. The zebrafish is an ideal vertebrate model for studying the development of the peripheral sensory nervous system, due to the transparent nature of its embryos and larvae, and the ease of generating fish expressing fluorescent reporters in a cell-specific fashion. Such transgenic lines would be very useful tools for developmental analysis due to the ease of carrying out in vivo imaging using epifluorescent and confocal microscopy. For optimal utility, a line for investigating the peripheral nervous system should express a marker in all afferent, but no efferent, neurons. This would allow for the selective investigation of sensory circuits without the confounding presence of the motor limbs of peripheral nerves. Currently, there are several lines that express fluorescent reporters in the peripheral nervous system, [e.g.,(Faucherre et al., 2009; Higashijima et al., 2000; Kucenas et al., 2006)]; however, labeling of all sensory circuits is not present in these lines, and in some the reporter is also expressed by motor neurons. Therefore, a transgenic line specifically labeling only the sensory limbs of all peripheral nerves is still needed. We now report the identification of an enhancer trap line in which the sensory, but not the motor, limb of all circuits- cranial nerves V, VII, VIII, IX and X, the anterior and posterior lateral line nerves, and the trunk nerves, are labeled. Herein, we describe the expression pattern of the reporter, the insertion site of the transgene and the expression pattern of the gene into which the transgene is inserted.
1.1. Characterization of enhancer trap reporter expression
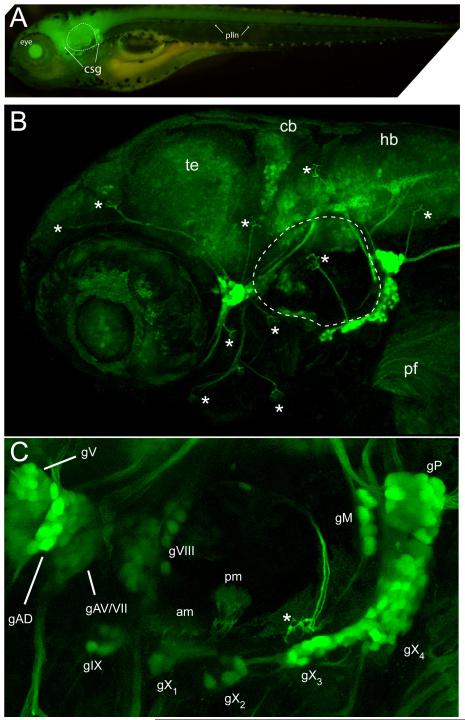
A panel of transgenes for reporting a variety of human promoters and candidate enhancers were screened for interesting expression patterns, including expression in peripheral neurons and cranial ganglia. One of these promoters was from the SKIV2l2 gene, which we had previously described as important for melanocyte regeneration (Yang et al., 2007). In an attempt to make a SKIV2L2:gfp reporter with the goal of marking intermediates in melanocyte regeneration, we found that one transgenic line, Tg(SKIV2L2:gfp)j1775, consistently showed bright expression in cranial ganglia, prompting us to investigate it further. Two independent insertions with this transposon were obtained, with the second line showing only a faint, ubiquitous GFP expression in the brain and spinal cord only. These two patterns suggest that the expression pattern seen in the Tg(SKIV2L2:gfp)j1775 line was not a property of the minimal SKIV2l2 promoter used in the construct, but may instead reflect a property of the chromosomal environment of the insertion. At 4 days post fertilization (dpf), Tg(SKIV2L2:gfp) j1775 larvae show high levels of GFP expression in sensory neurons (Figure 1A), with lower levels of GFP seen in CNS neurons and little to no expression seen in non-neuronal tissues. Confocal imaging revealed GFP expression in neurons of the trigeminal (gV), anterior lateral line complex (gAD/AV), statoacoustic (gVIII), glossopharyngeal (gIX), vagal (gX), and the medial (gM) and posterior (gP) lateral line ganglia (Fig 1B). The facial (gVII) ganglia was not extensively labeled, as it contained only a few GFP-expressing neurons (data not shown). The spatial relationships between the various cranial ganglia can be easily visualized (Fig. 1C) and illustrated that even though neurons of the gV, gAD and gAV are in close proximity, the ganglia themselves are individually distinct entities. In addition to somatic labeling, both the peripheral and central axonal branches of all these neurons were readily detectable. The ability to observe the finer details of these circuits is illustrated by the detection of gVIII axon terminals innervating the sensory maculae present in the otic vesicle (Fig. 1C), and of trigeminal axon branches in the periphery (data not shown). Lower levels of GFP expression were also observed in the CNS, as populations of neurons within the optic tectum, cerebellum and hindbrain were labeled. We could also detect GFP+ axons in the head and skin of adult fish.
Fig. 1.
GFP expression of SKIV2L2 enhancer trap line. (A) Whole mount epifluorescence image of Tg(SKIV2L2:gfp) at 4 days- note clusters of GFP+ cells surrounding otic vesicle (dotted line). (B) Confocal microscopy of GFP expression in the peripheral ganglia in the head at 4 days. Asterisks represent neuromasts. te, telencephalon, cb, cerebellum, hb, hindbrain. (C) A detailed view of the sensory ganglia labeled by the SKIV2L2 enhancer. am, anterior macula; gAD, dorsal anterior lateral line ganglia; gAV, ventral anterior lateral line ganglia; gV, trigeminal ganglia; gVII, facial ganglia; gVIII, statoacoustic ganglia; gIX, glossopharyngeal ganglia, gX1-4, vagal ganglia, gM, medial lateral line ganglia, gP, posterior lateral lined ganglia; plln, posterior lateral line; pm, posterior macula; * neuromast.
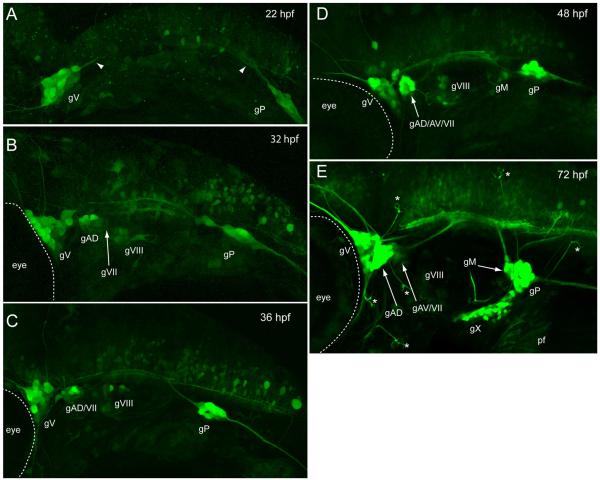
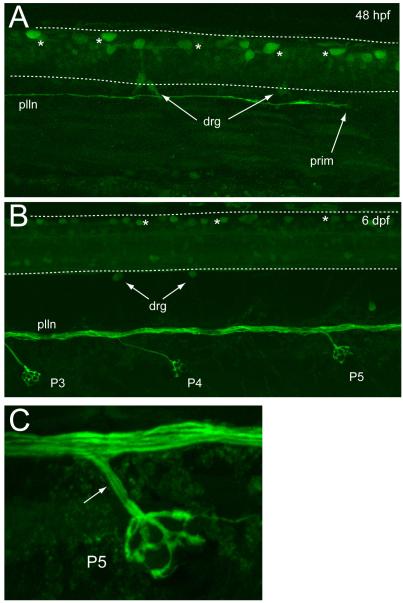
This reporter line will be a useful tool for developmental studies as GFP expression occurs in sensory neurons as they are beginning to extend their processes, and before they occupy their final anatomical locations. Figure 2 shows confocal images of reporter expression in the peripheral nervous system from 22 to 72 hpf. By 22 hpf, gV and gP are the only ganglia detected (Fig. 2A). The central projections of these ganglia are visible in the hindbrain, as are a number of unidentified central neurons. By 32 hpf, additional GFP expression included neurons forming gVIII and gAD (Fig. 2B), and within four hours those GFP+ neurons were coalescing to form discrete ganglia (Fig. 2C). At 48 hpf, the gIX, gM and gAV are beginning to form (Fig. 2D) and by 72 hpf (Fig. 2E), GFP expressing neurons are present in the vagal ganglia. Also at this stage, numerous lateral line afferent branches can be seen projecting to neuromasts positioned within the cranial epithelium, where they innervate the hair cells contained within. The central afferent limbs of each cranial sensory circuit can be readily distinguished as they extend to their final target regions in the hindbrain. In the trunk, the axons of gP are easily detected as they travel towards the tail, where they end in the lateral line primordium as it migrates posteriorly (Fig. 3A). The transient dorsal spinal sensory neurons, known as Rohon-Beard cells, also express GFP, as do nascent drg neurons. By 6 dpf (Fig. 3B), the axons forming the lateral line nerve have become separated into distinct fascicles, most likely due to the myelination that occurs around this time (Brosamle and Halpern, 2002). The contributions of individual axons to the lateral line neuromasts are also easily observed (Fig. 3C). Overall, the chronology of GFP expression in the head and trunk described here parallels the appearance of sensory neurons as reported in previous studies that have used histological markers [e.g., (Andermann et al., 2002; Andermann and Weinberg, 2001; Inoue et al., 1994; Metcalfe et al., 1990; Raible and Kruse, 2000)], thus validating this transgenic as a useful tool for in vivo study of the peripheral sensory circuit development.
Fig. 2.
Developmental expression of Tg(SKIV2L2:gfp) in the head. (A) 22 hpf. Trigeminal and posterior lateral line ganglia are labeled. (B) 32 hpf. The dorsal anterior lateral line ganglia and the statoacoustic ganglia have appeared. (C) 36 hpf. (D) 48 hpf. The ventral anterior lateral line ganglia and the medial lateral line ganglia have developed. (E) 60 hpf. All the ganglia that are labeled at 4 days are present and will coalesce and send out more processes over the 30 hrs to form the pattern seen in Fig 1. gAD, dorsal anterior lateral line ganglia; gAV, ventral anterior lateral line ganglia; gV, trigeminal ganglia; gVII, facial ganglia; gVIII, statoacoustic ganglia; gIX, glossopharyngeal ganglia, gX1-4, vagal ganglia, gM, medial lateral line ganglia, gP, posterior lateral lined ganglia; * neuromast Views are lateral, anterior to the left.
Fig. 3.
Developmental expression of Tg(SKIV2L2:gfp) in the trunk. (A) 48 hpf. GFP-labeled Rohon-Beard neurons (asterisks) can be seen in the spinal cord. Some dorsal root ganglia neurons (drg) are labeled at the dorsal side of the spinal cord. The posterior lateral line (plln) can be seen growing caudally (prim, lateral line primordium). (B) 6 days. Rohon-Beard neuronss are present (asterisks) and the plln has separated into distinct fascicles and can be seen to send off axonal branches to the neuromasts (P3, P4, P5). The drg neurons are more distinct. (C) A higher power image of neuromast P5 showing multiple axon fascicles (arrow) leaving the plln to innervate the neuromast.
1.2. Identification of enhancer trap reporter integration site reveals a novel lhfp-L4 gene
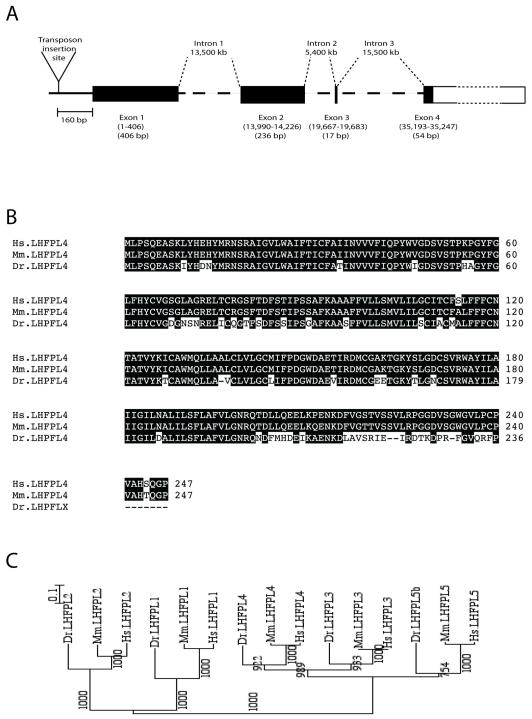
Identification of genomic DNA flanking Tg(SKIV2L2:gfp)j1775 showed that it integrated between two annotated genes in chromosome 6, at a position 160 bp 5′ of the predicted translational start site for ENSDARG00000078998, and around 13, 400 bp 5′ of the predicted transcriptional start of the zebrafish ortholog for MTMR14. The proximity of the integration site to the former gene lead us to explore it as the gene providing an enhancer or enhancers responsible for the specific expression pattern observed in Tg(SKIV2L2:gfp)j1775. We first note that although the integration site is 160 bp upstream of the translation initiation site, an EST (EST EB941707) is found immediately 5′ of this, suggesting that the transposon is inserted in the 5′ UTR of ENSDARG00000078998, further supporting the notion that Tg(SKV2l2:gfp)j1775 expression reflects some aspect of the regulation of ENSDARG00000078998. BLAST analysis of the predicted sequence of the protein encoded by the trapped gene indicated that it was most similar to mouse lipoma high mobility group protein isoform I-C fusion partner-like gene-4 (Lhfpl4). An alignment of the predicted zebrafish gene with the LHFPL4 proteins of human and mouse revealed 77% identity and is shown in Figure 4B. This gene is one of five family members (Lhfpl1-5) present in vertebrates (Longo-Guess et al., 2005; Petit et al., 1999), and is predicted to encode a protein containing four transmembrane domains (Longo-Guess et al., 2005). Phylogenetic analysis of the five known LHFPL proteins from human, mouse and zebrafish confirmed that the trapped gene encodes the zebrafish Lhfpl4 ortholog (Fig. 4C), and in silico analysis of the zebrafish genome indicated that it contains only a single copy of lhfpl4. Our data thus supports the identification of a novel zebrafish LHFPL4 gene.
Fig. 4.
Genomic analysis of enhancer trap line. (A) Insertion site of SKIV2L2:gfp on LG6, upstream of a novel Lhfpl4 gene. (B) Protein sequence alignment of human (Hs), mouse (Mm) and zebrafish (Dr) Lhfpl4 proteins. (C) Phylogenetic tree representing the evolutionary relationship between Lhfpl1-5 genes in zebrafish, human and mouse (numbers represent the bootstrap support for the nodes).
The finding that the insertion occurs in proximity to the start codon of lhfpl4 suggested the possibility that expression of this gene might be altered in transgenic fish. qPCR analysis showed no statistical difference (paired t-test) in expression levels of this gene between homozygous transgenic larvae (118 ± 16 arbitrary units, n=8) and their wild-type clutchmates (102 ± 19 arbitrary units, n=8), indicating that the insertion has no significant effect on gene expression.
1.3. Comparison of lhfpl4 expression with that of the transgenic reporter
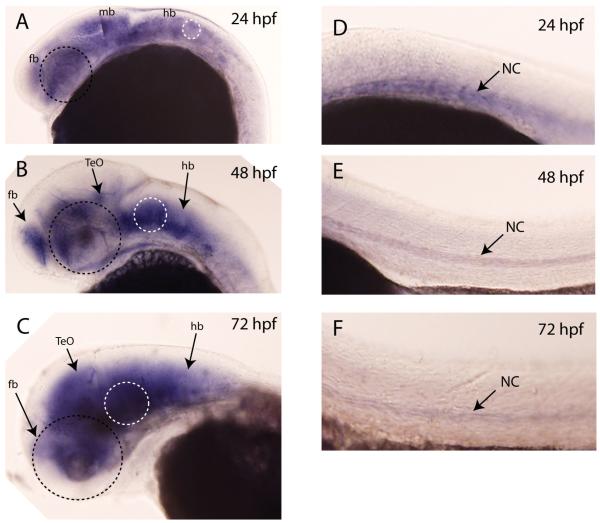
Our next step was to examine the spatiotemporal expression pattern of the lhfpl4 gene using whole mount in situ hybridization. Figure 5 shows that lhfpl4 expression in the head is limited to the brain between 24 and 72 hpf (Fig 5A-C) and that the levels of expression do not appear to change significantly between these times. Expression in the trunk (Figure 5 D-F) is restricted to the notochord, where mRNA levels were highest at 24 hpf and then decreased substantially by 72 hpf. There was no expression detected in the spinal cord or in the peripheral sensory ganglia. These results were somewhat surprising given the expression pattern of GFP in the transgenic line. The major differences are that GFP is present in the peripheral sensory ganglia of the head and trunk as well as in the spinal cord, both areas lacking lhfpl4 expression. These findings suggested the possibility that the genomic fragment used as the presumptive basal promoter in the transposon construct actually contained sequence(s) that could act as enhancers in these zebrafish tissues. If this were so, it would be expected that the Tg(SKIV2L2:gfp) j1775 expression pattern would resemble that of the native skiv2l2 gene. However, results from earlier studies (Yang et al., 2007) indicate that this is not the case: while skiv2l2 is expressed in the CNS, it does not have strong expression in the cranial ganglia nor in Rohon-Beard cells. In addition, we examined the transient expression patterns resulting from mosaic expression of the skiv2l2:gfp transposon in 3 dpf larvae from two different sets of injections. Sixty-one larvae had GFP+ cells in the CNS and in somatic tissues, with only three showing expression in any peripheral sensory neurons. In each of these positive cases, only 2 or 3 neurons were labeled and none showed expression in more than one type of ganglia: e.g., in one case expression was seen in a few cells of the anterior and posterior lateral line ganglia, but not in the trigeminal or epibranchial ganglia, and in another case two cells were detected in the trigeminal but none in the epibranchial or lateral line ganglia. These results support the interpretation that the observed expression pattern for the inserted transposon was not determined by any sequence present in the construct. This leaves a third possibility- that a sequence is present in the construct that acts as a cryptic enhancer element. The promoter fragment used was 243 bp long and obtained from the human SKIV2L2 gene. The terminal 204 bp are present in transcribed sequences corresponding to the 5′-untranslated region, suggesting that it is only the initial 39bp that contributes to the SKIV2L2 promoter. It is not known which part, or parts, of this genomic fragment might contain elements that contribute to the expression pattern seen for GFP, or whether a new or synthetic element was created at one of the junctions where the transposon was inserted. For instance, the transposon might separate activating (enhancer or promoter like sequences) from repressing sequences in the lhfpl4 gene, to reveal otherwise cryptic regulation of the lhfpl4 gene. Regardless of how the expression pattern is generated, our results demonstrate that this transgenic line will be a useful tool in the investigation of peripheral sensory circuit formation in vertebrates.
Fig. 5.
Expression pattern of lhfpl4 by whole mount in situ hybridization. (A) At 24 hpf, lhfpl4 is expressed throughout the brain (fb, forebrain, mb, midbrain, hb, hindbrain). At 48 hpf (B) and 72 hpf (C), its expression is still throughout the brain, especially in the forebrain, the optic tectum (TeO) and in the hindbrain. Expression in the trunk (D-F) is restricted to the notochord (nc). There is no staining evident in any peripheral ganglia, in the spinal cord or in the lateral line. Black dotted circle indicates position of the eye; white dotted circle indicates position of the ear.
2. Materials and methods
2.1. Zebrafish
Zebrafish and embryos were maintained as previously described (Kucenas et al., 2006). Embryos were treated with 0.003% phenylthiourea (PTU) to prevent pigmentation. The described line was constructed by a Tol2 transposon-mediated enhancer trap method (Fisher et al., 2006), using a fragment containing the eGFP gene and a 243 bp fragment of promoter and 5′-untranslated region from the human SKIV2L2 gene (superkiller viralicidic activity 2-like protein 2). Tg(SKIV2L2:gfp) was identified from a pool of four founders.
2.2. Identification of transposon integration site
Tail PCR was used to identify the integration site, as described by Liu and Whittier (Liu and Whittier, 1995). Identification of the integration site in the zebrafish genome was performed at the Sanger Institute web page (http://www.sanger.ac.uk) and at the Danio rerio ensembl blast web site (http://www.ensembl.org/Danio_rerio/blastview). Peptide alignments and phylogenetic tree analysis were performed using ClustalX software (http://www.clustal.org).
2.3. Whole mount in situ hybridization
Primers were designed from the predicted sequence of the zebrafish lhfpl4 cDNA (F: 5′-TGCTACCTTCACAAGAAGCCTCAA, R: 5′-CGGTTTCCGAGGACAAAAGCAAGA) and a 600 bp fragment was obtained by PCR from 4-day-old zebrafish cDNA. This fragment was cloned into the pGEM T vector (Promega). Digoxigenin-labeled RNA was prepared and in situ hybridization was performed as previously described (Kucenas et al., 2006).
2.4. Imaging
Epifluorescent and brightfield images were captured with a Nikon TE200 inverted microscope equipped with a CoolSnap HQ digital camera or an Olympus BX60 microscope equipped with a DP-71 digital camera. Confocal images were obtained using an Olympus FV1000 microscope using a long-working distance 20x water immersion lens. Images collected in the z-dimension were collapsed into one maximal intensity projection or were rendered into three-dimensions using FVW-AS2.1 software provided with the microscope.
2.5 q-PCR
4 dpf larvae from a single Tg(SKIV2L2:gfp) j1775/+ (AB) incross clutch were imaged by epifluorescence and separated into those with no GFP expression (wild-type), moderate GFP expression (heterozygotes) and strong GFP expression (homozygotes). RNA was then isolated from individual larvae using the RNeasy kit (Qiagen) and cDNA synthesized using Superscript II RT (Invitrogen). The individual cDNAs were then subjected to real-time PCR (BioRad Opticon2 thermocycler) using two primers that give a 100 bp product and were targeted to sequences present in exon 1 (atggcgctcttctttttctg, bp 336 – 356 of the ORF) and exon 2 (gtgctgggctgtttgatttt, bp 417-436 of the ORF) of the lhfpl4 gene. PCR products were observed in the tubes containing cDNA, but not in those containing the source RNA, and the C(t) values obtained were within the linear range of the standard curve.
Highlights.
Tg(SKIV2L2:gfp)j1775 is a zebrafish enhancer trap line that labels sensory neurons
GFP expression starts soon after neurogenesis and is maintained beyond 7 dpf
There is no labeling of efferent axons in the periphery
The transgene is inserted in the 5′ end of the lhfpl4 gene
The expression pattern is novel and does not arise solely from the trapped gene nor the basal promoter in the construct
Acknowledgements
We would like to thank Rachael Sheridan for help with fish husbandry, and Linda Cox and Dr. J. Chrivia for help with the qPCR experiments. This work was supported by grants from the NIH to A. S. McC. (GM71648), S.L.J. (GM56988) and M.M.V. (NS060074) and from the Saint Louis University School of Medicine to M.M.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andermann P, Ungos J, Raible DW. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002;251:45–58. doi: 10.1006/dbio.2002.0820. [DOI] [PubMed] [Google Scholar]
- Andermann P, Weinberg ES. Expression of zTlxA, a Hox11-like gene, in early differentiating embryonic neurons and cranial sensory ganglia of the zebrafish embryo. Dev Dyn. 2001;222:595–610. doi: 10.1002/dvdy.1239. [DOI] [PubMed] [Google Scholar]
- Brosamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Butler AB, Hodos W. Comparative Vertebrate Neuroanatomy. Second ed. John Wiley and Sons, Inc.; Hoboken, NJ: 2005. [Google Scholar]
- Faucherre A, Pujol-Marti J, Kawakami K, Lopez-Schier H. Afferent neurons of the zebrafish lateral line are strict selectors of hair-cell orientation. PloS one. 2009;4:e4477. doi: 10.1371/journal.pone.0004477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Soto F, Cox JA, Voigt MM. Selective labeling of central and peripheral sensory neurons in the developing zebrafish using P2X(3) receptor subunit transgenes. Neuroscience. 2006;138:641–652. doi: 10.1016/j.neuroscience.2005.11.058. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Longo-Guess CM, Gagnon LH, Cook SA, Wu J, Zheng QY, Johnson KR. A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7894–7899. doi: 10.1073/pnas.0500760102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe WK, Myers PZ, Trevarrow B, Bass MB, Kimmel CB. Primary neurons that express the L2/HNK-1 carbohydrate during early development in the zebrafish. Development. 1990;110:491–504. doi: 10.1242/dev.110.2.491. [DOI] [PubMed] [Google Scholar]
- Petit MM, Schoenmakers EF, Huysmans C, Geurts JM, Mandahl N, Van de Ven WJ. LHFP, a novel translocation partner gene of HMGIC in a lipoma, is a member of a new family of LHFP-like genes. Genomics. 1999;57:438–441. doi: 10.1006/geno.1999.5778. [DOI] [PubMed] [Google Scholar]
- Raible DW, Kruse GJ. Organization of the lateral line system in embryonic zebrafish. J Comp Neurol. 2000;421:189–198. [PubMed] [Google Scholar]
- Yang CT, Hindes AE, Hultman KA, Johnson SL. Mutations in gfpt1 and skiv2l2 cause distinct stage-specific defects in larval melanocyte regeneration in zebrafish. PLoS genetics. 2007;3:e88. doi: 10.1371/journal.pgen.0030088. [DOI] [PMC free article] [PubMed] [Google Scholar]