Abstract
Programmed methylation and demethylation of regulatory sequences has been proposed to play a central role in vertebrate development. We report here that the methylation status of the 5′ regions of a panel of tissue-specific genes could not be correlated with expression in tissues of fetal and newborn mice. Genes reported to be regulated by reversible methylation were not expressed ectopically or precociously in Dnmt1-deficient mouse embryos under conditions where demethylation caused biallelic expression of imprinted genes and activated transcription of endogenous retroviruses of the IAP class. These and other data suggest that the numerous published expression–methylation correlations may have described not a cause but a consequence of transcriptional activation. A model is proposed under which cytosine methylation represents a biochemical specialization of large genomes that participates in specialized biological functions such as allele-specific gene expression and the heritable transcriptional silencing of parasitic sequence elements, whereas cellular differentiation is controlled by conserved regulatory networks that do not depend on covalent modification of the genome.
Keywords: Methylation, demethylation, mammalian development, mouse embryos, gene expression, transcriptional silencing
Riggs (1975) and Holliday and Pugh (1975) predicted that programmed methylation and demethylation of DNA might be found to regulate the expression of genes during mammalian development. As originally envisioned, tissue-specific gene expression would depend on reversible sequence-specific methylation and demethylation that would control the interaction of regulatory sequences with the basal transcriptional apparatus. These hypotheses were original and ingenious, and their speculative nature could be legitimately justified “...in view of our almost complete ignorance of the mechanism for the unfolding of the genetic program during development” (Holliday and Pugh 1975).
A body of data is consistent with the methylation-development hypothesis. Correlations between transcriptional activation and demethylation of local 5′-CpG-3′ sites have been reported for a number of loci (for review, see Eden and Cedar 1994). It was also found that methylation patterns are replicated along with the DNA during S phase (Wigler et al. 1981) and that methylation of promoter regions inhibits transcription by recruitment of histone deacetylase and other transcriptional repressors (Jones et al. 1998; Nan et al. 1998). The heritability and strong repressive effects of cytosine methylation patterns are not inconsistent with a role in the regulation of tissue-specific genes, and there are numerous expression-methylation correlations, but decisive confirmatory evidence has been elusive.
In the years since 1975 it has become clear that development is controlled by regulatory networks that are conserved throughout the metazoa (for review, see Gerhart and Kirschner 1997). Many well-studied animals (including Drosophila melanogaster and Caenorhabditis elegans) have no detectable modified bases in their DNA; a complex regulatory system based on programmed methylation and demethylation of regulatory sequences would therefore have to be a specialization unique to vertebrates. This has come to appear increasingly unlikely with the rapid accumulation of evidence for strong conservation of regulatory networks.
In addition to the reservations created by comparative biology considerations, the regulatory role of cytosine methylation is also called into question by a body of experimental data. Although there are many published examples of tissue-specific methylation patterns, in no confirmed case has it been shown that a methylation pattern existing on a gene in a nonexpressing tissue will prevent transcription in a cell type normally capable of transcribing that gene (Hsiao et al. 1984). Several experiments that implicated cytosine methylation in the control of development relied on established lines of cultured cells, which frequently acquire methylation at sites in and around tissue-specific genes that are not methylated in nonexpressing tissues (Antequera et al. 1990; Jones et al. 1990). In a revealing experiment performed by Chiu and Blau (1985), it was found that amnionic fibroblasts and MRC-5 diploid lung fibroblasts expressed muscle-specific genes when fused to myotubes under conditions in which replication of the donor genomes did not occur, whereas the heteroploid HeLa cell line required pretreatment with the demethylating drug 5-azacytidine before becoming competent to express muscle-specific genes in the context of myotube cytoplasm. This experiment showed that cytosine methylation has the ability to suppress the expression of muscle-specific genes in cultured cells but does not do so in the organism in the case of the genes examined. Other heterokaryon experiments suggested that the activation of tissue-specific genes does not require DNA replication (Chiu and Blau 1984; Baron and Maniatis 1986), whereas current evidence indicates that replication is required for demethylation (Matsuo et al. 1998; Rougier et al. 1998).
Further evidence against a central role for cytosine methylation in the regulation of development stems from the finding that demethylating drugs are not notably teratogenic except when the effect can be attributed to the killing of proliferating embryonic cell populations (Rogers et al. 1994). The development of individuals with ICF (immunodeficiency, centromere instability, facial anomalies) syndrome, a human genetic disorder in which both repeated and single-copy sequences are undermethylated, is largely normal; the major clinical feature is variable immunodeficiency of unknown etiology (Miniou et al. 1994; Schuffenhauer et al. 1995). None of the heritable developmental abnormalities in vertebrates has been shown to be due to a primary defect in cytosine methylation, whereas many have been found to result from mutations in tissue-specific regulatory proteins. Furthermore, incipient developmental abnormalities are not observed in DNA methyltransferase-deficient mouse embryos (Li et al. 1992; Trasler et al. 1995); the phenotypes reported to date concern abnormal expression of imprinted genes (Li et al. 1993a), ectopic X inactivation (Panning and Jaenisch 1996), fulminating transcription of endogenous retroviruses of the intercisternal A particle (IAP) class (Walsh et al. 1998), and death caused by apoptosis in all tissues of the embryo (Li et al. 1992).
If methylation does regulate gene expression during development, tissue-specific genes should have densely methylated promoters in nonexpressing tissues and induced demethylation should activate tissue-specific genes ectopically or precociously. These predictions have been tested by analysis of methylation patterns of tissue-specific genes in nonexpressing tissues and by testing genes that have been reported to be regulated by cytosine methylation for ectopic activation in DNA methyltransferase-1 (Dnmt1)-deficient mouse embryos (Li et al. 1992). We report that the 5′ regions of a panel of tissue-specific genes are not densely methylated in nonexpressing tissues and that three genes reported previously to be regulated by promoter region methylation during mouse development are not expressed precociously in Dnmt1-deficient mouse embryos under conditions that cause biallelic expression of imprinted genes and activation of IAP endogenous retroviruses. These data suggest that previously observed correlations between demethylation and transcription at a number of loci might represent a consequence of transcriptional activation rather than a cause. Although a minor or augmenting role of cytosine methylation in gene control cannot be excluded completely, we propose a model under which cytosine methylation in mammals has roles in specialized processes such as allele-specific gene expression (Li et al. 1993a; Panning and Jaenisch 1996) and the heritable transcriptional silencing of parasitic sequence elements (Bestor 1990; Bestor and Coxon 1993; Yoder et al. 1997; Walsh et al. 1998), whereas developmental gene control relies on the conserved regulatory networks that direct the development of other metazoa.
Results
Methylation status of promoters of tissue-specific genes in nonexpressing tissues
A panel of previously characterized promoters was examined for evidence of dense cytosine methylation in nonexpressing tissues. DNA blot analysis after cleavage with methylation-sensitive restriction endonuclease was the method of choice, as this method is subject to fewer artifacts than alternative techniques. Restriction sites between −600 and +200 with respect to the cap site were tested. Genes were selected for analysis on the basis of well-characterized promoters, strict tissue-specific expression, restriction sites at spacings compatible with analysis by DNA blot hybridization, and a lack of repetitive sequence elements that would complicate hybridization analysis. Blots were stripped and rehybridized with a probe against mitochondrial DNA (mtDNA), which is abundant and not methylated at HpaII or HhaI sites, as a robust control for limit digestion.
The skeletal muscle α-actin genes (Cox and Buckingham 1992) from rat and mouse were examined first, as promoter methylation has been implicated in regulation of these genes (Yisraeli et al. 1986; Weiss et al. 1996). As shown in Figure 1A, HpaII restriction sites (5′-CCGG-3′) within 300 bp of the cap site of rat Acta1 were largely unmethylated in most tissues examined, with higher methylation in brain and liver and little methylation in kidney, lung, muscle, or spleen; the DNA in these latter tissues was as sensitive to HpaII as to MspI (a methylation-insensitive isoschizomer of HpaII). The mouse α-actin gene showed similar behavior, although a site upstream of the promoter displayed partial methylation (Fig. 1B). The HpaII sites closest to the cap site were not methylated in any tested tissue. The α-actin genes from both rat and mouse showed complete methylation at all tested sites in DNA from sperm. It is concluded that the promoter region of the α-actin gene displays little or no muscle-specific demethylation at the tested sites in rat or mouse, and that the methylation pattern in sperm does not predict the pattern to be found in nonexpressing somatic tissues. Muscle-specific active demethylation of the rat α-actin gene (Weiss et al. 1996; Yisraeli et al. 1986) seems unlikely, as the sites examined in those studies are not differentially methylated in muscle versus nonmuscle tissues (Fig. 1A). The discrepancy is likely to stem from the use of cultured cells, in which tissue-specific genes tend to undergo de novo methylation (Antequera et al. 1990; Jones et al. 1990).
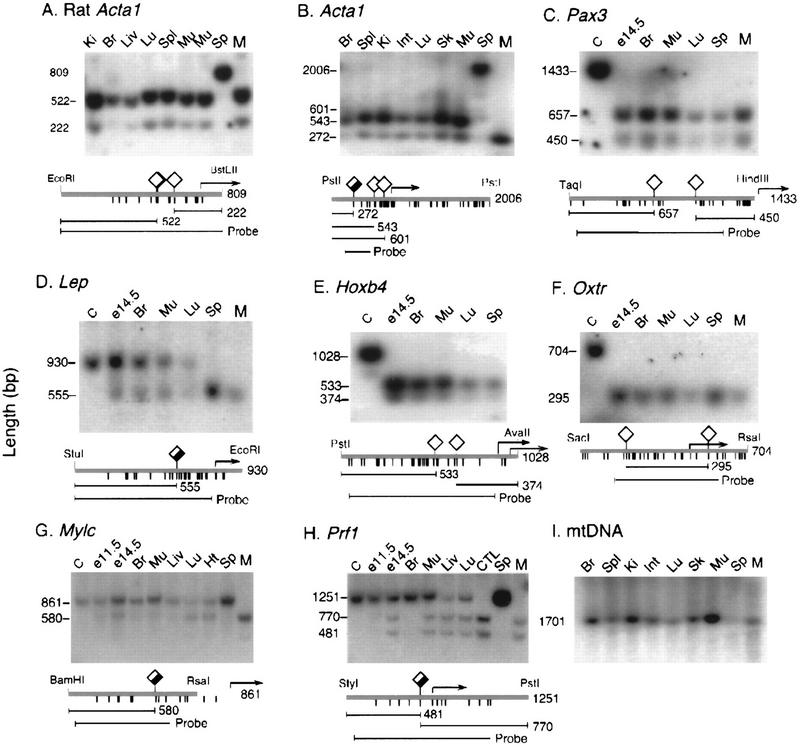
Figure 1.
Methylation analysis of the 5′ regions of tissue-specific genes. Diamonds above the heavy line indicate HpaII sites for all but E, in which HhaI sites (5′-GCGC-3′) are denoted. The amount of fill in the diamond symbol represents average methylation level in nonexpressing tissues. Tick marks below the heavy line indicate the positions of each CpG dinucleotide. Probes and predicted fragment sizes are shown at the bottom of each panel, and transcription start sites are indicated by arrows. Lanes C contained DNA not cleaved with methylation-sensitive enzymes; lanes M contained DNA digested with MspI (absent in E, which contained HhaI digests). DNA of sperm is completely methylated at tested sites in A, B, G, and H and completely unmethylated in C, D, E, and F. Except for Lep in D, none of the latter genes show detectable methylation in somatic tissues. Prf1 in I is completely unmethylated in Prf1-expressing cytotoxic T lymphocytes and largely unmethylated in lung, liver, and whole E14.5 embryos, none of which express appreciable levels of Prf1. Methylation levels of non-CpG island genes are generally higher in brain and whole E11.5 embryos than in E14.5 embryos and adult organs. (E11.5 and E14.5) Whole mouse embryos at 11.5 and 14.5 days postcoitum; (Ki) kidney; (Br) brain; (Liv) liver; (Spl) spleen; (Mu) skeletal muscle from body wall; (Sp) sperm; (Ht) heart; (CTL) cytotoxic T cells purified by flow sorting mouse lymphocytes stained with antibody to CD8.
The methylation status of a panel of promoters whose activity is restricted to specific cell types was evaluated by DNA blot hybridization after cleavage with methylation-sensitive restriction endonucleases. Pax3, which is expressed in precursors of limb muscles and regions of the central nervous system (Natoli et al. 1997), is not appreciably methylated in any tissue or in sperm; the same is true of Oxtr [oxytocin receptor, expressed in myometrium, endometrium, mammary gland, and ovary (Kubota et al. 1996)], and Hoxb4 [lung, kidney, testes; not present in liver, heart, or spleen (Gutman et al. 1994)]. Complete demethylation in sperm and partial methylation in somatic tissues was observed only for Lep [leptin; expressed primarily in adipocytes (Hoggard et al. 1997; Mason et al. 1998), whereas complete methylation in sperm DNA and partial methylation in somatic tissues was observed for Mylc [alkaline myosin light chain; ventricular myocardium and slow skeletal muscle (Barton et al. 1985)] and Prf1 [pore-forming protein; cytotoxic T lymphocytes and natural killer cells (Youn et al. 1991; Lichtenheld et al. 1995)]. In the case of Prf1 it was possible to analyze the first CpG dinucleotide upstream of the transcription start site. Methylation immediately upstream of the promoter has been reported to repress transcription most strongly (Busslinger et al. 1983). This region was only partially methylated in tissues that do not express Prf1 (Fig. 1H).
It is concluded that promoter methylation status is not closely associated with transcriptional activity for any of the randomly selected tissue-specific genes examined here. It should be noted that complete methylation was observed only in certain promoters in sperm DNA; somatic tissues showed either no methylation or partial methylation. The Prf1 promoter showed complete demethylation of the first CpG dinucleotide upstream of the transcription start site in cytotoxic T lymphocytes (which express the gene), but this site was largely demethylated in lung and liver, which do not express Prf1. It is also noted that partial methylation is observed only within domains that have low CpG densities and are less sensitive to cytosine methylation (Hsieh 1994; Kass et al. 1997).
Methylation history of CpG islands
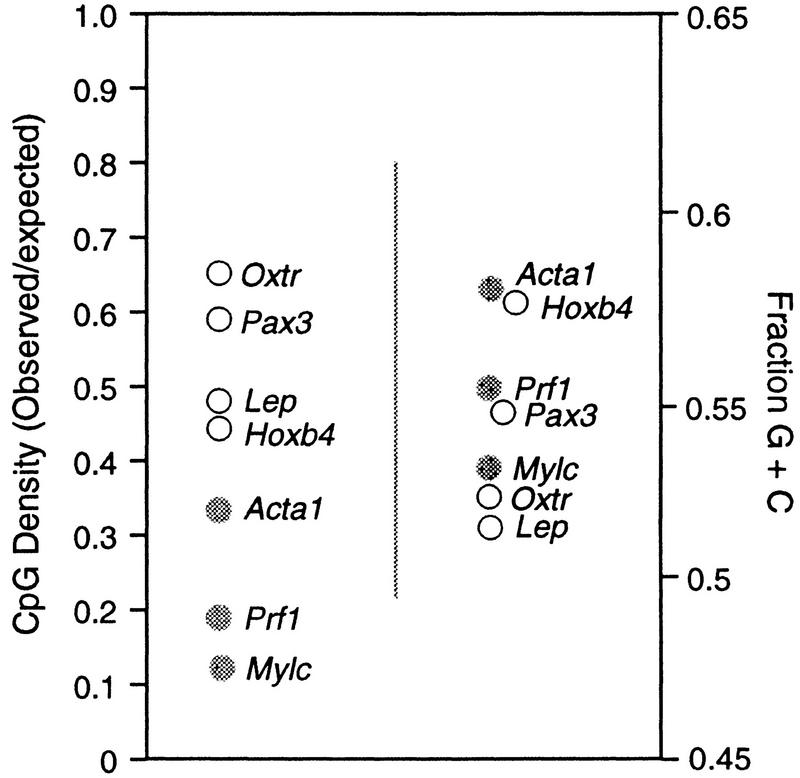
CpG islands are associated with both housekeeping and tissue-specific genes and are not normally methylated at any developmental stage (Cooper et al. 1983; Tyckocinski and Max 1984), except when associated with certain imprinted genes and genes subject to X inactivation. Of the seven promoters tested in this study, three (Acta1, Mylc, and Prf1) were methylated at all tested sites in DNA of sperm, whereas the other four (Hoxb4, Lep, Pax3, and Oxtr) were not detectably methylated in sperm and showed lower levels of partial methylation in somatic tissues. Inspection of dinucleotide frequencies around the cap site (Fig. 2) showed that the genes that were unmethylated in sperm DNA were CpG island genes with mean observed/expected CpG densities of 0.54 (range 0.44–0.65) whereas the mean value for promoters that were methylated was 0.19 (range 0.11–0.33; P < 0.03). These data confirm that CpG island domains were not subject to methylation in the male germ line, whereas genes that lack islands are heavily methylated in the spermatozoon but undergo demethylation during early development. A lack of promoter methylation in sperm DNA is proposed as an objective criterion for the designation of sequence domains as CpG islands.
Figure 2.
CpG densities predict methylation status of 5′ regions of tissue-specific genes in sperm DNA. The 5′ regions of the genes in Fig. 1 were analyzed for CpG density, G+C content, and methylation status. (•) Methylated sequences; (○) unmethylated sequences. Notice that sequences with higher observed/expected values for CpG densities tend to be unmethylated in sperm DNA. CpG densities (left) are a more accurate predictor of methylation status in sperm DNA than are G+C contents (right).
Normal expression of tissue-specific genes but biallelic expression of imprinted genes and activation of IAP elements in Dnmt1N mutant mouse embryos
Dnmt1N is a targeted allele of Dnmt1 that reduces Dnmt1 protein levels by 95% and allows homozygous embryos to develop to day 10.5 postcoitum (Li et al. 1992), at which time methylation levels fall to ∼30% of wild type. Mutant embryos display normal morphology but die as a result of apoptosis in all tissues (Li et al. 1992; Trasler et al. 1995). The S and C alleles of Dnmt1 are severe or null alleles that stop development at embryonic day 8.5 (E8.5) or earlier and show more severe developmental asynchrony. Whereas 5-methylcytosine levels are lower at the time of death in Dnmt1S and Dnmt1C homozygotes than in Dnmt1N homozygotes, in none of the mutants do they approach zero, and some imprinted genes are not reactivated in any Dnmt1 mutant background (Caspary et al. 1998). Dnmt1N homozygotes had been shown previously to express the H19 gene from both the maternal and paternal alleles (it is normally expressed only from the allele of maternal origin) and to express Igf2 from neither allele (the paternal allele is normally expressed in most tissues; Li et al. 1993a;b). Dnmt1N homozygotes also display ectopic X inactivation due to demethylation of Xist (Panning and Jaenisch 1996) and transcribe IAP endogenous retroviruses at very high levels (Walsh et al. 1998).
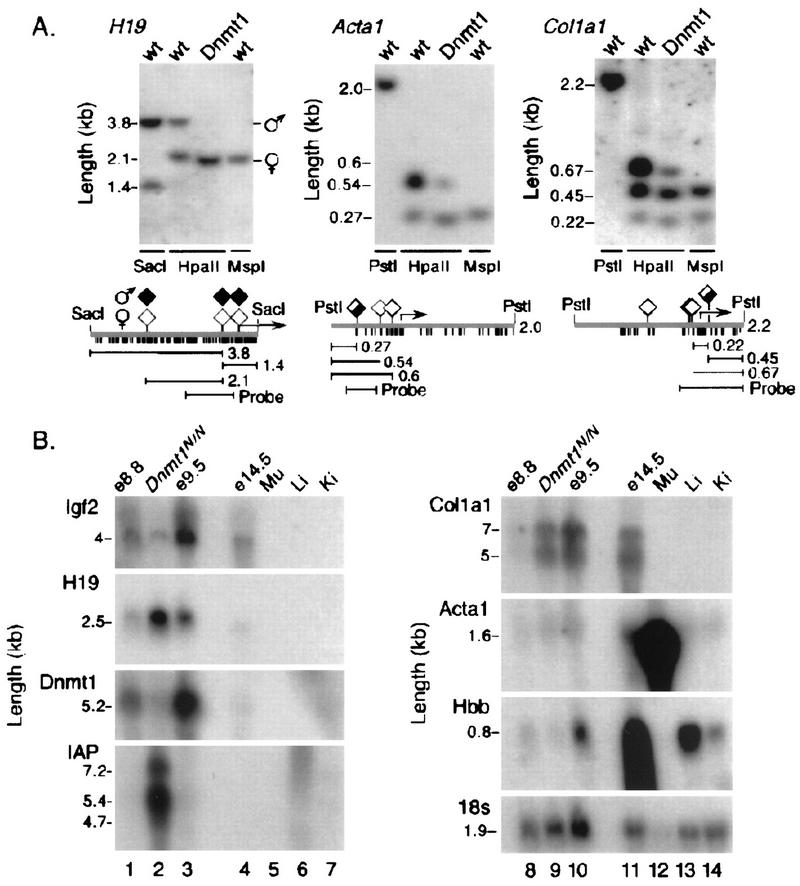
A single RNA blot containing DNA from Dnmt1N homozygotes and wild-type embryos was hybridized consecutively with cDNA probes for H19, Igf2, Dnmt1, IAP elements, and three tissue-specific genes that had been reported to be regulated by methylation of promoter regions and are activated at ∼9.5 days of gestation. Dnmt1N was chosen over other alleles because Dnmt1S and Dnmt1C homozygotes show greater developmental asynchrony, and precise staging, which is essential for the purposes of this experiment, is more difficult than for Dnmt1N homozygotes. RNA from Dnmt1N mutant embryos was flanked by RNA samples from normal E8.8 and E9.5 embryos to control for any developmental asynchrony among the embryos. Southern blot analysis showed that the paternal allele of H19 is demethylated in mutant embryos (Fig. 3A) and H19 RNA can be seen to increase in amount while Igf2 decreases (Fig. 3B) as reported previously (Li et al. 1993a). Dnmt1 mRNA is barely detectable in the mutant embryos, whereas IAP transcripts are expressed at greatly increased levels (Fig. 3B; Walsh et al. 1998). The genes for Iα1 collagen (Col1a1; Rhodes et al. 1994), skeletal muscle α-actin (Acta1; Weiss et al. 1996), and β-globin (Hbb; Loo and Cauchi 1992) have been reported to be regulated by promoter methylation, and transcripts of all three genes are first detectable around day E10. However, the promoters of Acta1 and Col1a1 are largely unmethylated in control DNA (Fig. 3A), and Figure 3B shows that there is no up-regulation of these genes under conditions that caused further demethylation. The decrease in β-globin mRNA in the mutants is consistent with the reduced number of erythroid cells observed in histological analysis of embryos homozygous for Dnmt1N (Li et al. 1992). Furthermore, the β-globin promoter contains only one CpG dinucleotide, whose methylation status is not associated with transcriptional silencing when present as a transgene (Garrick et al. 1996). These results indicate that under conditions where demethylation has caused dysregulation of imprinted loci and IAP retroviral LTR promoters there is no detectable precocious expression of genes that have been reported to be repressed in non expressing tissues by methylation of promoter sequences.
Figure 3.
Normal expression of tissue-specific genes in DNA methyltransferase-deficient mouse embryos. (A) The genes of interest were largely unmethylated in control samples and underwent additional demethylation in Dnmt1-deficient mouse embryos at day E9.5. (B) A single RNA blot was hybridized consecutively with the eight probes shown. H19 and IAP retroviral transcripts can be seen to increase in the mutants; Igf2 declines and Dnmt1 mRNA is barely detectable. Col1a1, Acta1, and Hbb mRNAs are largely unaffected or reduced in amount in the mutant embryos, although all three genes had been reported to be repressed by cytosine methylation.
Discussion
It was noted recently that most (∼90%) of the ∼3 × 107 5-methylcytosine residues in human DNA actually lie within the retroposons and endogenous retroviruses that represent at least 35% of the mammalian genome (Yoder et al. 1997). Furthermore, stringent allele-specific gene expression has to date been documented only in organisms that bear modified bases in their DNA (Bestor 1998). Most cytosine methylation therefore appears to be involved in host defense against the activation of retroposon promoters (Bestor 1990; Yoder et al. 1997; Walsh et al. 1998), with smaller fractions having key roles in genomic imprinting (Li et al. 1993a) and X chromosome inactivation (Panning and Jaenisch 1996). The central role attributed to sequence-specific promoter methylation and demethylation in developmental gene control does not enjoy a comparable body of supporting evidence, and we report here a reevaluation of the issue.
The role of cytosine methylation in developmental gene control was addressed directly by examination of a panel of tissue-specific genes for evidence of tissue-specific methylation differences. No strong correlations between methylation density and transcriptional activity were observed, and promoter regions were found to be partially or completely unmethylated at tested sites in all tissues. Partial methylation was observed only within domains that had low CpG densities, and according to the findings of Hsieh (1994) and Kass et al. (1997) the observed levels of methylation in the partially methylated examples would not be expected to cause transcriptional silencing. Cytosine methylation has been implicated in the normal transcriptional regulation of the Acta1 gene in cell culture systems (Yisraeli et al. 1986; Weiss et al. 1996). Although the promoter of this gene is heavily methylated in sperm DNA, the promoter region is largely unmethylated at tested sites in both muscle and non-muscle tissues, as reported previously for rat Acta1 by Shani et al. (1984). Further demethylation in Dnmt1-deficient mouse embryos did not increase expression levels of the α-actin gene. The lack of discernible tissue-specific methylation patterns and the insensitivity to demethylating conditions indicate that the α-actin gene is unlikely to be regulated by cytosine methylation. The collagen Iα1 and β-globin genes have also been reported to be regulated by cytosine methylation (Loo and Cauchi 1992; Rhodes et al. 1994), but activation of these genes was not observed in Dnmt1-deficient mouse embryos under conditions that induced IAP retrovirus transcription and caused biallelic expression of the imprinted genes Igf2 and H19. A central role for cytosine methylation in development is also unlikely in view of the lack of teratology or incipient developmental defects in Dnmt1-deficient mouse embryos, which die as a result of widespread apoptosis rather than defects in morphogenesis (Li et al. 1992).
Most of the experimental support for the methylation and development hypotheses has been in the form of methylation–expression correlations. Even though there is likely to be a reporting bias in favor of genes that show strong tissue-specific methylation differences, it appears that none of the published examples have met a key criterion: demonstration that a methylation pattern observed in a nonexpressing tissue can prevent transcription in a cell type normally capable of transcribing the gene of interest. A recent example is the CD43 (leukosialin) gene, which is heavily methylated at two HpaII sites in nonexpressing cells and unmethylated at those sites in expressing cells (Kudo and Fukuda 1995). The gene was concluded to be regulated by cytosine methylation even though in vitro methylation of the HpaII sites did not repress transcription (Kudo and Fukuda 1995). A similar conclusion was reached for the galectin-1 gene (Salvatore et al. 1998), which careful genomic sequencing showed to be largely unmethylated 5′ of the transcription start site in expressing tissues and lightly and variably methylated in nonexpressing tissues (both active and inactive genes were methylated at a site next to a transposon in the first intron; see below). There was no evidence that the observed methylation patterns were capable of repressing transcription of the galectin-1 gene, and the observed density of methylation would not be expected to trigger transcriptional silencing (Hsieh 1994; Kass et al. 1997).
The observed association between increased methylation levels and transcriptional inactivity in earlier reports may be explained by expansion of methylated domains that flank many promoter regions. Ono and colleagues observed methylation patterns encroaching on the promoters of housekeeping genes in aging mice (Uehara et al. 1989). The proximity of transposable elements and retroviral DNA, which represent >35% of the genome and are heavily methylated (Yoder et al. 1997), can increase the likelihood of de novo methylation in flanking cellular sequences. This was first observed for cellular sequences in the vicinity of a retrovirus integration site (Jähner and Jaenisch 1985). Magewu and Jones (1994) found that an exon of the TP53 gene that is especially prone to de novo methylation was adjacent to Alu elements in a flanking intron and that de novo methylation of the exon depended on the nearby Alu elements. A recent genomic sequencing study has shown that a single CpG site in the first intron of the galectin-1 gene is heavily methylated in both expressing and nonexpressing tissues (Salvatore et al. 1998), and inspection of this neighboring sequence with RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) revealed that the only transposable element in this region [a MIR (mammalian-wide interspersed repeat) element] is immediately adjacent to the methylated CpG site.
Encroachment of methylation patterns from flanking retroposon sequence into regulatory sequences of endogenous genes results in light and variable methylation at the promoters of non-CpG island genes, which is unlikely to prevent normal activation. The binding and activity of Sp1 and some other transcription factors is insensitive to cytosine methylation (Brandeis et al. 1994; Macleod et al. 1994; Matsuo et al. 1998), and transcription factor binding can induce the demethylation of local CpG sites in a replication-dependent manner (Matsuo et al. 1998). This result suggests that tissue-specific transcription factors might overcome and then induce erasure of methylation patterns in the vicinity of specific binding sites to produce the impression of regulated tissue-specific methylation. According to this explanation, many of the observed tissue-specific methylation patterns within regulatory regions are a consequence, rather than a cause, of transcriptional activation.
Given the length of time that has elapsed since the methylation–development hypotheses were first put forward and the still-ambiguous nature of the data that support them, it seems an appropriate time to introduce an alternative model. We propose a model under which cytosine methylation has only a very minor role in the regulation of mammalian development, which instead depends on regulatory networks homologous to those that regulate the development of other metazoa. This model suggests that most cytosine methylation in mammals participates in highly specialized functions such as allele-specific gene expression (as seen in genomic imprinting and X chromosome inactivation) and the heritable transcriptional silencing of parasitic sequences.
Materials and methods
Isolation of genomic DNA
C57BL/6J mice were obtained from the Jackson Laboratories. The Dnmt1N allele (Li et al. 1992, 1993b) was backcrossed to mice of strain C57BL/6J for >15 generations. Animals were sacrificed at the indicated ages, with E0.5 being the day the plug was observed. Tissues were digested overnight in 50 mm Tris-HCl (pH 8.0), 100 mm EDTA, 0.5% SDS, and 100 μg/ml proteinase K at 50°C, and extracted twice with phenol/chloroform/isoamylalcohol (25:24:1). Sperm were collected from the cauda epididymus of adult males, washed once in PBS, resuspended in 10 mm Tris-HCl (pH 7.5), 10 mm EDTA, 2% SDS, 140 mm 2-mercaptoethanol, and 100 μg/ml proteinase K, and incubated overnight at 50°C before phenol extraction. DNA concentrations were determined using a DNA fluorometer and Hoechst 33258 (Hoefer Scientific Instruments, San Francisco, CA). Rat tissues were the gift of B. Tycko, and DNA from CD8+ cytotoxic T lymphocytes was the gift of G. Siu (both from Columbia University).
Probes for DNA blot hybridization analyses
Unless noted otherwise, the probes were generated by PCR with the indicated primer pairs cloned into the plasmid vector pCR2.1 (Invitrogen), and confirmed by DNA sequencing. All numbering is with respect to the start sites in the respective GenBank or EMBL entries.
Acta1: Plasmid pBSαactin containing 809 bp of the rat promoter (Yisraeli et al. 1986) was the kind gift of J. Swisher and A.M. Pyle (Columbia University). The full promoter sequence was obtained by combining data on pBS α-actin with EMBL accession number V01218. For the mouse gene, the sequence from −623 to +339 bp (GenBank accession no. M12347) was amplified using the primers TCCTACACTGACTGACAGCC and TGGTAATGGATAGATGTCAG. Pax3: Primers TGAACAACTACACGCACGCC and CCTCTACATGAGATTCAGCC were used to amplify and subclone the region from −1389 to −292 bp (GenBank accession no. U61230). Lep: Primers GTCTGGCTGGTTCTGAGGAT and GTGCCACTTGCGCAACTGTC were used to amplify and subclone the region from −852 to −43bp (GenBank accession no. U36238). Hoxb4: Primers TCTTGTGCGTGTGATGTTGG and CCTACTTACTGTCAAGTGAAC were used to amplify and subclone the region from −817 to +205 bp with respect to the first transcriptional start site (−895 to +127 with respect to the second) (GenBank accession no. X71912). Oxtr: Primers CCAAGGTTCCTATATCTCTG and GAAGCGAGACTGCAGGTTAG were used to amplify and subclone the region from −374 to +109 bp (GenBank accession no. D86631). Mylc: Primers GGATAACAGGAGTAGTAGAGG and TGCATTGTCTGGTGTCGCTG were used to amplify and subclone the region from −1201 to −549 bp (GenBank accession no. X12972). Prf1: Primers ACAGTAACCTCAGGCAGAAC and ATGTGCTCCATACTTGGCTC were used to amplify and subclone the region from −571 to +356 bp (GenBank accession no. G200293). mtDNA: A fragment of the mouse mtDNA genome corresponding to nucleotides 901–1620 on the standard map (GenBank accession no. J01420) generated using the primers ACACACCGCCCGTCACCCTCC and GGCTGCTTTTAGGCCTACAATGG was kindly donated by E. Schon (Columbia University). H19: The plasmid pNotBEco containing a fragment of the mouse gene spanning the BaglII–EcoRI fragment immediately upstream of the promoter (−1992 to −53; GenBank accession no. AF049091) in pBluescript KS was a kind gift of L. Dandolo (Columbia University). Col1a1: The plasmid pSTBB2.6 contained genomic sequence encompassing the region from −220 to +2359 bp (including exons 1–5) and was generously donated by M. Breindl (Rhodes et al. 1994).
Probes for RNA analyses
Igf2: Plasmid p27c containing the rat cDNA (545 bp) was a kind gift of A. Efstratiadis (Columbia University). H19: A PCR product spanning a region from the fourth to fifth exons was a kind gift of B. Tycko. Dnmt1: The cDNA clone used was pMG and contained 5 kb of the 5.2-kb sequence (Bestor et al. 1988; GenBank accession no. X14805). IAP elements: The IΔ1 IAP probe used was as described previously (Walsh et al. 1998) and was kindly provided by T. Vasicek and S. Tilghman (Princeton University, NJ). Col1a1: A probe for the 3′-untranslated region of the gene that does not cross-hybridize to other collagen mRNAs (Metsäranta et al. 1991) was generated using the primers GGATCCCGGACTAGACATTGGC and CTCGAGTGGTAAGGTTGAATGC, which contain a BamHI and XhoI site, respectively. Acta1: The primers GTGAGCCTTGGAGCCAG and CAACCAAGGCTCAATAGC were used to generate a probe for the first noncoding exon of the gene, which has been shown previously not to cross-hybridize to other actin mRNAs (Sassoon et al. 1988). Hbb: A plasmid containing a mouse β-globin cDNA (Rougeon and Mach 1977) fragment was the kind gift of F. Costantini (Columbia University). 18s: The oligonucleo tide ACGGTATCTGATCGTCTTCGAACC was end-labeled with [γ-32P]ATP and polynucleotide kinase and used to visualize 18S rRNA as a means of normalizing RNA loadings.
Blot hybridization of Genomic DNA
Genomic DNA (20 μg) was digested for at least 4 hr with a fivefold excess of restriction enzymes in the appropriate buffer supplemented with 1 mm spermidine. After electrophoresis (1% agarose) the DNA was transferred to a Nytran Plus nylon membrane (Schleicher & Schuell) by capillary transfer using the manufacturer’s protocol. Membranes were UV cross-linked, heated to 80°C for 2 hr in a vacuum oven, and prehybridized in 6× SSC, 5× Denhardt’s reagent, and 1% SDS for 1–2 hr and hybridized in fresh solution containing 9 mg/ml herring testes DNA (Sigma) and 1 × 107 cpm/ml of probe [made with Highprime (Boehringer Mannheim) according to the manufacturer’s recommendations] at 65°C overnight. Blots were washed twice with 0.1× SSC, 1% SDS, at 65°C and exposed to film overnight or Molecular Dynamics storage Phosphor plates. Membranes were stripped as recommended by the manufacturer and reprobed with mitochondrial probes to confirm limit digestion.
Preparation and analysis of RNA
RNA was prepared as described previously (Chomczynski and Sacchi 1987) from wild-type and Dnmt1N/N mutant embryos. Only embryos that showed beating hearts and intact RNA were used for RNA blot analysis. Purified RNA was fractionated by electrophoresis on gels containing 18% formaldehyde, 0.8% agarose, 1× MOPS buffer and blotted to Nytran Plus (Schleicher & Schuell) in 20× SSC. Hybridization was performed under the conditions described by Church and Gilbert (1984). Membranes were stripped in accordance with the manufacturer’s instructions. 18S rRNA hybridizations were carried out using an end-labeled oligonucleotide hybridized in 6× SSC, 5× Denhardt’s reagent, 1% SDS, 0.05% sodium pyrophosphate, and 9 mg/ml herring testes DNA at 42°C. Filters were washed twice for 30 min each with 6× SSC, 0.05% pyrophosphate, and 1% SDS at 55°C.
Acknowledgments
We thank the colleagues named in Materials and Methods for gifts of probes and DNA, K.V. Anderson for comments on the manuscript, and B. Tycko for helpful discussions. This work was supported by fellowships from Cancerfonden of Sweden and the Leukemia Foundation (to C.P.W.) and by grants from the National Institutes of Health and the Leukemia Society of America (to T.H.B.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL THB12@columbia.edu; FAX (212) 740-0992.
References
- Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;6:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Baron MH, Maniatis T. Rapid reprogramming of globin gene expression in transient heterokaryons. Cell. 1986;46:591–602. doi: 10.1016/0092-8674(86)90885-8. [DOI] [PubMed] [Google Scholar]
- Barton PJ, Cohen L, Robert B, Fiszman MY, Bonhomme F, Guenet JL, Leader DP, Buckingham ME. The myosin alkali light chains of mouse ventricular and slow skeletal muscle are indistinguishable and are encoded by the same gene. J Biol Chem. 1985;260:8578–8584. [PubMed] [Google Scholar]
- Bestor TH, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzyme is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bestor TH. DNA methylation: How a bacterial immune function has evolved into a regulator of gene expression and genome structure in higher eukaryotes. Phil Trans R Soc Lond Biol Sci. 1990;326:179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- ————— Cytosine methylation and the unequal developmental potentials of the oocyte and sperm genomes. Am J Hum Genet. 1998;62:1269–1273. doi: 10.1086/301891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH, Coxon A. The pros and cons of DNA methylation. Curr Biol. 1993;3:384–386. doi: 10.1016/0960-9822(93)90209-7. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- Busslinger M, Hurst J, Flavell RA. DNA methylation and the regulation of globin gene expression. Cell. 1983;34:197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chiu CP, Blau HM. Reprogramming cell differentiation in the absence of DNA synthesis. Cell. 1984;37:879–887. doi: 10.1016/0092-8674(84)90423-9. [DOI] [PubMed] [Google Scholar]
- ————— 5-Azacytidine permits gene activation in a previously noninducible cell type. Cell. 1985;40:417–424. doi: 10.1016/0092-8674(85)90155-2. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Taggart MH, Bird AP. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983;11:647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RD, Buckingham ME. Actin and myosin genes are transcriptionally regulated during mouse skeletal muscle development. Dev Biol. 1992;149:228–234. doi: 10.1016/0012-1606(92)90279-p. [DOI] [PubMed] [Google Scholar]
- Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Devel. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- Garrick D, Sutherland H, Robertson G, Whitelaw E. Variegated expression of a globin transgene correlates with chromatin accessibility but not methylation status. Nucleic Acids Res. 1996;24:4902–4909. doi: 10.1093/nar/24.24.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. Cells, embryos, and evolution. New York, NY: Blackwell; 1997. [Google Scholar]
- Gutman A, Gilthorpe J, Rigby PWJ. Multiple positive and negative regulatory elements in the promoter of the mouse homeobox gene Hoxb4. Mol Cell Biol. 1994;14:8143–8154. doi: 10.1128/mcb.14.12.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard N, Hunter L, Duncan JS, Williams LM, Trayhurn P, Mercer JG. Leptin and leptin receptor mRNA and protein expression in the murine fetus and placenta. Proc Natl Acad Sci. 1997;94:11073–11078. doi: 10.1073/pnas.94.20.11073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Hsiao WL, Gattoni-Celli S, Kirschmeier P, Weinstein IB. Effects of 5-azacytidine on methylation and expression of specific DNA sequences in C3H 10T1/2 cells. Mol Cell Biol. 1984;4:634–641. doi: 10.1128/mcb.4.4.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jähner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- Jones PA, Wolkowicz MJ, Rideout WM, III, Gonzales FA, Marziasz CM, Coetzee GA, Tapscott SJ. De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proc Natl Acad Sci. 1990;87:6117–6121. doi: 10.1073/pnas.87.16.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Kimura T, Hashimoto K, Tokugawa Y, Nobunaga K, Azuma C, Saji F, Murata Y. Structure and expression of the mouse oxytocin receptor gene. Mol Cell Endocrinol. 1996;124:25–32. doi: 10.1016/s0303-7207(96)03923-8. [DOI] [PubMed] [Google Scholar]
- Kudo S, Fukuda M. Tissue specific transciptional regulation of human leukosialin (CD43) gene is achieved by DNA methylation. J Biol Chem. 1995;270:13298–13302. doi: 10.1074/jbc.270.22.13298. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993a;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Forster AC, Bestor TH, Jaenisch R. DNA methylation, genomic imprinting, and mammalian development. Cold Spring Harb Symp Quant Biol. 1993b;58:297–305. doi: 10.1101/sqb.1993.058.01.035. [DOI] [PubMed] [Google Scholar]
- Lichtenheld MG, Podack ER, Levy RB. Transgenic control of perforin gene expression. Functional evidence for two separate control regions. J Immunol. 1995;154:2153–2163. [PubMed] [Google Scholar]
- Loo LS, Cauchi MN. DNA methylation patterns of the γ-δ-β genes in human fetal and adult erythroid tissues Am. J Hematol. 1992;39:289–293. doi: 10.1002/ajh.2830390410. [DOI] [PubMed] [Google Scholar]
- Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes & Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- Magewu AN, Jones PA. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol Cell Biol. 1994;14:4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MM, He Y, Chen H, Quon MJ, Reitman M. Regulation of leptin promoter function by Sp1, C/EBP, and a novel factor. Endocrinology. 1998;139:1013–1022. doi: 10.1210/endo.139.3.5792. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsáranta M, Toman D, De Crombrugghe B, Vuorio E. Specific hybridization probes for mouse type I, II, III and IX collagen mRNAs. Biochim Biophys Acta. 1991;1089:241–243. doi: 10.1016/0167-4781(91)90014-d. [DOI] [PubMed] [Google Scholar]
- Miniou P, Jeanpierre M, Blanquet V, Sibella V, Bonneau D, Herbelin C, Fischer A, Niveleau A, Viegas-Pequignot E. Abnormal methylation pattern in constitutive and facultative (X inactive chromosome) heterochromatin of ICF patients. Hum Mol Genet. 1994;3:2093–2102. doi: 10.1093/hmg/3.12.2093. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Natoli TA, Ellsworth MK, Wu C, Gross KW, Pruitt SC. Positive and negative DNA sequence elements are required to establish the pattern of Pax3 expression. Development. 1997;124:617–626. doi: 10.1242/dev.124.3.617. [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes & Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- Rhodes K, Rippe RA, Umezawa A, Nehls M, Brenner DA, Breindl M. DNA methylation represses the murine 1 α (I) collagen promoter by an indirect mechanism. Mol Cell Biol. 1994;14:5950–5960. doi: 10.1128/mcb.14.9.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Rogers JM, Francis BM, Sulik KK, Alles AJ, Massaro EJ, Zucker RM, Elstein KH, Rosen MB, Chernoff N. Cell death and cell cycle perturbation in the developmental toxicity of the demethylating agent, 5-aza-2′-deoxycytidine. Teratology. 1994;50:332–339. doi: 10.1002/tera.1420500504. [DOI] [PubMed] [Google Scholar]
- Rougeon F, Mach B. Cloning and amplification of α and β mouse globin gene sequences synthesised in vitro. Gene. 1977;1:229–239. doi: 10.1016/0378-1119(77)90047-6. [DOI] [PubMed] [Google Scholar]
- Rougier N, Bourc’his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes & Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore P, Benvenuto G, Caporaso M, Bruni CB, Chiariotti L. High resolution methylation analysis of the galectin-1 gene promoter region in expressing and nonexpressing tissues. FEBS Lett. 1998;421:152–158. doi: 10.1016/s0014-5793(97)01553-6. [DOI] [PubMed] [Google Scholar]
- Sassoon DA, Garner I, Bockingham W. Transcripts of α-cardiac and α-skeletal actins are early markers for myogenesis in the mouse embryo. Development. 1988;104:155–164. doi: 10.1242/dev.104.1.155. [DOI] [PubMed] [Google Scholar]
- Schuffenhauer S, Bartsch O, Stumm M, Buchholz T, Petropoulou T, Kraft S, Belohradsky B, Hinkel GK, Meitinger T, Wegner RD. DNA, FISH and complementation studies in ICF syndrome: DNA hypomethylation of repetitive and single copy loci and evidence for a trans acting factor. Hum Genet. 1995;96:562–571. doi: 10.1007/BF00197412. [DOI] [PubMed] [Google Scholar]
- Shani M, Admon S, Yaffe D. The methylation state of 2 muscle-specific genes: Restriction enzyme analysis did not detect a correlation with expression. Nucleic Acids Res. 1984;12:7225–7234. doi: 10.1093/nar/12.18.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasler JM, Trasler DG, Bestor TH, Li E, Ghibu F. DNA methyltransferase in normal and DnmtN/DnmtN mouse embryos. Dev Dyn. 1995;206:239–247. doi: 10.1002/(SICI)1097-0177(199607)206:3<239::AID-AJA2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Tykocinski ML, Max EE. CG dinucleotide clusters in MHC genes and in 5′ demethylated genes. Nucleic Acids Res. 1984;12:4385–4396. doi: 10.1093/nar/12.10.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara Y, Ono T, Kurishita A, Kokuryu H, Okada S. Age-dependent and tissue-specific changes of DNA methylation within and around the c-fos gene in mice. Oncogene. 1989;4:1023–1028. [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retrovirus is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: Involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- Yisraeli J, Adelstein RS, Melloul D, Nudel U, Yaffe D, Cedar H. Muscle-specific activation of a methylated chimeric actin gene. Cell. 1986;46:409–416. doi: 10.1016/0092-8674(86)90661-6. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Youn BS, Liu CC, Kim KK, Young JD, Kwon MH, Kwon BS. Structure of the mouse pore-forming protein (perforin) gene: Analysis of transcription initiation site, 5′ flanking sequence, and alternative splicing of 5′ untranslated regions. J Exp Med. 1991;173:813–822. doi: 10.1084/jem.173.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]