Abstract
Setleis Syndrome (OMIM ID: 227260) is a rare autosomal recessive disease characterized by abnormal facial development. Recently, we have reported that two nonsense mutations (c.486C>T [Q119X] and c.324C>T [Q65X]) of the basic helix-loop-helix (bHLH) transcription factor TWIST2 cause Setleis Syndrome. Here we show that periostin, a cell adhesion protein involved in connective tissue development and maintenance, is down-regulated in Setleis Syndrome patient fibroblast cells and that periostin positively responds to manipulations in TWIST2 levels, suggesting that TWIST2 is a transactivator of periostin. Functional analysis of the TWIST2 mutant form (Q119X) revealed that it maintains the ability to localize to the nucleus, forms homo and heterodimers with the ubiquitous bHLH protein E12, and binds to dsDNA. Reporter gene assays using deletion constructs of the human periostin promoter also reveal that TWIST2 can activate this gene more specifically than Twist1, while the Q119X mutant results in no significant transactivation. Chromatin immunoprecipitation assays show that both wild-type TWIST2 and the Q119X mutant bind the periostin promoter, however only wild-type TWIST2 is associated with higher levels of histone acetylation across the 5′-regulatory region of periostin. Taken together, these data suggest that the C-terminal domain of TWIST2, which is missing in the Q119X mutant form of TWIST2, is responsible for proper transactivation of the periostin gene. Improper regulation of periostin by the mutant form of TWIST2 could help explain some of the soft tissue abnormalities seen in these patients therefore providing a genotype-phenotype relationship for Setleis Syndrome.
Keywords: TWIST1, Dermo-1, Focal Facial Dermal Dysplasia, skin development
1. Introduction
Setleis Syndrome is a rare autosomal recessive disorder characterized as a focal facial dermal dysplasia type III [1]. Patients exhibit dysmorphic facial characteristics such as forceps-like lesions at the temples, eyelash and eyebrow abnormalities, wrinkled facial skin, bulbous nose, thick protruding lips and chin defects, giving them a leonine appearance [2,3]. Recently, we demonstrated that Setleis Syndrome is caused by two nonsense mutations of the basic helix-loop-helix (bHLH) transcription factor TWIST2 (c.324C>T [p.Q65X] and c.486C>T [p.Q119X]) resulting in truncated forms of the protein [4].
TWIST2 is an important regulator of gene expression during the differentiation of mesenchymal cell lineages. It is expressed mainly in the craniofacial mesenchyme, chondrogenic precursors, and in the dermis of the skin, therefore it directs the development of dermal and chondrogenic tissues [5,6]. TWIST2 belongs to the Twist subfamily of transcription factors that share identity and conservation in the bHLH domain that include TWIST1, PARAXIS, SCLERAXIS, HAND-1, and HAND-2 [7]. Their tissue specific expression pattern classifies them as Class B bHLH transcription factors, which are known to heterodimerize with the ubiquitously expressed Class A bHLH factors, such as E12/E47 [8]. Dimerization of bHLH factors is mediated through the helix-loop-helix domain, allowing the basic domain to bind specifically to conserved E-box sequences (5′-CANNTG-3′) that are found throughout the regulatory regions of many lineage-specific genes [9,10]. Originally, TWIST2 was described as a repressor of gene activation capable of only forming heterodimers with E12 [11,12]. However, we now provide evidence that TWIST2 functions as an activator of gene expression through the formation of functional homodimers.
Periostin (POSTN) is a 90 kDa secreted homophilic cell adhesion protein involved in connective tissue development and maintenance [13]. Based on its expression pattern and function during development and wound repair, POSTN has been characterized as a matricellular protein that has the ability to modulate cell matrix interactions affecting aspects of cell morphology and differentiation [14]. It is expressed in many tissues such as fibroblasts, skin, osteoblasts, periosteum, periodontal ligament, and the endocardial cushions of the heart [14–16]. Periostin also regulates collagen I fibrillogenesis and contributes to the mechanical properties of connective tissues in skin and heart [17]. Because of its role in cell adhesion, migration and maintenance of connective tissue, POSTN has been implicated in cancer progression and invasiveness [18,19].
It is precisely the role of POSTN in connective tissue physiology that makes it a good target gene to help understand some of the pathogenesis involved in the facial abnormalities described for Setleis Syndrome patients. Previous reports have shown that POSTN is positively regulated by Twist1, a close family member of TWIST2 [20,21]. Although human TWIST2 and TWIST1 encode highly homologous proteins, the finding that TWIST2 recessive mutations cause Setleis Syndrome and dominant TWIST1 mutations cause Saethre-Chotzen Syndrome, a form of craniocynostosis, suggests that they may function independently in skin and bone formation [4,6]. Here we establish evidence for a genotype-phenotype relationship in Setleis Syndrome by demonstrating that TWIST2 is a transactivator of the human POSTN gene, and that a nonsense mutation (Q119X) found in Setleis Syndrome patients may function as a dominant negative mutation either by sequestering E proteins and/or by binding to E-box targets. Finally, a possible mechanism for TWIST2 transactivation is proposed where the C-terminal domain of TWIST2 allows for the acetylation of histones necessary for an open chromatin conformation characteristic of actively transcribed genes.
2. Materials and methods
2.1 Microarray analysis
Skin punch biopsies from the forearms of Setleis Syndrome patients homozygous for the TWIST2 Q119X mutation and normal controls from the same ethnic background were used to generate primary fibroblasts using standard protocols [4]. RNA samples isolated from cultured skin fibroblasts were prepared using the Qiagen RNeasy kit. RNA labeling and microarray hybridizations were performed by the Mount Sinai School of Medicine Microarray Facility. Principal components analysis (PCA) was applied to the expression microarray data set to evaluate variability. A summarized data set from each of the two groups of samples was obtained using Robust Multi-Chip Average (RMA) analysis for the background correction and normalization. We combined the Affymetrix data in order to assess a consistent difference between the mutant and wild-type profiles and used the following three criteria to find candidate genes: a) the modified t-statistic was no less than 3; b) the average fold-change was no less than a two-fold; c) the maximum log2 based signal intensity across the arrays was greater than 3.5, which is approximately twenty percentile of all log2 based signal intensities. The software used was LIMMA [22], a Bioconductor [23] package for the analysis of gene expression microarray data that uses linear models for the assessment of differential expression. The candidate genes identified in this manner are all those that satisfied the above criteria, which were ranked by average fold-change.
Real Time RT-PCR validation of differentially regulated genes from the microarray data was performed using RNAs extracted from primary fibroblast cell lines. derived from Setleis syndrome patients and controls. Total RNA was isolated using the QIAGEN RNAeasy kit following manufacturer’s instructions. Reaction conditions were designed and validated for the POSTN and three housekeeping genes (ACTB, RPS11 and TUBA1). Primers pairs are indicated in Supplementary Table S1. Reactions were done in 20 μl volumes using 10 ng of total RNA and 40 cycles of amplification using the QuantiTect SYBR Green RT-PCR Kit in a BioRad ICycler. The fold change of detected amplicons were calculated using the ΔΔCt method [24] by comparing the average threshold cycles (Ct) of the three housekeeping genes to that of the tested genes.
2.2. Plasmid transfection, silencing, and real time RT-PCR
Six-well tissue culture plates were seeded with 2×105 per well with fibroblast cells (GM00637) and grown to 80% confluence in 15% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin at 37 °C. Cells were transfected using N-terminally myc-tagged pCINEO TWIST2 wild-type and mutant constructs using the Lipofectamine 2000 reagent (Invitrogen) at a ratio of 2 μg DNA in 10 μl per well. As a negative control, a second set of fibroblasts were transfected with the pEGFP-N1 plasmid (Clontech) that contains the GFP gene driven by the same CMV promoter as the TWIST2 plasmid. Supplementary Figure S2 contains immunofluorescence images of fibroblast cells expressing GFP. For gene silencing, cells were transfected with pre-designed SMARTpool siTWIST2 (Dharmacon) targeted to human TWIST2 mRNA using DharmaFECT transfection reagents (Dharmacon), following the manufacturer’s protocol. A second set of fibroblast cells were transfected with non-targeting scrambled siRNA (Dharmacon) as a negative control for comparison with the experimental group. At 24–72 hrs post transfection, cells were harvested for RNA extraction using the RNAeasy kit (Qiagen). Transfection efficiency was determined by microscopy using a fluorescently labeled non-targeting siRNA (siGLO Dharmacon) transfected into the cells. Real time PCR reactions were carried out with 50 ng of total RNA template/reaction using the QuantiTect SYBR Green RT-PCR Kit (Qiagen, Inc., Valencia, CA). PCR primer sets for human POSTN and TWIST2 as well as the housekeeping genes were purchased from the QuantiTect Primer Assay collection (Qiagen, Valencia, CA).
2.3. Immunofluorescence microscopy and immunoblotting
HeLa cells were grown on round cover slips placed inside 6 well plates and grown to 70% confluence. Cells were transfected using pCINEO TWIST2 constructs using the Lipofectamine 2000 reagent (Invitrogen) as described above. At 24 hrs post transfection, cells were fixed with 4% paraformaldehyde for 1 hour, permeabilized with 0.2% Triton X100 for 10 min, and incubated with mouse monoclonal anti-Myc antibody (Santa Cruz, Sc-40) at a dilution of 1:1000 for visualization of TWIST2, and anti-H-81 at a dilution of 1:500 (Santa Cruz). Cells were then incubated with donkey anti-goat antibody conjugated with fluorescein isothiocyanate (FITC) and Cy3-conjugated goat anti-mouse antibody for TWIST2 visualization. DAPI staining was used to delineate the nucleus. Fluorescence microscopy images were taken with an Olympus BX40 fluorescence microscope at 100X magnification.
Preparation of nuclear and cytoplasmic fractions was performed 24 hrs post transfection using a Nuclear Extraction Kit (Active Motif). Equal volumes of the protein fractions were resolved in 12% denaturing polyacrylamide gels, transferred to a PVDF membrane and incubated overnight with anti-Myc mouse monoclonal antibody (1/1000, Santa Cruz). A secondary goat anti-mouse antibody conjugated to horseradish peroxidase (Thermo Scientific) was used to detect a chemiluminescent signal using Kodak Biomax MS X-ray film.
2.4. Yeast two-hybrid assays
Wild-type and mutant forms of the TWIST2 coding region were cloned into the EcoRI and BamHI sites of the pGBKT7 vector (Clontech) to produce TWIST2 proteins fused to the GAL4 DNA binding domain at the N-terminus. The E12, TWIST1 and TWIST2 coding sequences were cloned into the EcoRI and BamHI sites of the pGADT7 vector (Clontech) to produce proteins fused to the GAL4 activation domain. The yeast-two-hybrid assays were performed as recommended by the manufacturer (Matchmaker Gal4 Two-Hybrid System3, Clontech). Briefly, S. cerevisiae strain AH-109 was co-transformed with the respective plasmids using the standard lithium acetate method. Double transformants were grown on plates with synthetic media lacking leucine and tryptophan, and subsequently re-plated on selective media lacking leucine, tryptophan, and histidine. Positive interactions resulted in transactivation of the HIS3 reporter gene, which allows for growth in media lacking histidine.
2.5. Electrophoretic mobility shift assays
TWIST2, Q119X, and E12 were produced by in vitro transcription coupled translation (TnT) using the Quick Coupled TnT System (Promega). To confirm protein expression, duplicate side reactions using 35S-methionine were analyzed by SDS-PAGE and subsequent autoradiography. Aliquots of 1–2 μl of TnT products were incubated for 5 minutes with gel shift binding buffer (20 mM Hepes pH 7.9, 60 mM KCl, 1 mM MgCl2, 0.5 mM DTT, 1 μg poly(dI-dC)-poly(dI-dC), 50 ng denatured salmon sperm DNA, 10% glycerol). Regions containing E-boxes in the POSTN promoter (−3000 to +1) were identified using the Transcription Element Search Software (TESS, http://www.cbil.upenn.edu/cgi-bin/tess). Primers were designed to amplify ~100 bp of the target E-box, with the E-box located in the middle of the PCR product. PCR products were purified and end- labeled using polynucleotide kinase (New England Biolabs), γ-32P ATP, and purified by spin column chromatography (Illustra ProbeQuant G-50 Micro Columns, GE Health Care). These were added to the reaction mixture and incubated for 20 min (20,000 cpm/reaction). Reactions were then loaded on a 6% polyacrylamide (29:1) native gel containing 2.5% glycerol and 0.5X TBE buffer, which was previously pre-run for 2 hrs. Samples were run for 6–7 hrs at 4°C using 0.5X TBE running buffer. Specific competition reactions were done with >50 fold excess of unlabeled probe. Gels were dried under vacuum at 80°C for 1 hr and exposed to X-ray film at −80°C for 1–2 days.
2.6. Reporter gene assays
Deletion constructs of the 5′ flanking region of POSTN containing E-boxes identified using the Transcription Element Search Software were cloned into the pGL4 vector (Promega) that contains the firefly luciferase reporter gene. Approximately 1.7×105 mouse mesenchymal cells (C3H10T1/2) per well were seeded in 12 well plates. Cells were nucleofected with 500 ng SV-40 renilla pGL4, 2 ug of POSTN promoter construct and 3 ug plasmid (GFP, TWIST1, TWIST2 or Q119X), according to AMAXA technology protocol using reagent V and program T-020. Protein lysates were analyzed 4 days post nucleofection using the Dual-Luciferase reporter assay system (Promega). GFP was co-expressed to assay for nucleofection efficiency and relative luciferase units were calculated by normalizing against renilla luciferase units.
2.7. Chromatin immunoprecipitation
Aliquots containing 2 × 106 GM00637 fibroblasts were seeded in 10 cm plates 24 hrs prior to transfection with either TWIST2 or Q119X myc-tagged pCINEO constructs. Cells were transfected with 10 μg of plasmid DNA using the Fugene HD transfection reagent (Roche) at a ratio of 6 μl of Fugene to 2 μg DNA. Western blot analysis of whole cell extracts was performed to ensure proper expression of TWIST2 at 24 hrs post transfection. Cells were fixed in 1% formaldehyde, and ChIP was performed using the Chromatin Immunoprecipitation Assay Kit according to manufacturer’s instructions (Millipore). Briefly, cell lysate from approximately 9×106 cells per condition (TWIST2, Q119X, and Untreated) were sonicated using a bath sonicator (Bioruptor). Precleared lysates equivalent to 3×106 cells per antibody were incubated with anti-myc antibody (Santa Cruz, # Sc-40) or normal mouse IgG overnight at 4°C. For histone acetylation, anti-histone H3K9/14 (Millipore, #06-599), anti-histone H3 (Active Motif, #39163), or normal rabbit IgG was used. Immunocomplexes were collected, and purified DNA was resuspended in 50 μl of water. For ChIP analysis by Real Time PCR, 2 μl of the purified DNA was used for each PCR reaction. For this PCR, 10% input was utilized and its value was adjusted to 100% for normalization of the results. Each ChIP DNA threshold cycle number (CT) was normalized to the input DNA fraction CT value to account for differences in chromatin sample preparation. Percent input was calculated as follows: ΔCT= Input (CT) − IP (CT) and ΔΔCT = IPΔCT − IgGΔCT, finally %Input = 2ΔΔCT. Percent input of the sample was then divided by the percent input of the nonspecific antibody for calculation of the relative protein association. For histone ChIPs a corrected percent input was generated by subtracting the percent input of the nonspecific antibody from the percent input of total histone H3 and of acetylated histone H3. Finally, the corrected percent input of acetylated histone H3 was divided by the corrected percent input of total histone H3 to obtain % input relative to H3.
2.8. Statistical analysis
The real-time PCR data for ChIP assay was analyzed by two-way ANOVA with a Bonferroni post-test to analyze which means were statistically significant from one another. Gene expression and luciferase assay data were analyzed by one-way ANOVA followed by Tukey’s range test to compare with the non-treated control of experimental group. This analysis was performed using Prism 4 Graph Pad Software. Differences of P< 0.05 were considered to be significant. Error bars represent standard error of the mean.
3. Results
3.1. Dimerization and subcellular localization of TWIST2 and its mutant forms
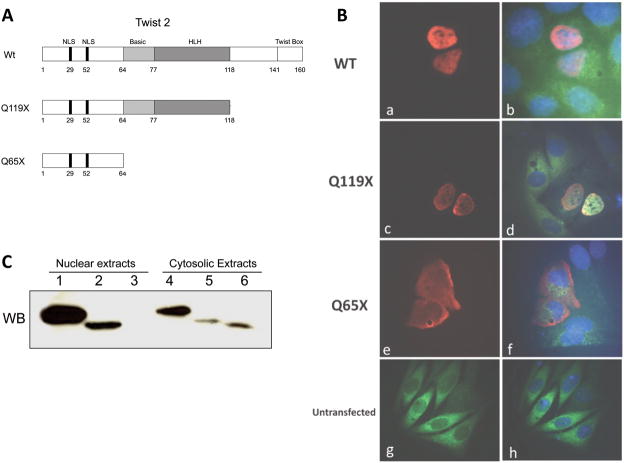
Nucleo-cytoplasmic protein trafficking of bHLH proteins is an important regulatory checkpoint in the control of target gene expression [25]. Mutations in TWIST2 such as the ones described for Setleis Syndrome could affect their nuclear localization, and therefore alter their DNA binding capabilities [4]. In order to address this issue, plasmid constructs expressing myc-tagged TWIST2 and its mutant forms were transfected into HeLa cells and visualized through immunofluorescence microscopy (Fig. 1). As seen in figure 1A, the Q119X mutation, originally found in patients of Puerto Rican ancestry, results in a truncated protein that is missing the C-terminal domain, while retaining the nuclear localization signals (NLS), basic, and helix-loop-helix domains (HLH). A more severe mutation originally found in patients from the United Arab Emirates (Q65X) results in a truncated protein that lacks the basic, HLH, and C-terminal domains but retains the NLS. Strong nuclear staining is observed in cells transfected with either wild-type or Q119X forms of TWIST2, which suggests that the functional defects of the Q119X mutant protein occur after nuclear localization (Fig. 1B). In contrast, strong cytoplasmic staining is observed in cells overexpressing the Q65X mutant, suggesting that the presence of the nuclear localization signals is not sufficient for proper import of this mutant protein. Fractionation of transfected cells into nuclear and cytoplasmic protein extracts was used for immunoblotting analysis. Wild-type TWIST2 and the Q119X mutant protein were detected in both the nuclear and cytoplasmic fractions, however, the Q65X mutant was only detected in the cytoplasmic fraction, consistent with the microscopy results (Fig. 1C). Other studies have proposed that heterodimerization of HLH proteins takes place in the cytoplasm prior to nuclear import [26,27], and that dimerization of TWIST2 is necessary for DNA binding, therefore, the capabilities of TWIST2 and its mutant forms to dimerize were assayed.
Figure 1.
Subcellular immunolocalization of TWIST2 and its Q119X mutant form reveal nuclear staining. A. Diagram of the TWIST2 protein and mutant forms described for Setleis Syndrome. B. Fluorescence microscopy of transfected HeLa cells with myc-tagged TWIST2 expression vectors. Anti-myc antibody conjugated with Cy3 was used to visualize TWIST2 localization. DAPI stain was used to delineate the nuclei. (a–d) Wt and Q119X exhibit strong nuclear staining (red), whereas Q65X is localized mainly in the cytosol (e–f). C. Western blot of nuclear and cytosolic fractions of transfected HeLa cells. Wt TWIST2 is in lanes 1 and 4, Q119X in lanes 2 and 5, and Q65X in lanes 3 and 6. The presence of the Q65X mutant protein is observed only in the cytosolic extract consistent with microscopy results.
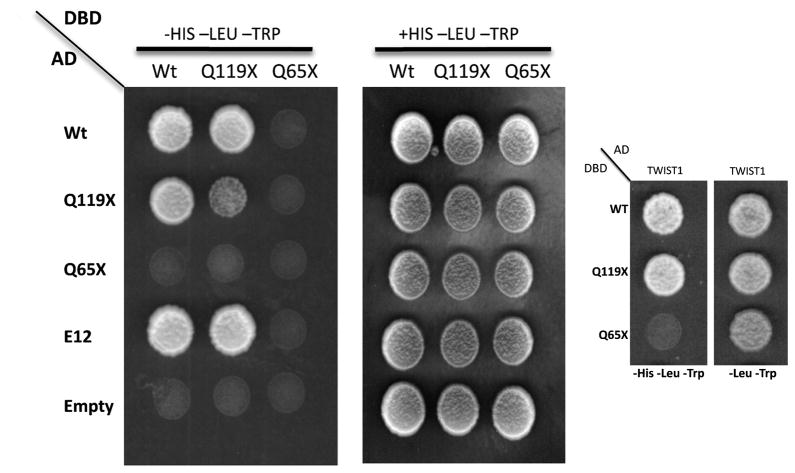
In order to assess the ability of the mutant forms of TWIST2 to form functional dimers, we used the in vivo yeast two-hybrid system. The coding sequence of TWIST2 and its mutant forms were expressed as fusion proteins to either the GAL4 activation domain (pGADT7 by Clontech) or the GAL4 DNA binding domain (pGBKT7 by Clontech) for homodimerization, while E12 and TWIST1 were expressed as a yeast fusion to the GAL4 activation domain to test for heterodimerization. Previous reports have demonstrated that heterodimerization of Twist1 is mediated through the HLH motif as nonsense mutations truncating the Twist1 HLH domain hampered heterodimer formation [27]. Considering that TWIST1 and TWIST2 are 98% identical in the bHLH region, we would expect the same outcome for the TWIST2 mutants. Two-hybrid analysis indicated that wild-type TWIST2 and Q119X have the ability to interact with each other, suggesting the potential for homodimer formation (Fig 2 and Suppl. Fig. S1). Protein-protein interaction was seen to occur between E12, wild-type TWIST2 and the Q119X mutant protein, suggesting that the Q119X mutant form of TWIST2 maintains the ability for heterodimerization. However, the Q65X mutant protein could not interact with any of the bHLH proteins tested (Fig 2 and Suppl. Fig. S1). Although the paradigm of dimerization for Class B bHLH proteins is that they heterodimerize with Class A bHLH proteins, recent studies have shown that heterodimerization between Class B bHLH factors is also possible between Twist1 and Hand2 [28,29]. Here we show another example of Class B bHLH protein-protein interaction between TWIST2 and TWIST1, adding another level of complexity to the potential dimer partners for these proteins (Fig 2). Taken together, these data suggest that the Q65X mutation of TWIST2 most likely represents a true loss of function mutation due to its inability to dimerize and localize to the nucleus. In contrast, the Q119X mutant could cause aberrant expression of target genes by retaining the ability to localize to the nucleus and dimerize with other bHLH proteins.
Figure 2.
Protein-protein interactions between TWIST2, Q119X, E12, and TWIST1. Growth on the –Leu-Trp selection plates demonstrates proper yeast transformation. Growth on the –Leu-Trp-His selection plates shows proper protein interaction. Wt TWIST2 and Q119X maintain the ability to interact with each other, with E12 and Twist1. However, the Q65X mutant cannot form competent protein-protein interactions with any of the proteins tested.
3.2. TWIST2 is a transactivator of POSTN in human fibroblasts
Microarray analysis was performed in order to determine novel TWIST2 gene targets, and gain insight into the possible regulatory networks of TWIST2. Gene expression profiles of RNAs derived from adult normal and Setleis Syndrome patient cultured skin fibroblasts were examined by microarray analysis using Affymetrix gene chips. Table 1 presents a partial list of diferentally regulated genes combined with the results of real time RT-PCR validation. Out of the top 287 differentially regulated probes (those with a fold change > 2.0 and a modified t value > 6.0), 189 were down-regulated and 98 were up-regulated. The complete set of microarray data files can be found at the Gene Expression Omnibus website at the National Center for Biotechnology Information (NCBI GEO #GES16524). One of the most down regulated genes in Setleis Syndrome patients is periostin, and real time RT-PCR validation shows a 134-fold down regulation consistent with the microarray data. Hence, POSTN was chosen as a potential target gene for the study of TWIST2 transactivation due to its key role in dermal tissue deposition and development and the prominent dermal phenotypes seen in Setleis Syndrome patients.
Table 1.
Selected differentially regulated genes between Setleis syndrome patient fibroblasts and normal control Fibroblasts using human Affymetrix 3′IVT expression arrays. Microarray data files can be found at the National Center for Biotechnology Information (NCBI GEO #GES16524).
| Gene | MA Fold change | Modified t/p-value | Median RT-PCR fold change | Function |
|---|---|---|---|---|
| LAMA4, laminin α4 | +3.7 | +19.9/0.009 | +4.7 | Major non-collagenous constituent of basement membranes |
| POSTN, periostin | −61 | −8.8/0.009 | −134 | Cell adhesion protein. Has been shown to be regulated by Twist1. |
| COL15A1, collagen15A1 | −62 | −13.12/0.011 | −4.7 | Adheres to basement membranes and to underlying connective tissue stroma. |
| IL7, interleukin 7 | +11 | +35.8/0.015 | +2.7 | Plays an essential role in lymphoid cell survival. |
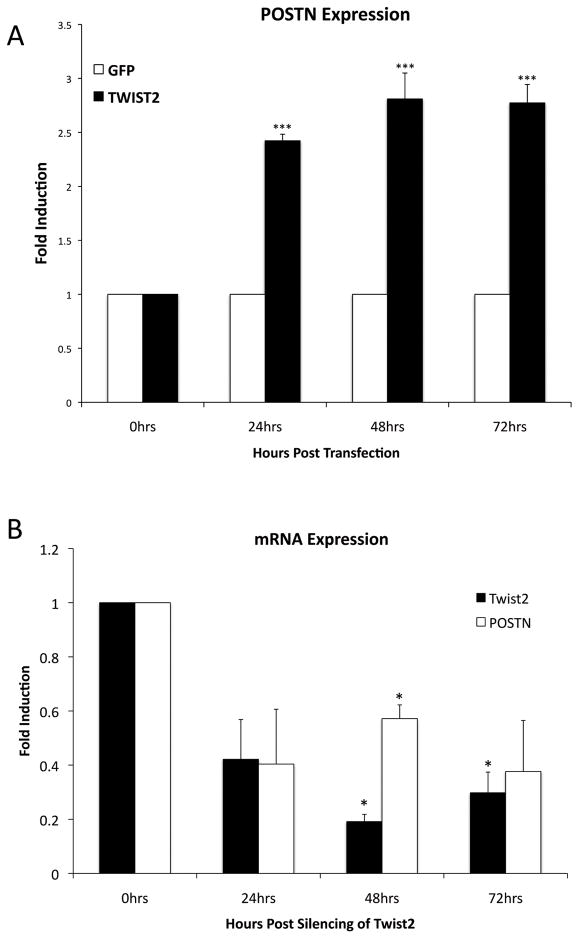
In order to confirm TWIST2 as a transactivator of POSTN, we measured endogenous POSTN mRNA levels in response to manipulations in the levels of TWIST2. Overexpression of TWIST2 in human fibroblasts resulted in increased POSTN expression over a 72 hr time frame (Fig. 3A). Conversely, in order to determine if decreased levels of TWIST2 resulted in decreased POSTN expression, siRNA directed against TWIST2 was transfected into human fibroblast cells. Knockdown of TWIST2 via RNA interference resulted in decreased POSTN expression over a 72 hr time frame when compared to fibroblasts transfected with a non-targeting scrambled siRNA (Fig. 3B). These results support the hypothesis that TWIST2 functions as an activator of POSTN gene expression in human skin fibroblasts.
Figure 3.
TWIST2 positively regulates POSTN expression in human fibroblasts. A. Overexpression of TWIST2 by transfection increases endogenous POSTN mRNA levels B. Conversely, RNAi mediated knockdown of TWIST2 results in decreased levels of endogenous periostin when compared to cells treated with non-targeting siRNA. Error bars represent standard error, and means significantly different from the untreated controls are delineated by asterisks (p<0.05 = *, p<0.01=**, p<0.001=***).
3.3. TWIST2 and the Q119X mutant can bind to specific regions in the POSTN promoter
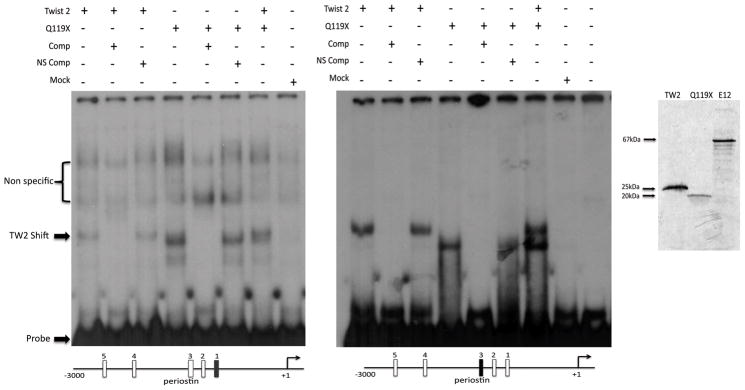
The Q119X mutation of TWIST2 has been shown to possess the ability to localize to the nucleus and form functional dimers, therefore, in order to assay for the DNA binding capabilities of Q119X, we performed electrophoretic mobility shift assays (EMSA) using segments from the POSTN promoter as target sequences. Five putative Twist binding sites (E-Boxes) were identified within the 3000 base pairs located upstream of the 5′ flanking region of the POSTN gene using the Transcription Element Search Software (Supplementary Figure S3). Two of these sites were assayed individually to determine if TWIST2 and the Q119X mutant could bind. Both TWIST2 and Q119X were found to specifically bind to these E-boxes on the POSTN promoter suggesting a direct mechanism of gene activation (Fig 4A). Because bHLH transcription factors require dimerization for DNA binding, these results suggest homodimeric binding of TWIST2 and Q119X to the promoter region of POSTN.
Figure 4.
Electrophoretic mobility shift assays reveal binding of TWIST2 and the Q119X mutant to the POSTN promoter. Left: Gel Shift shows binding of TWIST2 and the Q119X mutant to the 1st and 3rd E-Boxes in the POSTN promoter. Specific competitions (Comp) in 50-fold excess of the unlabeled probe completely ablated the shifted band, while non-specific unlabeled probe competitions (NS Comp) had no effect. Mock reaction represent the un-programmed rabbit reticulocyte lysate used for the production of the TWIST2 proteins. Right. To confirm protein expression and stability, duplicate in vitro TnT reactions were analyzed by SDS-PAGE resulting in bands of expected weight and intensity.
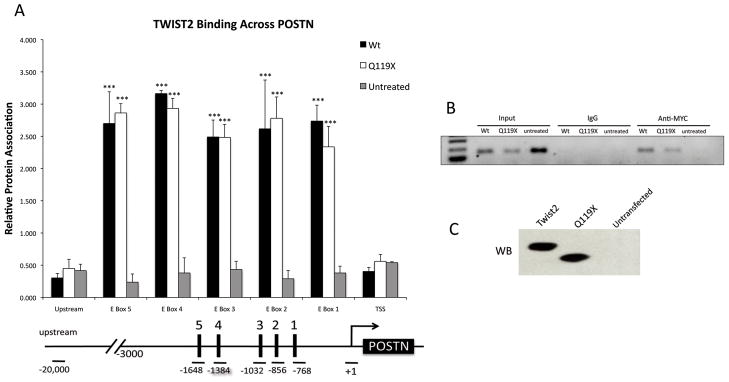
Chromatin immunoprecipitation (ChIP) was performed as an in vivo confirmation of the electrophoretic mobility shift assay results. As observed in Figure 5, both TWIST2 and Q119X bound specifically to the POSTN promoter on all five putative binding sites for TWIST2. Relative protein association was observed to be similar between TWIST2 and Q119X suggesting that the mutant protein retains the ability to bind DNA, similar to that of the wild-type protein (Fig. 5A). It must be noted that the ChIP signals detected for TWIST2 and Q119X may reflect both the affinity of these proteins for their E-box recognition sequences, as well as their affinity for endogenous interacting proteins. Also, ChIP signals present at multiple E-box elements may reflect the average affinities of the proteins for the E-box being assayed as well as neighboring elements because the distance between individual elements may be smaller than the standard sonicated DNA fragment size produced during ChIP assays (Suppl. Fig. S3). Nonetheless, the ability of Q119X to bind to the POSTN promoter suggests that it may function possibly as a dominant negative mutation either by occupying available DNA binding sites without mediating proper transactivation or by sequestering available bHLH binding partners.
Figure 5.
ChIP of fibroblasts transfected with either TWIST2, Q119X or untransfected. A. Real time PCR of selected regions shows TWIST2 and Q119X binding to all five E-boxes found in the promoter region of POSTN with similar intensity. Asterisks represent means that are significantly different (p<0.001) from the untransfected control. Error bars represent standard error of the mean. Below. Diagram of the primer locations used to probe the E-boxes, the transcription start site (TSS), and a “gene desert” region of genomic DNA approximately 20 kb upstream of the POSTN promoter. B. Representative semi-qPCR of ChIP DNA corresponding to E-box 1 of the POSTN promoter. C. Immunoblot to confirm proper transfection and expression of TWIST2 and Q119X proteins.
3.4. TWIST2 but not Q119X transactivates and affects the acetylation state of the POSTN promoter
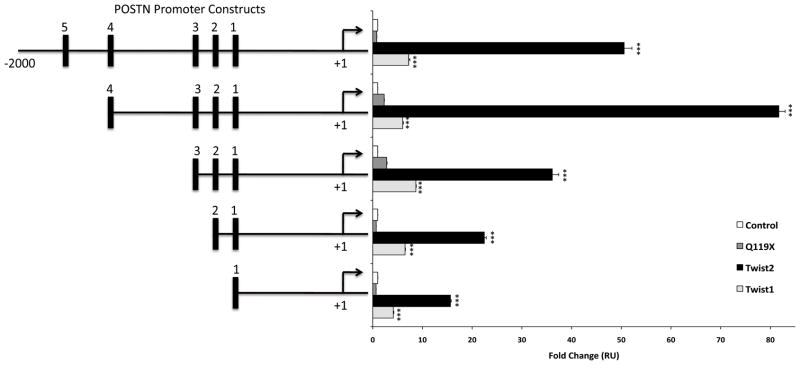
Reporter gene assays were employed in order to demonstrate that TWIST2 directly up-regulates POSTN and to assay which region of the promoter was sufficient and necessary for this activation. Previous reports have used reporter gene assays to show that Twist1 regulates periostin, however these experiments were conducted using the mouse POSTN promoter that differs from the human promoter [20]. As seen in Figure 6, wild-type TWIST2, but not Q119X can activate the reporter gene using all five deletion constructs of the POSTN promoter with the maximum activity observed for the promoter containing 4 out of the 5 binding sites. Although we have seen that TWIST1 can bind to the POSTN promoter region through EMSA analysis (data not shown), we observed that activation of the reporter gene was significantly greater for TWIST2 than TWIST1 showing specificity for TWIST2.
Figure 6.
TWIST2 transcriptionally activates the POSTN promoter. Deletion constructs of the POSTN promoter was transfected by nucleofection (AMAXA) together with plasmids encoding TWIST1, TWIST2, or Q119X. TWIST2 significantly activates the POSTN promoter when compared to Q119X and Twist1. Maximum activation is observed for the promoter with 4 out of the 5 E-boxes in place. Error bars represent standard deviation, and means that were statistically significant from the control were highlighted with asterisks (p<0.001=***).
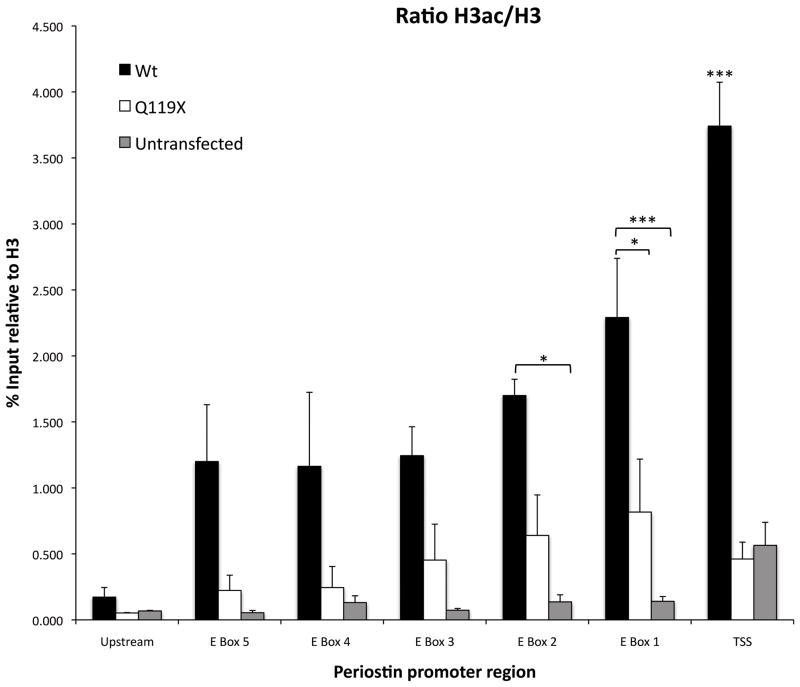
In order to assay for a possible mechanism of POSTN activation by TWIST2, chromatin immunoprecipitation assays were used to determine the acetylation states of histone H3 in the POSTN promoter of cells treated with TWIST2 or the Q119X mutant (Fig. 7). The acetylation of histone H3 at lysines 9 and 14 (H3K9/14) is usually associated with actively transcribed genes, and it was observed that fibroblast cells treated with wild-type TWIST2 had greater acetylation of H3K9/14 within the 5′ flanking region of POSTN when compared to Q119X and untreated controls (Fig. 7), and that acetylation increases at closer distances to the transcription start site. It is therefore proposed that the C-terminal domain missing in the Q119X mutant is necessary for proper acetylation of transcriptional targets.
Figure 7.
TWIST2 facilitates histone acetylation of the POSTN promoter. ChIP of fibroblasts transfected with either TWIST2, Q119X or untransfected. Anti-acetylated histone H3, anti-histone H3, and IgG (negative control) antibodies were used in probing for POSTN promoter DNA pulldown. Real time PCR of selected regions shows that cells treated with TWIST2 resulted in higher acetylation across the POSTN promoter while Q119X mutant is not associated with histone acetylation. Acetylation decreases at greater distances 5′ from the transcription start site. Primers used for the amplification of DNA are depicted in figure 5B. Asterisks represent means that are significantly different (p<0.001) from one another. Error bars represent standard error of the mean.
4. Discussion
The present study was undertaken to determine how the mutations of TWIST2 found in Setleis syndrome patients affected protein function and the transactivational activity of this transcription factor. Our data indicate that the Q65X mutation of TWIST2 most likely represents a true loss of function mutation due to its inability to form dimers and localize to the nucleus. However, the Q119X mutant protein is expressed, can localize to the nucleus, form functional dimers, and bind DNA. Thus, the loss of transactivational activity associated with the truncation of the TWIST2 C-terminal domain cannot be attributed to protein misfolding or instability. Instead, we propose that the C-terminal domain missing in this mutant allows for histone acetylation of chromatin on the POSTN gene resulting in an open conformation consistent with transcriptional activation. The last 20 amino acids of Twist1 have been characterized as an activation domain, and this region was shown to be sufficient and necessary for proper gene activation [31]. The C-terminal domain of TWIST2 retains 100% identity with TWIST1, therefore, the loss of transactivation activity of Q119X could be attributed to the loss of this activation domain. Moreover, this activation domain, termed Twist Box, is conserved between TWIST1 and TWIST2 throughout the animal kingdom and not found in any of the other Twist subfamily members [31].
Many of the target genes of mammalian TWIST2 have multiple E-boxes in their promoter regions. One of the hallmarks of bHLH protein function is that dimer selection affects E-box preference, and that combinatorial binding of dimer pairs results in different transcriptional outcomes. Partner choice of these proteins is mainly influenced by the availability of other bHLH proteins within the cell and the phosphorylation state of these proteins [6]. In the present study, we observed that POSTN gene expression responded positively to TWIST2, and that TWIST2 was able to bind to specific E-boxes within the POSTN promoter as a homodimer. It is noted that Twist1 was also proposed to be an activator of POSTN transcription as a homodimer within the cranial sutures of mice [21]. Although both POSTN and Twist proteins are known to play a central role in bone morphogenesis, no major cranial bone phenotypes are observed for Setleis Syndrome patients consistent with the lack of TWIST2 expression in the developing skull [4,5]. Recently we have suggested that unlike TWIST1, TWIST2 does not function in the cranial neural crest and that TWIST2 is instead involved in cranial dermal mesenchyme development [4]. We do not however discount the possibility that some compensatory functionality between TWIST1 and TWIST2 may exist in the regulation of certain developmental programs, therefore, future dissection of TWIST1 and TWIST2 regulatory dynamics is still warranted.
One of the most significant phenotypes of Setleis Syndrome patients is their hypoplastic dermis and forceps-like lesions located at the temporal lobes [4]. Periostin is known to be highly expressed in collagen-rich tissues including the epidermis and fibroblasts in the dermis, and plays a central role in collagen fibrillogenesis [15]. This is consistent with the overlapping expression patterns of TWIST2 with type I collagen during embryogenesis [5]. Consequently, dysregulation of POSTN by mutant TWIST2 can alter the composition of the extracellular matrix and contribute to the development of the severe skin and facial abnormalities observed in Setleis Syndrome.
Supplementary Material
Acknowledgments
We would like to dedicate a special thanks to Dr. Carmelo Carmona for his valuable guidance with the yeast-two-hybrid and Immunofluorescence microscopy experiments. We would also like to extend our immense gratitude towards Dr. Boyko S. Atanassov and Dr. Calley L. Hirsch from the Sharon Dent Lab at the MD Anderson Cancer Center for their instruction and guidance with the chromatin immunoprecipitation assays. Finally we would like to thank Dr. Douglas Spicer for his helpful discussions and advice.
Funding
Supported by the National Institutes of Health: NCRR grant P20-RR016470, RCMI Grants NCRR-G12RR03051 and 5P20RR011126, as well as NIGMS R25GM061838 and DK073781 and P20 RR15555 (to RF) and the Deanship of Biomedical Sciences at University of Puerto Rico Medical Sciences Campus.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Setleis H, Kramer B, Valcarcel M, Einhorn AH. Congenital ectodermal dysplasia of the face. Pediatrics. 1963;32:540–548. [PubMed] [Google Scholar]
- 2.McGaughran J, Aftimos S. Setleis syndrome: three new cases and a review of the literature. Am J Med Genet. 2002;111:376–80. doi: 10.1002/ajmg.10632. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg JG, Drolet BA. What syndrome is this? Setleis syndrome. Pediatr Dermatol. 2004;21:82–83. doi: 10.1111/j.0736-8046.2004.21119.x. [DOI] [PubMed] [Google Scholar]
- 4.Tukel T, Sosic D, Al-Gazali L, Erazo M, Casasnovas J, Franco HL, Richardson JA, Olson EN, Cadilla CL, Desnick RJ. Homozygous nonsense mutations in TWIST2 cause Setleis Syndrome. Am J Hum Genet. 2010;87(2):289–296. doi: 10.1016/j.ajhg.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Cserjiesi P, Olson EN. Dermo-1: A novel twist related bHLH protein expressed in the developing dermis. Dev Biol. 1995;172:280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- 6.Franco HL, Casasnovas J, Rodriguez-Medina JR, Cadilla CL. Redundant of separate entities? – roles of Twist1 and TWIST2 as molecular swiches during gene transcription. NAR. 2010:1–10. doi: 10.1093/nar/gkq890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int J Dev Biol. 2009;52:17–24. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 9.Wilon-Rawls J, Rhee JM, Rawls A. Paraxis is a Basic Helix-Loop-Helix Protein That Positively Regulates Transcription through Binding to Specific E-Box Elements. JBC. 2004;279:37685–37692. doi: 10.1074/jbc.M401319200. [DOI] [PubMed] [Google Scholar]
- 10.Ellenberger T, Fass D, Arnaud M. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Gen & Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 11.Gong X, Li L. Dermo-1, a Multifunctional Basic Helix-Loop-Helix Protein, Represses MyoD Transactivation via the HLH Domain, MEF2 Interaction, and Chromatin Deacetylation. JBC. 2002;277:12310–12317. doi: 10.1074/jbc.M110228200. [DOI] [PubMed] [Google Scholar]
- 12.Lee YS, Lee HH, Park J, Yoo EJ, Glackin CA, Choi YI, Jeon SH, Seong RH, Park SD, Kim JB. TWIST2, a novel ADD1/SREBP1c interacting protein, represses the transcriptional activity of ADD1/SREBP1c. NAR. 2003;31:7165–7174. doi: 10.1093/nar/gkg934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeshita S, Kikuno R, Tezuka K, Amann E. Osteoblast-specific factor 2: Cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem J. 1993;294:271–278. doi: 10.1042/bj2940271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton DW. Functional Role of POSTN in development and wound repair: implications for connective tissue disease. J Cell Comm Signal. 2008 doi: 10.1007/s12079-008-0023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson-Boeters L, Wen W, Hamilton DW. Periostin localizes to cell in normal skin, but is associated with the extracellular matrix during wound repair. J Cell Comm Signal. 2009 doi: 10.1007/s12079-009-0057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afanador E, Masahiko Y, Oba Y, Kitase Y, Takahashi T, Kudo A, Moriyama K. Messenger RNA expression of POSTN and Twist transiently decrease by occlusal hypofunction in mouse periodontal ligament. Ach Oral Bio. 2005;50:1023–1031. doi: 10.1016/j.archoralbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Norris RA, Damon B, Mironov V, Kasyanov V, Ramamurthi A, Moreno-Rodriguez R, Trusk T, Potts JD, Goodwin RL, Davis J, et al. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem. 2007;101:195–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puglisi F, Puppin C, Pegolo E, Andreetta C, Pascoletti G, D’Aurizio F, Pandolfi M, Fasola G, Piga A, Damante G, Di Loreto C. Expression of Periostin in Human Breast Cancer. J Clin Pathol. 2008;61:494–8. doi: 10.1136/jcp.2007.052506. [DOI] [PubMed] [Google Scholar]
- 19.Ruan K, Boa S, Ouyang G. The multifaceted fole of POSTN in tumorigenesis. Cell Mol Life Sci. 2009;66:2219–2230. doi: 10.1007/s00018-009-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshima A, Tanabe H, Yan T, Lowe GN, Glackin CA, Kudo A. A Novel Mechanism for the Regulation of Osteoblast Differentiation: Transcription of Periostin, a Member of the Fasciclin I Family, is Regulated by the bHLH Transcription Factor, Twist. Jorn Cell Biochem. 2002;86:792–804. doi: 10.1002/jcb.10272. [DOI] [PubMed] [Google Scholar]
- 21.Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. Twist1 Dimer Selection Regulates Cranial Suture Patterning and Fusion. Dev Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Gen Mol Biol. 2004;3(1):Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:10. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michael W. Pfaffl. A new mathematical model for relative quantification real-time RT-PCR. NAR. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MS, Lowe G, Flanagan S, Kuchler K, Glackin CA. Human Dermo-1 has Attributes Similar to Twist in Early Bone Development. Bone. 2000;27:591–602. doi: 10.1016/s8756-3282(00)00380-x. [DOI] [PubMed] [Google Scholar]
- 26.Goldfard AN, Lewandowska K. Nuclear redirection of a cytoplasmic helix-loop-helix protein via heterodimerization with a nuclear localizing partner. Exp Cell Res. 1994;214:481–485. doi: 10.1006/excr.1994.1285. [DOI] [PubMed] [Google Scholar]
- 27.Ghouzzi VE, Legeai-Mallet L, Aresta S, Benoist C, Munnich A, Gunzburg J, Bonaventure J. Saethre-Chotzen Mutations Cause TWIST Protein Degredation or Impaired Nuclear Location. Hum Mol Genet. 2000;9:813–819. doi: 10.1093/hmg/9.5.813. [DOI] [PubMed] [Google Scholar]
- 28.Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Altered Twist1 and Hand2 dimerization is associated with saethre-chotzen syndrome and limb abnormalities Nat. Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Firulli BA, Howard MJ, McDaid JR, McIlreavey L, Dionne KM, Centonze VE, Cserjesi P, Virshup DM, Firulli AB. PKA, PKC, and the protein phosphatase 2A influence HAND factor function: A mechanism for tissue-specific transcriptional regulation. Mol Cell. 2003;12:1225–1237. doi: 10.1016/s1097-2765(03)00425-8. [DOI] [PubMed] [Google Scholar]
- 30.Connerney J, Andreeva V, Leshem Y, Mercado MA, Dowell K, Yang X, Lindner V, Friesel RE, Spicer DB. Twist1 homodimers enhance FGF responsiveness of the cranial sutures and promote suture closure. Devl Bio. 2008;318:323–34. doi: 10.1016/j.ydbio.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laursen KB, Mielke E, Iannaccone P, Füchtbauer EM. Mechanism of Transcriptional Activation by the Proto-oncogene Twist1. JBC. 2007;282:34623–34633. doi: 10.1074/jbc.M707085200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.