Abstract
Progressive mitochondrial failure is tightly associated with the onset of many age-related human pathologies. This tight connection results from the double-edged sword of mitochondrial respiration, which is responsible for generating both ATP and ROS, as well as from risks that are inherent to mitochondrial biogenesis. To prevent and treat these diseases, a precise understanding of the mechanisms that maintain functional mitochondria is necessary. Mitochondrial protein quality control is one of the mechanisms that protect mitochondrial integrity, and increasing evidence implicates the cytosolic ubiquitin/proteasome system (UPS) as part of this surveillance network. In this review, we will discuss our current understanding of UPS-dependent mitochondrial protein degradation, its roles in diseases progression, and insights into future studies.
Keywords: ubiquitin, proteasome, mitochondria, mitochondrial protein quality control
Introduction
Mitochondria play fundamentally important roles in various cellular processes including ATP production, ion storage, apoptosis and critical anabolic and catabolic metabolism. Many studies have indicated that mitochondrial dysfunction is central to the development of most age-related human diseases including neurodegenerative diseases, cancer, and type 2 diabetes [1]. Accordingly, a comprehensive understanding of the mechanisms that enable preservation of functional mitochondria would inform the prevention and treatment of those diseases.
While mitochondria have their own genome, about 99% of the roughly 1000 mitochondrial proteins are encoded in the nuclear genome [1–2,4]. Most mitochondrial proteins are therefore synthesized in the cytoplasm, unfolded, transported across one or both mitochondrial membranes, then refolded and/or assembled into complexes [3]. Failure of this complex series of events generates unfolded or misfolded proteins within mitochondria, often disrupting critical functions [3].
Mitochondrial oxidative phosphorylation generates usable cellular energy in the form of ATP, but also produces reactive oxygen species (ROS) as a byproduct [1–4]. ROS tend to react quickly, so their predominant sites of damage are mitochondrial macromolecules that are localized nearby the source ROS production. Exposure to oxidative stress facilitates misfolding and aggregation of these mitochondrial proteins, leading to disassembly of protein complexes and eventual loss of mitochondrial integrity [3].
Because of this inherent source of damage, clearance of misfolded and aggregated proteins is constantly needed to maintain functional mitochondria. There are several systems promoting this turnover. Mitophagy, a selective mitochondrial autophagy, mediates a bulk removal of damaged mitochondria. The detailed mitophagic process will be discussed in more detail below. In addition to mitophagy, mitochondria intrinsically contain proteases in each of their compartments and these proteases recognize misfolded mitochondrial proteins and mediate their degradation [5]. Interestingly, accumulating evidence shows that the ubiquitin proteasome system (UPS) plays an important role in mitochondrial protein degradation. At various cellular sites, the UPS is involved in protein degradation. With the help of ubiquitin E1-E2-E3 enzyme cascades, target proteins destined for destruction are marked by conjugation of K48-linked poly-ubiquitin chain. This poly-ubiquitinated protein is then targeted to the proteasome for degradation [4].
In this review, we will discuss recent advances in our knowledge of mitochondrial protein quality control systems, specifically focusing on UPS-dependent protein degradation.
The UPS and mitochondrial protein degradation
Increasing evidence shows that the UPS is a part of the mitochondrial protein quality control system. One of the first observations connecting the cytoplasmic proteasome and mitochondrial protein degradation was the detection of ubiquitin-conjugated proteins in purified mitochondria by mass spectrometry [4]. Moreover, cells treated with proteasome inhibitors exhibit elevated levels of ubiquitinated mitochondrial proteins, suggesting the potentially important roles of the proteasome on mitochondrial protein degradation [6]. Consistent with this, many studies have identified mitochondrial substrates of the UPS. Fzo1, an outer mitochondrial membrane (OMM) protein involved in mitochondrial fusion, is partially dependent on the proteasome for its degradation in yeast [4]. The F box protein Mdm30 mediates ubiquitination of Fzo1 by Skp1-Cullin-F-boxMdm30 ligase, which leads to proteasomal degradation [4]. The UPS has also been implicated in mitochondrial protein degradation in higher organisms. In mammals, the OMM proteins mitofusin 1 and 2 (Mfn1/2; the mammalian orthologs of Fzo1) and Mcl1 are polyubiquitinated and degraded by the proteasome [7-9] (See Table 1). VDAC1, Tom20 and Tom70 were also suggested as targets of proteasomal degradation as they are stabilized by proteasome inhibition [8]. Additionally, inactivation of the proteasome induces accumulation of intermembrane space (IMS) proteins and, consistent with this, the proteasome plays a role in degradation of the IMS protein, Endonuclease G [10].
Table 1.
List of known mitochondrial ubiquitin E3 ligases and their known substrates
Surprisingly, turnover of some inner mitochondrial membrane (IMM) proteins is also dependent upon the proteasome. Uncoupling proteins (UCPs) 2 and 3 exhibit an unusually short half-life compared with other IMM proteins, and Brand and colleagues showed that inactivation of the proteasome prevents their turnover in vivo and in a reconstituted in vitro system [11]. Finally, mitochondrial matrix proteins can also be degraded by the proteasome. For example, OSCP, oligomycin sensitivity conferral protein, is stabilized upon inactivation of the proteasome [6]. When import of newly synthesized OSCP was blocked, inhibition of the proteasome induced accumulation of OSCP in the OMM, suggesting that matrix-localized OSCP was exported to the OMM for proteasomal degradation. Taken together, the evidence is compelling that the UPS participates in protein quality control in all mitochondrial compartments.
How does the cytosolic UPS recognize and degrade mitochondrial proteins, particularly those that are not exposed to the cytosol? It has been reported that the proteasomal subunit Rpt4 can directly dislocate and degrade an ER-associated protein degradation (ERAD) substrate in the absence of other cofactors [12]. By analogy, one possibility is that the proteasome per se can directly detect and degrade mitochondrial substrates without the need for cofactors. A second possibility would be the existence of factor(s) that retrotranslocate substrates from the mitochondria to the cytoplasm for delivery to the proteasome. Consistent with this idea, several studies have shown that Cdc48 (the yeast ortholog of p97 or VCP in mammals) participates in such an activity [7,9,13]. Cdc48 is a component of the UPS involved in protein degradation at a variety of cellular sites. Its role has been extensively studied in ERAD, where it mediates the dislocation of ER membrane-localized proteins to the cytoplasm for presentation to the proteasome using energy from ATP hydrolysis [4,13]. Interestingly, the association of Cdc48/p97 and mitochondria has been reported in several studies. Xu et al. (2011) and Tanaka et al. (2010) have shown that p97 mediates the proteasomal degradation of polyubiquitin-conjugated Mfn1/2 [7]. Dissipation of the mitochondrial membrane potential causes p97 translocation to ubiquitin-rich mitochondria and a p97 mutant lacking ATPase activity shows impaired degradation of Mfn1/2. Xu et al. provided further evidence for the involvement of p97 in proteasome-mediated degradation of two OMM proteins, Mfn1 and Mcl1 [9]. Depletion of p97 as well as inhibition of the proteasome stabilizes these proteins. Further, a p97 mutant exhibits impaired retrotranslocation of Mcl1 from mitochondria to the cytoplasm as well as stabilization of Mcl1. Taken together, it is clear that Cdc48/p97 is a crucial component bridging mitochondrial substrates and the cytoplasmic proteasome.
Cdc48/p97 is involved in many cellular processes through its role in protein degradation and is targeted to different subcellular sites by adaptor proteins. For example, Cdc48/p97 is recruited to the endoplasmic reticulum with the help of two adaptor proteins, Npl4 and Ufd1 [14]. This implies the existence of specific adaptors that recruit Cdc48/p97 to mitochondria. Consistent with this notion, we recently identified a mitochondrial adaptor protein for Cdc48, which we named Vms1 (VCP/Cdc48-associated mitochondrial stress responsive 1) [13]. Interestingly, Vms1 interacts with Cdc48/p97 and Npl4, but not with Ufd1. This implies that the Cdc48/p97-Npl4-Ufd1 complex functions in ER protein degradation while the Vms1-Cdc48/p97-Npl4 complex acts in mitochondria. In agreement with this notion, overexpression of Cdc48 or Npl4 rescues the vms1 mutant phenotype while Ufd1 has no effect.
Normally, Vms1 is cytoplasmic. Upon mitochondrial stress, however, Vms1 recruits Cdc48 and Npl4 to mitochondria. In agreement with the role of Cdc48/p97 in OMM protein degradation, loss of the Vms1 system results in accumulation of ubiquitin-conjugated proteins in purified mitochondria as well as stabilization of Fzo1 under mitochondrial stress conditions. Accumulation of damaged and misfolded mitochondrial proteins disturbs the normal physiology of the mitochondria, leading to mitochondrial dysfunction. As expected, the vms1 mutants progressively lose mitochondrial respiratory activity, eventually leading to cell death. The VMS1 gene is broadly conserved in eukaryotes, implying an important functional role in a wide range of organisms. The C. elegans Vms1 homolog exhibits a similar pattern of mitochondrial stress responsive translocation and is required for normal lifespan. Additionally, mammalian Vms1 also forms a stable complex with p97. Combining these observations, we conclude that Vms1 is a conserved component of the UPS-dependent mitochondrial protein quality control system. This system senses mitochondrial stress, recruits Cdc48/p97 to damaged mitochondria, and mediates proteasomal degradation of damaged mitochondrial proteins.
The UPS regulates mitochondrial dynamics and initiation of mitophagy
Mitochondria undergo continuous fission and fusion events and they utilize this dynamic procedure to maintain their function [7–8]. When damage is moderate, fusion combines mitochondrial pools, leading to dilution of damaged structures. If damage is more severe, fission facilitates removal of impaired portions from the healthy mitochondrial network by fragmentation, followed by their disposal through mitophagy[15].
Accumulating evidence shows that the UPS plays essential roles in regulating mitochondrial dynamics. Mfn1/2, Fis1 and Drp1, major players regulating mitochondrial fusion and fission, are degraded by the proteasome [7–8,16]. MITOL, a mitochondrial E3 ubiquitin ligase, is required for Drp1-dependent mitochondrial fission as depletion or inactivation of MITOL blocks mitochondrial fragmentation [17]. Moreover, knockdown of USP30, an OMM-localized deubiquitinating enzyme, induces an elongated mitochondrial morphology, suggesting a defect in fission [18]. Although the underlying mechanisms linking the UPS to the regulation of mitochondrial dynamics remain unclear, these observations demonstrate that these processes are tightly linked.
Cells mark damaged mitochondria by polyubiquitination of OMM proteins. Parkin, an ubiquitin E3 ligase, and the PINK1 protein kinase are the most characterized players in this process [7]. When mitochondria are depolarized, PINK1 is selectively stabilized leading to recruitment of cytoplasmic Parkin to damaged mitochondria [7,19]. Following recruitment, Parkin marks the mitochondria with K-63 or K-27-linked, proteasome-independent polyubiquitination and mediates their transport to the perinuclear region where they fuse to the autophagosome with the help of p62 and HDAC6 [7,20]. Loss of function mutations in either Parkin or PINK cause Parkinson's disease, a devastating neurodegenerative disease characterized by movement disorder [7–8], revealing the importance of this mitochondrial quality control mechanism in maintaining normal neuronal function.
Increasing evidence links the UPS to the initiation of Parkin-dependent mitophagy [7–8]. Youle and colleagues demonstrated that Parkin is the E3 ligase responsible for K-48 ubiquitination-mediated proteasomal degradation of Mfn1/2 as they are stabilized either by depletion or inactivation of Parkin [7]. Moreover, degradation of mitofusins was only observed in the presence of PINK1 and mitochondrial membrane depolarization by treatment with the mitochondrial uncoupler, CCCP, which are required for Parkin recruitment. Depletion of mitofusins facilitates mitochondrial fragmentation, a process required for initiating mitophagy. Lu and colleagues further demonstrated the involvement of Parkin in modulation of mitochondrial fusion/fission dynamics as overexpression of Parkin facilitates mitochondrial fission in mammalian primary neurons [21]. Parkin, therefore, might facilitate mitophagy by altering mitochondrial dynamics. Consistent with this notion, Parkin-mediated proteasomal degradation of mitofusins is a prerequisite for the initiation of mitophagy. Stabilization of Mfn1/2 by inhibition of the proteasome abolished the CCCP-driven mitophagy [7]. Further, blocking K48-linked ubiquitination caused a deficit in mitophagy [8]. Taken together, it is apparent that the UPS is part of a regulatory system involved in mitochondrial dynamics and is required for the tight regulation of Parkin-dependent mitophagy (Figure 1).
Figure 1. The UPS regulates mitochondrial dynamics.
Major proteins involved in mitochondrial fission or fusion (e.g. Mfn1/2, Drp1 and Fis1) are degraded by the UPS. Through this regulatory process, the UPS controls mitochondrial dynamics. Parkin, an E3 ligase involved in mitophagy, utilizes the UPS to enhance mitochondrial fission through degradation of components of the fusion machinery (blue arrow). By facilitating fragmentation of damaged mitochondria, which is essential for initiation of mitophagy, Parkin stimulates mitophagy.
Mitochondrial protein quality control and human diseases
Accumulation of aberrant proteins within mitochondria often disturbs mitochondrial function and threatens cell survival. This has been extensively studied in neurodegenerative diseases wherein aberrant pathological proteins accumulate throughout the cell, including sites in mitochondria (See Table 2). Amyloid precursor protein (APP), a protein associated with Alzheimer's disease, accumulates within mitochondria and is implicated in blockade of mitochondrial protein import [22]. Aβ, a neurotoxic APP cleavage product, can also facilitate the formation of the mitochondrial permeability transition pore (mPTP) by binding to mPTP components VDAC1, CypD and ANT, which provokes cell death [23–25]. α-synuclein, a protein associated with the development of Parkinson's disease, is targeted to the IMM where it binds to the mitochondrial respiratory complex I and impairs its function [26]. α-synuclein also interferes with mitochondrial dynamics as its unique interaction with the mitochondrial membrane disturbs the fusion process [27]. Finally, in Huntington's disease, increased association of the mutant huntingtin protein with mitochondria can impair mitochondrial trafficking [28–29]. Moreover, accumulation of mutant huntingtin protein disrupts cristae structure and facilitates mitochondrial fragmentation by activation of Drp1 [28, 30]. These examples demonstrate the crucial importance of prompt removal of dysfunctional and/or aberrant proteins in maintaining functional mitochondria.
Table 2.
List of human diseases caused by accumulation of proteins in mitochondria.
Future outlook
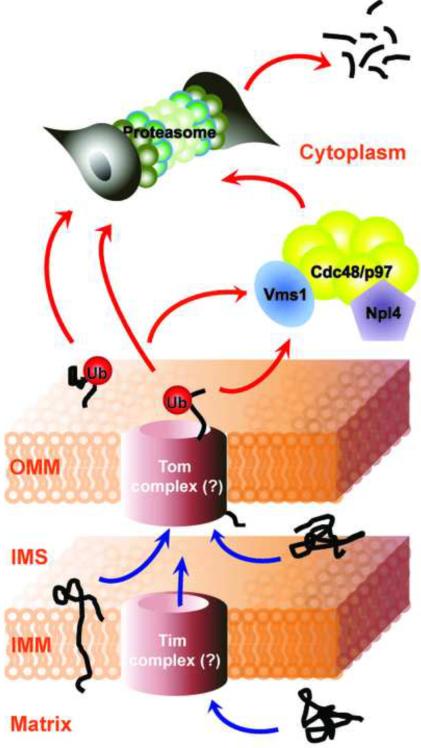
Discoveries regarding the involvement of the UPS in the mitochondrial protein quality control process provide new insights into normal mitochondrial and cellular physiology. This linkage also raises several questions. First, several ubiquitin E3 ligases including MITOL and MULAN have been identified in the OMM [4]. Yanagi and collegues have shown that MITOL regulates mitochondrial dynamics by ubiquitination-mediated degradation of mitochondrial fission proteins, hFis1 and Drp1 [31]. While MULAN has been also implicated in modulation of mitochondrial dynamics [32], its substrates remain unknown. Therefore, it will be of great interest to determine the specific substrates of these ligases and the role of ubiquitination in regulating their functions. Second, the involvement of the UPS in degrading intra-mitochondrial proteins implies the existence of special machinery involved in the retrotranslocation and export of these proteins (See Figure 2). One possible explanation is that mitochondrial proteases recognize and dislocate mitochondrial proteins to the surface. Distinct from their proteolytic activity, some mitochondrial proteases contain a chaperone-like function. Yme1, an IMM protease, can mediate the translocation of cytoplasmic PNPase to the IMS without degradation [33]. Yta10 and Yta12, IMM proteases facing the matrix side, are involved in complex assembly of cytochrome c oxidase [34]. Additionally, a more detailed understanding of the Vms1 system is necessary. What is the signal that recruits Vms1 to mitochondria? What are the mitochondrial substrates for the Vms1 system? Are there other auxiliary factors involved in this pathway? Finally, some proteins are targets of the UPS while others are proteolyzed by intrinsic mitochondrial proteases, but the process for making this decision is not known.
Figure 2. UPS-mediated mitochondrial protein degradation.
Misfolded and/or damaged mitochondrial proteins destined for proteasomal degradation in the cytosol are recruited to the outer mitochondrial membrane (OMM) from each mitochondrial compartment by unknown mechanisms (blue arrows). Upon reaching the OMM, these proteins are presented to the proteasome through a series of events. They are K48 polyubiquitinated by the cytoplasmic (e.g. Parkin) or mitochondrial ubiquitin E3 ligases. For proteasomal degradation, polyubiquitinated mitochondrial substrate proteins need to be retrotranslocated to the cytoplasm, probably, either by the proteasome per se or by the help of UPS components such as Vms1, Cdc48/p97 and Npl4. Following dislocation to the cytoplasm, these substrate proteins are degraded by the proteasome (red arrows).
Treatment of diseases that arise from defects in protein quality control will depend on greater depth in our understanding of this process, which could contribute to the development of novel therapeutic approaches. For instance, both mutant SOD1, a misfolded mitochondrial protein associated with the onset of amyotrophic lateral sclerosis, and polyglutamine expanded ataxin-3, a pathogenic protein causing Machado-Joseph disease, are ubiquitinated by MITOL and then degraded by the proteasome [35–36]. Facilitating the proteasomal degradation of these aberrant proteins might therefore efficiently control diseases progression and, eventually, cure the diseases. Answering these questions would partially unveil the mysterious physiology of mitochondria, which, in turn, would facilitate the development of therapeutics to prevent and cure devastating human diseases.
Acknowledgements
We thank Dr. Tim Formosa, Dr. Dennis Winge and Dr. Janet Shaw for critical reading of the manuscript. The work in the Rutter laboratory related to this topic is supported by NIH grant GM087346.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Finley LWS, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Aging Research Reviews. 2009;8:173–188. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tatsuta T. Protein quality control in mitochondria. J. Biochem. 2009;149:455–461. doi: 10.1093/jb/mvp122. [DOI] [PubMed] [Google Scholar]
- [4].Livnat-Levanon N, Glickman MH. Ubiquitin-Proteasome System and mitochondria – Reciprocity. Biochimica et Biophysica Acta. 2010;1809:80–87. doi: 10.1016/j.bbagrm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- [5].Martinelli P, Rugarli EI. Emerging roles of mitochondrial proteases in neurodegeneration. Biochim. Biophys. Acta. 2010;1797:1–10. doi: 10.1016/j.bbabio.2009.07.013. [DOI] [PubMed] [Google Scholar]
- [6].Margineantu DH, Emerson CB, Diaz D, Hockenbery DM. Hsp90 Inhibition Decreases Mitochondrial Protein Turnover. PLoS ONE. 2007;10:e1066. doi: 10.1371/journal.pone.0001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;197:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RLJ, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011 Feb 4; doi: 10.1093/hmg/ddr048. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol. Biol. Cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Radke S, Chander H, Schäfer P, Meiss G, Krüger R, Schulz JB, Germain D. Mitochondrial Protein Quality Control by the Proteasome Involves Ubiquitination and the Protease Omi. J. Biol. Chem. 2008;283:12681–12685. doi: 10.1074/jbc.C800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Azzu V, Brand M. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 2009;123:578–585. doi: 10.1242/jcs.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lipson C, Alalouf G, Bajorek M, Rabinovich E, Atir-Lande A, Glickman M, Bar-Nun S. A Proteasomal ATPase Contributes to Dislocation of Endoplasmic Reticulum-associated Degradation (ERAD) Substrates. J. Biol. Chem. 2008;283:7166–7175. doi: 10.1074/jbc.M705893200. [DOI] [PubMed] [Google Scholar]
- [13].Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, Ashrafi K, Glickman MH, Rutter J. A Stress-Responsive System for Mitochondrial Protein Degradation. Mol. Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shcherbik N, Haines DS. Cdc48pNpl4p/Ufd1p Binds and Segregates Membrane-Anchored/Tethered Complexes via a Polyubiquitin Signal Present on the Anchors. Mol. Cell. 2007;25:385–397. doi: 10.1016/j.molcel.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-Anchored Receptor Atg32 Mediates Degradation of Mitochondria via Selective Autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- [16].Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, Zhou J, Chen Q. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson's disease. J. Biol. Chem. 2011:M110. doi: 10.1074/jbc.M110.144238. 144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell. Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nakamura N, Hirose S. Regulation of Mitochondrial Morphology by USP30, a Deubiquitinating Enzyme Present in the Mitochondrial Outer Membrane. Mol. Biol. Cell. 2008;19:1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- [21].Yu W, Sun Y, Guo S, Lu B. The PINK1/Parkin pathway regulates mitochondrial dynamics and function in mammalian hippocampal and dopaminergic neurons. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr235. doi: 10.1093/hmg/ddr235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of Amyloid Precursor Protein in the Mitochondrial Import Channels of Human Alzheimer's Disease Brain Is Associated with Mitochondrial Dysfunction. J. Neuroscience. 2006;26:9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Marin DR, Ramírez CM, González M, González-Muñoz E, Zorzano A, Camps M, Alonso R, Díaz M. Voltage-dependent anion channel (VDAC) participates in amyloid beta-induced toxicity and interacts with plasma membrane estrogen receptor α in septal and hippocampal neurons. Mol. Membr. Biol. 2007;24:148–160. doi: 10.1080/09687860601055559. [DOI] [PubMed] [Google Scholar]
- [24].Du H, Guo L, Fang F, Chen D, Sosunov AA, McKhann GM, Yan Y, Wang C, Zhang H, Molkentin JD, Gunn-Moore FJ, Vonsattel JP, Arancio O, Chen JX, Yan SD. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer's disease. Nat. Med. 2008;14:1097–1105. doi: 10.1038/nm.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Singh P, Suman S, Chandna S, Das TK. Possible role of amyloid-beta, adenine nucleotide translocase and cyclophilin-D interaction in mitochondrial dysfunction of Alzheimer's disease. Bioinformation. 2009;3:440–445. doi: 10.6026/97320630003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Devi L, Raghavendran V, Prabhu BM, Avadhani NG, Anandatheerthavarada HK. Mitochondrial Import and Accumulation of α-Synuclein Impair Complex I in Human Dopaminergic Neuronal Cultures and Parkinson Disease Brain. J. Biol. Chem. 2008;283:9089–9100. doi: 10.1074/jbc.M710012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kamp F, Exner N, Lutz AK, ender N, Hegermann J, Brunner B, Nuscher B, Bartels T, Giese A, Beyer K, Eimer S, Winklhofer KF, Haass C. Inhibition of mitochondrial fusion by α-synuclein is rescued by PINK1, Parkin and DJ-1. The EMBO. 2010;29:3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shirendeb U, Reddy AP, Manczak M, Calkins MJ, Mao P, Tagle DA, Reddy PH. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington's disease: implications for selective neuronal damage. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr024. doi: 10.1093/hmg/ddr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-Terminal Mutant Huntingtin Associates with Mitochondria and Impairs Mitochondrial Trafficking. J. Neuroscience. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, Inatome R, Yanagi S. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CAP. Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle's Dynamics and Signaling. Plos One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM. A New Function in Translocation for the Mitochondrial i-AAA Protease Yme1: Import of Polynucleotide Phosphorylase into the Intermembrane Space. Mol. Cell. Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12 Complex, an AAA Protease with Chaperone-like Activity in the Inner Membrane of Mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- [35].Yonashiro R, Sugiura A, Miyachi M, Fukuda T, Matsushita N, Inatome R, Ogata Y, Suzuki T, Dohmae N, Yanagi S. Mitochondrial Ubiquitin Ligase MITOL Ubiquitinates Mutant SOD1 and Attenuates Mutant SOD1-induced Reactive Oxygen Species Generation. Mol. Biol. Cell. 2009;20:4524–4530. doi: 10.1091/mbc.E09-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sugiuraa A, Yonashiroa R, Fukudaa T, Matsushitaa N, Nagashimaa S, Inatomea R, Yanagilow S. A mitochondrial ubiquitin ligase MITOL controls cell toxicity of polyglutamine-expanded protein. Mitochondrion. 2011;11:139–146. doi: 10.1016/j.mito.2010.09.001. [DOI] [PubMed] [Google Scholar]
- [37].Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, Xiao Y, Christie AL, Aster J, Settleman J, Gygi SP, Kung AL, Look T, Nakayama KI, DePinho RA, Wei W. SCFFBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]