Abstract
Tau is a neuronal-specific microtubule-associated protein that plays an important role in establishing neuronal polarity and maintaining the axonal cytoskeleton. Aggregated tau is the major component of neurofibrillary tangles (NFTs), structures present in the brains of people affected by neurodegenerative diseases called tauopathies. Tauopathies include Alzheimer’s disease (AD), frontotemporal dementia with Parkinsonism (FTDP-17), the early onset dementia observed in Down syndrome (DS; trisomy 21) and the dementia component of myotonic dystrophy type 1 (DM1). Splicing misregulation of adult-specific exon 10, which codes for a microtubule binding domain, results in expression of abnormal ratios of tau isoforms, leading to FTDP-17. Positions 3 to 19 of the intron downstream of exon 10 define a hotspot of splicing regulation: the region diverges between humans and rodents, and point mutations within it result in tauopathies. In this study, we investigated three regulators of exon 10 splicing: serine/arginine-rich protein SRp75 and heterogeneous nuclear ribonucleoproteins hnRNPG and hnRNPE2. SRp75 and hnRNPG inhibit splicing of exon 10 whereas hnRNPE2 activates it. Using co-transfections, co-immunoprecipitations and RNAi we discovered that SRp75 binds to the proximal downstream intron of tau exon 10 at the FTDP-17 hotspot region; and that hnRNPG and hnRNPE2 interact with SRp75. Thus, increased exon 10 inclusion in FTDP mutants may arise from weakened SRp75 binding. This work provides insights into the splicing regulation of the tau gene and into possible strategies for correcting the imbalance in tauopathies caused by changes in the ratio of exon 10.
Keywords: MAP tau, Exon 10 and tangle dementia, Isoform ratios, Alternative splicing regulation, Splicing factor SRp75, Heterogeneous nuclear ribonucleoproteins G and E2
1. Introduction
Tau is a microtubule-associated protein (MAP) enriched in axons of mature and growing neurons. Tau establishes neuronal polarity, organizes axonal microtubules and is involved in axonal transport. Hyperphosphorylated, microtubule-dissociated tau is the major component of neurofibrillary tangles (NFTs), a hallmark of many neurodegenerative diseases (Goedert and Jakes, 2005). Null tau mice, though viable, show morphological and cognitive defects (Ikegami et al., 2000). Additionally, human pedigrees that contain microdeletions and microduplications in the tau locus show developmental defects and learning disabilities (Shaw-Smith et al., 2006; Kirchoff et al., 2007).
The human tau gene undergoes extensive alternative splicing that is regulated spatially and temporally (Andreadis, 2005; Liu and Gong, 2008). Exon 10 modulates the C-terminus of the tau protein and encodes a microtubule binding domain. Exon 10 is adult-specific in rodents and humans but shows a crucial difference relevant to neurodegeneration: in adult rodents, exon 10 becomes constitutive. In contrast, in adult humans exon 10 remains regulated in the central nervous system where the 10+ and 10− isoforms are present in a 1:1 ratio (Andreadis, 2005).
Misregulation of tau exon 10 splicing that disturbs the 1:1 ratio causes neurodegeneration whether the cause is cis or trans: Mutations in exon 10 that are silent on the protein level nevertheless result in tangle-only dementias grouped under the term “tauopathies” (represented by inherited frontotemporal dementia with Parkinsonism, FTDP-17; Goedert and Jakes, 2005); changes in factors that influence exon 10 splicing result in the cognitive defects associated with myotonic dystrophy 1 (DM1; Jiang et al., 2004; Hernández-Hernández et al., 2006). The correct ratio of tau exon 10 is also disturbed in Alzheimer’s disease (AD; Glatz et al., 2005; Conrad et al., 2007) and Down syndrome (DS; Mehta et al., 1999; Shi et al., 2008).
Bioinformatics analysis of the human genome indicates that almost all human genes are alternatively spliced (Pan et al., 2008). Alternative splicing plays a critical role in controlling differentiation and development (Stamm et al., 2005), and misregulation of alternative splicing is the cause of many life-threatening human diseases (Tazi et al., 2009). Despite the high fidelity of exon recognition in vivo, it is currently impossible to accurately predict alternative exons; it appears that combinatorial control and “weighing” of splice element strength are used to enable precise recognition of the short and degenerate splice sites (Hertel, 2008).
Exonic and intronic enhancers and silencers are involved in splicing regulation (Wang and Burge, 2008). These cis elements are regulated by trans-acting factors that mostly belong to two superfamilies, the SR/SR-like and hnRNP proteins (Long and Cáceres, 2009; Martinez-Contreras et al., 2007). Several mammalian splicing factors are enhanced in or restricted to neurons. Nevertheless, it appears that the exquisite calibration of mammalian alternative splicing is primarily achieved by spatial and temporal variation in the expression and activity levels of quasi-ubiquitous splicing regulators (Hertel, 2008).
Exon 10 splicing is affected by exonic and intronic enhancers and silencers as well as by several trans factors and their phosphorylation (Andreadis, 2005; Gao et al., 2007; Novoyatleva et al., 2008; Shi et al., 2008; Wang et al., 2010). Investigations of dementia pedigrees have established that the proximal downstream intron of exon 10 is a hotspot for tauopathy mutations (reviewed in Andreadis, 2005; Liu and Gong, 2008). We previously established that the SR protein 9G8 inhibits exon 10 splicing, whereas hnRNP protein E3 activates it, both by interacting with this region of the exon (residues 14 and 19–21, respectively; Fig. 1A; Gao et al., 2007; Wang et al., 2010).
Fig. 1.

SRp75 inhibits splicing of tau exon 10. (A) Alignment of human (H) and mouse (M) of tau exon 10 and its downstream proximal intron, up to position +30. The exon is in uppercase, the intron in lowercase. The boundaries of the three riboprobe constructs are indicated. The CACAC motif that is the likely site of the SRp75 interaction is underlined. (B) Schematic representation of construct SP/10L. P and T represent the vector promoter and terminator. Introns and exon junctions in the vector portions of the constructs are not shown for the sake of clarity. The numbers on each side of exon 10 show (above) how many kilobases of flanking introns are present and (below) the extent of native introns flanking exon 10. The major splicing product is shown by solid, the minor one by dashed lines. (C) RT-PCR of SP/10L in COS cells in the absence and presence of SRp75. The RT-PCR products come from 1:1 co-transfections of SP/10L and FLAG-SRp75. % exon inclusion was calculated by scanning the bands from three independent transfections and measuring their areas. Primer pair: SPL-LS/SPL-LN. In all figures showing co-transfections, the asterisks show constructs in which inhibition of exon 10 by SRp75 approaches [(*)=p<0.1] or reaches statistical significance [*=p<0.05, **=p<0.01].
In this report we show that SRp75 also inhibits exon 10 splicing by binding in this region of the exon, although it appears to exert its effect independently of 9G8. Instead, its partners are hnRNPG, which strongly inhibits splicing of exon 10 (Wang et al., 2004), and hnRNPE2, which modestly activates splicing of exon 10 and interacts with hnRNPE3 (Broderick et al., 2004; Wang et al., 2010). This work adds detail to our earlier findings of the actions of these factors in this region (Broderick et al., 2004; Wang et al., 2004).
2. Materials and methods
2.1. Plasmid Construction and Mutagenesis
The starting construct was SP/10L (Fig. 1B), which contains human tau exon 10 plus 471 bp of its upstream intron and 408 bp of its downstream intron inserted into the EcoRI site of pSPL3 (Invitrogen). Deletions within the 30 bp downstream of exon 10 (I10-Δ3/10, I10-Δ11/18, I10-Δ19/26 and I10-Δ23/29) and point mutations reproducing FTDP-17 pedigree mutations (M11, M12, M13, M14, M16 and M19) were previously described (Gao et al., 2007; Wang et al., 2004). The mutations are diagrammed in Fig. 2A (deletions) and Fig. 3A (point mutations).
Fig. 2.
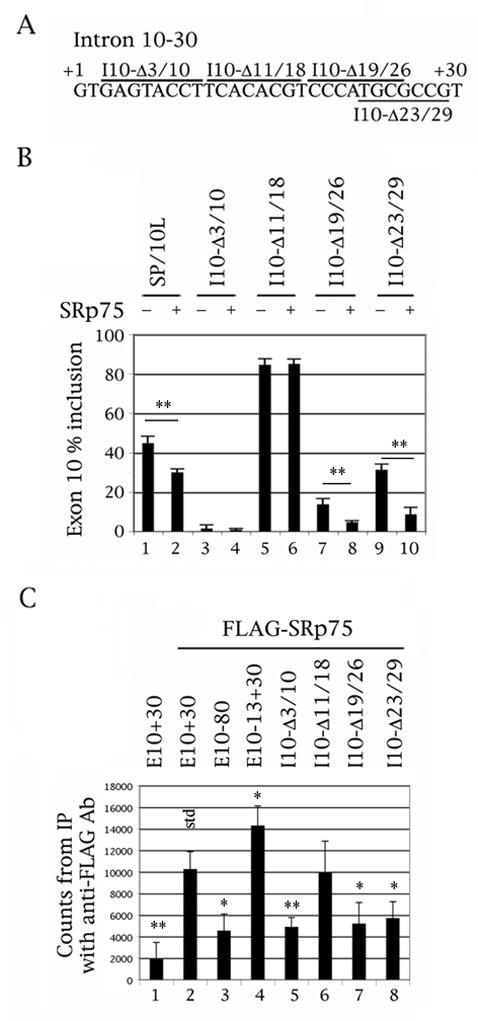
Deletion of the CACAC motif in the proximal downstream intron abolishes the ability of SRp75 to inhibit exon 10 splicing, although that deletion still binds SRp75. (A) Nucleotides +1 to +30 of the intron downstream of tau exon 10. The deletions are demarcated by bars above or below the sequence. (B) RT-PCR of wild-type and deleted SP/10L in COS cells in the absence and presence of SRp75. The RT-PCR products come from 1:1 co-transfections of tau constructs and FLAG- SRp75. Exon ratio calculations, primers, graph conventions and p-value notations are as in Fig. 1C. (C) 32P-labeled riboprobes containing wild-type or deleted exon 10 were incubated with extracts from COS cells transfected with FLAG- SRp75 and were immunoprecipitated by anti-FLAG monoclonal antibody M2. Amounts of riboprobe bound to SRp75 were calculated by measuring the counts retained after washing (means ± SD of three analyses). The asterisks show riboprobes in which SRp75 binding is significantly different from that of the wild-type riboprobe (std); the p-value notations are as in Fig. 1C. Equal amounts of FLAG- SRp75 were present in each experiment and the counts were normalized for starting probe counts and C-content.
Fig. 3.

Mutation of residues 12 and 13 of the intron downstream of tau exon 10, which affect an intronic silencer, abolishes SRp75 inhibition and binding. (A) The last three nucleotides of exon 10 and nucleotides +1 to +30 of the intron downstream of tau exon 10. The point mutations are indicated. (B) RT-PCR of wild-type and point-mutated SP/10L in COS cells in the absence and presence of SRp75. The RT-PCR products come from 1:1 co-transfections of tau constructs and FLAG- SRp75. Co-transfections were not done with mutants M19 and M29, because they exhibit no exon 10 inclusion and would not show additional inhibition by SRp75. Exon ratio calculations, primers, graph conventions and p-value notations are as in Fig. 1C. (C) 32P-labeled riboprobes containing wild-type or point-mutated exon 10 were incubated with extracts from COS cells transfected with FLAG- SRp75 and were immunoprecipitated by anti-FLAG monoclonal antibody M2. Conditions, calculations and notations are as in Fig. 2C.
For protein/RNA pulldowns we used previously described riboprobes E10+30, E10-80 and E10-13+30. E10+30 contains human tau exon 10 plus 30 bp of its downstream intron. E10-80 contains the 80 5′most bp of exon 10. E10-13+30 contains the 13 3′most bp of exon 10 plus 30 bp of its downstream intron (Gao et al., 2007; Fig. 1A). The deletions and point mutations created in SP/10L were also recreated in E10+30 as previously described (Gao et al., 2007).
For co-transfections and RNA pull-downs with exon 10 constructs, we cloned SRp75 into FLAG vector Tag2A (Stratagene). For co-immunoprecipitations (co-IPs) with other splicing factors, we cloned SRp75 into 3xMyc vector Tag3A (Stratagene). SRp75 deletion variants (diagrammed in Fig. 4A) were generated by PCR and cloned into Tag2A, B or C (Stratagene) depending on their reading frame. The primers used to create the variants are listed in Table 1.
Fig. 4.
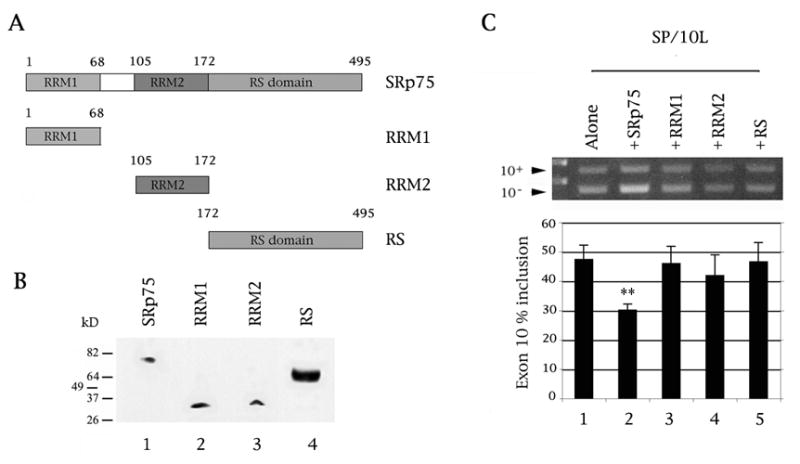
Both the RRM and RS domains of SRp75 are required for inhibition of exon 10 splicing. (A) Diagram of SRp75 variants used in this study. The domains and the amino acids that code for them are indicated. Total length of hnRNPE3 is 495 amino acids. (B) The SRp75 variants express as FLAG fusion proteins in COS cells. The proteins on the Western blot were detected by anti-FLAG monoclonal antibody M2. Protein markers are indicated on the left of the panel. The cell lysates were normalized for protein content. (C) RT-PCR of SP/10L in COS cells in the presence of full-length and single-domain variants of SRp75. The RT-PCR products come from 1:1 co-transfections of SP/10L and FLAG- SRp75 fusion constructs. Exon ratio calculations, primers, graph conventions and p-value notations are as in Fig. 1C.
Table 1.
Primers
| Name | Nt | Strand | Location | Sequence |
|---|---|---|---|---|
|
SRp75 deletion constructs & PCR of endogenous and transfected SRp75
| ||||
| 75-PS | 27 | S | Start of ORF | CCGCGGGTGTACATCGGCCGCCTGAGC |
| 75RRM-1N | 27 | A
|
End of RRM1 | CTCAACAATTACTCGCTCACCACAAAG |
| 75RRM-2S | 27 | S | Start of RRM2 | AGACTTATTGTGGAGAATTTGTCAAGT |
| 75RRM-2N | 27 | A
|
End of RRM2 | TAATCTGATTTTTCTCCCATTGACTTC |
| 75RS-S | 27 | S
|
Start of RS domain | GAAGTCAATGGGAGAAAAATCAGATTA |
| 75-DN | 27 | A | End of ORF | TTAGGACCTTGAGTGGGACCTAGATCT |
|
| ||||
|
PCR of endogenous and transfected tau exon 10
| ||||
| HT7S3 | 24 | S | In tau exon 7 | CAACGCCACCAGGATTCCAGCAAA |
| HT11N | 24 | A | In tau exon 11 | ATGTTGCCTAATGAGCCACACTTG |
| SPL3-LS | 27 | S | In SPL3 vector | TCTGAGTCACCTGGACAACCTCAAAGG |
| SPL3-LN | 27 | A | In SPL3 vector | ATCTCAGTGGTATTTGTGAGCCAGGGC |
|
| ||||
|
SRp75 RNAi constructs
| ||||
| In pFIV
| ||||
| 75i-1S | 23 | S | Near start of ORF | AAAGTGTGGAGCGCTTCTTTAAG |
| 75i-1N | 23 | A | Near start of ORF | AAAACTTAAAGAAGCGCTCCACA |
| 75i-2S | 23 | S | Within RS domain | AAAGGCCGCAGCAAGAGCAAAGA |
| 75i-2N | 23 | A | Within RS domain | AAAATCTTTGCTCTTGCTGCGGC |
| 75i-3S | 23 | S | Near end of ORF | AAAGTCTAGGTCCCACTCAAGGT |
| 75i-3N | 23 | A | Near end of ORF | AAAAACCTTGAGTGGGACCTAGA |
|
| ||||
| In SM2c
| ||||
| SM75-1 | 19 | S | Within ORF | GTGATTGAATTTGTATCTT (Open Biosystems V2HS_56694) |
| SM75-2 | 19 | S | Within ORF | CAGTGGATATGGTTATAGA (Open Biosystems V2HS_56696) |
| SM75-3 | 19 | S | Within ORF | GGGAACATGCCAAGTCTGA (Open Biosystems V2HS_56698) |
Nt=Length in nucleotides. S=sense, A=antisense.
We had previously cloned 9G8, Nova1 and hnRNPE2 in FLAG vectors from Sigma or Stratagene (pCMV-6 and Tag2, respectively; Gao et al., 2007; Wang et al., 2010) and hnRNPG into GFP vector EGFP-C2 from Promega (Wang et al., 2004).
Three pairs of siRNA oligonucleotides for SRp75 were designed and synthesized (Table 1). We annealed and cloned each pair into the pFIV-H1/U6 vector (SBI). We also obtained three Open Biosystems plasmids containing shRNA for SRp75 from the UMMS SiRNA Core (Table 1) as well as a mix of three SRp75 siRNAs (sc-38344) from Santa Cruz Biotechnology.
2.2. Cell Culture and Transfections
We cultured monkey kidney (COS) and human epithelioma (HeLa) as previously described (Wang et al., 2010). We prepared plasmid DNA by Qiagen Tip-100s or Promega PureYield midi columns. We grew the cells on 60 mm or 100 mm plates (for RT-PCR and co-IPs, respectively) and transfected them when they reached confluence of 50% (if using LT1 from Mirus) or 80% (if using Lipofectamine 2000 from Invitrogen).
For siRNA work, we transfected the siRNA first, followed ~12 h later by the tau DNA, because the optimal ratios of RNA and DNA differ greatly vis-à-vis lipofectamine. We changed the medium 16 hours after transfection and harvested the cells 48 hours after transfection.
2.3. RNA Preparation Reverse Transcription and PCR
We isolated total RNA from transfected cells by the TRIzol method (Invitrogen). We performed reverse transcription and PCR as previously described (Wang et al., 2010). To analyze the endogenous SRp75 levels, we used the Ambion Quantum kit with a ratio of 3:7 18S primers to 18S competimers as the internal control. For the RNAi experiments, vectors pFIV and SM2c-GFP or RNA sc-37007 (Santa Cruz Biotechnology) acted as the negative controls.
For PCR, we used the following primer pairs (shown in Table 1): SPL-LS/SPL-LN for transfected tau, HT7S3/HT11N for endogenous tau, 75-PS/75RRM-1N for endogenous SRp75. We did 27 PCR cycles if analyzing transfected constructs (denaturation 94 °C/1 min, annealing 62 °C/1 min, extension 72 °C/1 min); 30 cycles if analyzing endogenous SRp75 in cells (denaturation 94 °C/1 min, annealing 58 °C/1 min, extension 72 °C/1 min); and 21 cycles if analyzing endogenous SRp75 in polyA+ RNA (fetal brain, adult whole brain, cerebellum, hippocampus, spinal cord, skeletal muscle, heart, and liver; Clontech).
We calculated RNA ratios by scanning and averaging the bands from three independent transfections using the ImageJ program. The exception was endogenous SRp75 in polyA+ RNA, which we performed only once.
2.4. Protein Expression in Cells and co-IP Assays
We verified expression of the FLAG, Myc and GFP factor constructs by using antibodies against FLAG (mouse M2, Sigma), GFP (mouse, Invitrogen) or myc (rabbit, GeneScript) on Western blots of protein lysates from cells transfected with the constructs (Fig. 4B, 5A–D).
Fig. 5.
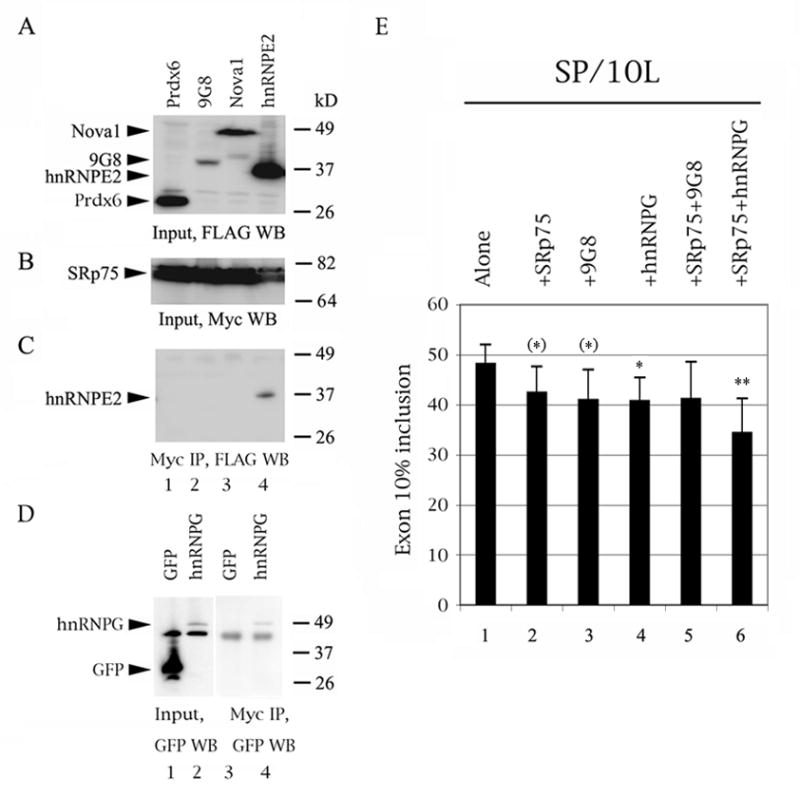
SRp75 interacts with hnRNPE2 and hnRNPG, the former a moderate activator and the latter a strong inhibitor of exon 10 splicing and SRp75 and hnRNPG may act synergistically. (AD) Western blots of protein lysate inputs and co-IPs from 1:1 co-transfections of myc-SRp75 and FLAG- or EGFP-factors in COS cells. Protein marker sizes are indicated on the right of each panel. The positions of factors are indicated on the left. (A) Input and (C) co-IP, factors detected by anti-FLAG mouse antibody M2. (B) Input SRp75 detected by anti-myc rabbit antibody. (D) Input (left) and co-IP (right), hnRNPG detected by anti-GFP mouse antibody. (E) RT-PCR of SP/10L in COS cells co-transfected with various combinations of FLAG-SRp75, FLAG-9G8 and EGFP-hnRNPG constructs. Exon ratio calculations, primers, graph conventions and p-value notations are as in Fig. 1C.
We prepared cell lysates from transfected cells using RIPA lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Nonidet P-40 [NP-40], 0.5% deoxycholate, 0.1% SDS) containing 1x Protease Inhibitor Cocktail Tablets (Roche Molecular Biochemicals). Lysates were nutated at 4 °C for 1 h, cleared by centrifugation at 13,200 rpm for 15 min at 4 °C and boiled for 10 min prior to running on SDS-PAGE gels.
For protein co-IPs, we prepared protein lysates from cells co-transfected with Myc-SRp75 plus EGFP-hnRNPG or FLAG-9G8, -Nova1 and -hnRNPE2. EGFP-C2 and FLAG-Prdx6 (peroxiredoxin 6) acted as the respective negative controls. After the centrifugation clearing step, we added rabbit anti-myc antibody agarose beads (Sigma) to the supernatants, incubated them with nutation at 4 °C overnight and rinsed the beads 4x with wash buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl).
We analyzed the complexes by Western blotting using rabbit anti-myc (input) and mouse anti-FLAG (input and IP) antibodies (Fig. 5A-D) as detailed in Wang et al. (2010). For detection we used Opti-4CN (Roche). We used primary and secondary antibodies at 1:5,000 dilution.
2.5. Riboprobe Generation and RNA-protein Immunoprecipitation
We linearized all riboprobes at the unique MluI site downstream of the inserts and phenol-extracted them. We made radiolabeled riboprobes from these templates using T7 RNA polymerase (Promega) with a Promega in vitro transcription kit and [32P]-CTP (Amersham).
For RNA-protein immunoprecipitation, we mixed the riboprobes with cell lysates transfected with FLAG-SRp75 for 20 min at RT. Then we added mouse anti-FLAG-antibody agarose beads (Sigma) and incubated with nutation at 4 °C overnight. The reactions were exposed to 254 nm UV light for 15 min on ice. We then washed the beads 4x with wash buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl) and counted the radioactivity they retained. We did the IPs in triplicate and averaged the results, normalizing for starting probe counts and C content.
3. Results
3.1. SRp75 inhibits splicing of exon 10 by binding to its downstream proximal intron
The splicing behavior of our mutants (Figs. 2B, 3B, odd-numbered lanes), defines region 11–18 as an intronic splicing silencer (ISS) in agreement with the expression patterns in FTDP-17 pedigrees and results from other laboratories (Andreadis, 2005 and references therein). This is the region that shows significant divergence between human and mouse tau (Fig. 1A).
Previous work from our laboratory also showed that SRp75 and hnRNPG inhibit exon 10 splicing (Fig. 1C; Wang et al., 2004) whereas hnRNPE2 modestly activates it (Broderick et al., 2004). We and other groups also showed that hnRNPG does not require its RRM domain to inhibit exon 10 splicing (Heinrich et al., 2009; Wang et al., 2004), strongly suggesting that it exerts its effect by binding to another splicing factor. Additionally, an expression cloning study identified SRp75 as one of the factors influencing exon 10 splicing (Wu et al., 2006).
SR proteins most commonly exert their effects on exons or exon-intron junctions (Long and Cáceres, 2009). SRp75 continues to inhibit exon 10 splicing when co-transfected with FTDP-17 point mutations that define exonic silencers within exon 10 (data not shown). We therefore decided to test the possibility that SRp75 may exert its effect at or near the ISS within the proximal downstream intron.
The co-transfections show that SRp75 can no longer inhibit splicing of exon 10 in mutant I10-Δ11/18, which essentially contains the ISS (Fig. 2B, lane 5 versus 6). SRp75 is also unable to inhibit splicing of FTDP-17 reproducing point mutants M12, M13 and M14 (Fig. 3B, lanes 5–10) but still inhibits splicing of mutants M11 and M16 (Fig. 3B, lanes 3, 4, 11 and 12).
The pulldowns of the equivalent constructs with FLAG-SRp75 show that SRp75 interacts directly with exon 10 (Fig. 2C, lane 1 versus 2). SRp75 binds to the intronic construct, E10-13+30, more strongly than it binds to E10+30, whereas it binds very weakly to the exonic one, E10-80 (Fig. 2C, lanes 2 to 4).
In the deletion pulldowns, SRp75 binds to I10-Δ11/18 (Fig. 2C, lane 6) as strongly as it binds to the full-length E10+30 but weakly to the other three (Fig. 2C, lanes 5, 7 and 8). In the point mutation pulldowns, SRp75 binds to M11, M14 and M16 as strongly as it does to E10-13+30 (Fig. 3C, lanes 3, 6 and 7), but very weakly to M12 and M13 (Fig. 3C, lanes 4 and 5).
These results indicate that SRp75 regulates splicing of exon 10 by binding to the proximal downstream intron and that intron residues 12 and 13 are critical for this interaction.
3.2. Both the RRM and RS domains of SRp75 are required to inhibit exon 10 splicing
To establish which domains of SRp75 are required for regulation of exon 10, we created three SRp75 single-domain constructs as FLAG fusions: Two contain the RNA recognition motif (RRM) domains, and one the serine/arginine-rich (RS) domain (Fig. 4A, 4B). None of them inhibits splicing of exon 10 (Fig. 4C). Also, the RS variant of SRp75 does not boost inclusion of exon 10 (Fig. 4C, lane 5), as is the case with equivalent constructs of other splicing factors (Wang et al., 2004, 2005).
This strongly implies that SR75 exerts its influence through direct binding to the exon 10 RNA but that it also requires interactions with additional factors to discharge its function.
3.3. SRp75 interacts with hnRNPG, another inhibitor of exon 10 splicing, and the two inhibitors appear to act synergistically
Since the behavior of the SRp75 variants indicates that it needs partners, we tested the ability of SRp75 to interact with factors known (9G8, hnRNPG, hnRNPE2, hnRNPE3) or suspected (Nova1) to influence exon 10 splicing via its proximal downstream intron (Fig. 5A–D). Of these, hnRNPE2 and hnRNPE3, members of the hnNRP family, modestly activate splicing of exon 10 (Broderick et al., 2004; Wang et al., 2010), whereas the other three factors inhibit splicing of exon 10 strongly (9G8, hnRNPG) or moderately (Nova1; Gao et al., 2007; Wang et al., 2004).
We tested interactions of SRp75 with other splicing factors with co-IPs. These show that SR75 does not interact with 9G8 (Fig. 5C, lane 2) or hnRNPE3 (Wang et al., 2010). However, it co-precipitates strongly with hnRNPG (Fig. 5D, lane 4) and hnRNPE2 (Fig. 5C, lane 4) and very weakly with Nova1 (Fig. 5C, lane 3).
To test whether the inhibitors that seem to interact by co-IP also enhance each other’s inhibition of exon 10 splicing, we co-transfected SP/10L with FLAG-SRp75, FLAG-9G8 and EGFP-hnRNPG combinations (Fig. 5E). These are triple transfections, so the inhibition effect decreases compared to the double transfections of the other experiments. As a result, addition of FLAG-SRp75 or FLAG-9G8 plus EGFP just misses attaining statistical significance (Fig. 5E, lanes 2 and 3). Addition of FLAG-SRp75 plus FLAG-9G8 shows no inhibition (Fig. 5E, lane 5), suggesting that the two factors may compete for an RNA region – a possibility, given that they apparently interact with residues 12/13 and 14, respectively. However, addition of FLAG-SRp75 plus EGFP-hnRNPG attains high statistical significance (Fig. 5E, lane 6), suggesting that the two factors may act synergistically, a finding congruent with the co-IP results.
The co-IP and co-transfection results suggest the possibility that SRp75 and hnRNPG may act in concert to inhibit exon 10 splicing and also that hnRNPE2 may antagonize the action of SRp75 by either titration or steric interference.
3.4. SRp75 knockdown reverses the inhibition of exon 10 splicing
To investigate if SRp75 is a native inhibitor of tau exon 10 splicing, we tested three siRNA plasmids, three shRNA plasmids and a mixture of siRNAs against it (75i-1, -2 and -3; SM75-1, -2 and -3; and si75, sc-38344 from Santa Cruz; Table 1). We chose HeLa because it expresses SRp75 as well as tau mRNA with 40% exon 10 inclusion.
We had great difficulty suppressing SRp75 in HeLa cells, perhaps because it is abundantly expressed in them (Zahler et al., 1993). Neither the siRNA nor the shRNA plasmids decreased endogenous SRp75 reproducibly in HeLa cells. Si75, a mixture of three SRp75 siRNAs, has little effect on endogenous exon 10; however, it decreases the expression levels of endogenous SRp75 and concommitantly increases inclusion of exon 10 in co-transfected SP/10L, although both changes just miss attaining statistical significance, possibly because the effect is variable (Fig. 6A, 6B, lanes 1 versus 2 and 3 versus 4).
Fig. 6.
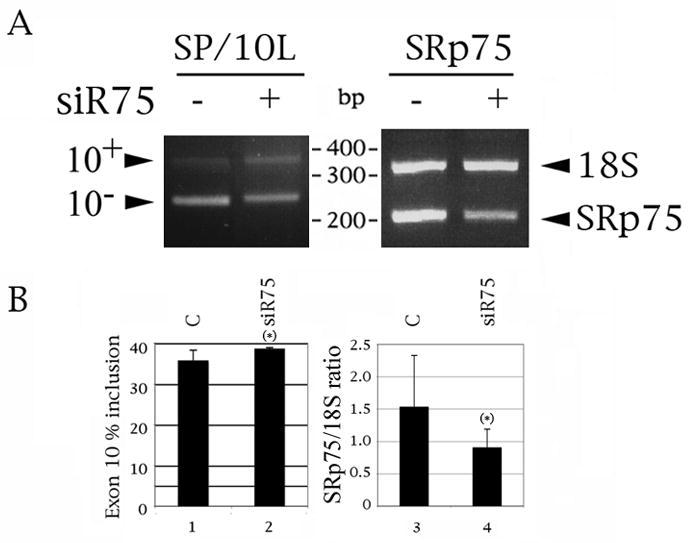
Suppression of SRp75 slightly increases inclusion of exon 10, although the changes in both SRp75 and exon 10 just miss attaining statistical significance. 1 ug of SP/10L was transfected into HeLa cells following transfection of control or Si75 RNA (Santa Cruz). (A) A representative result of the effect of si75 on transfected tau (left) and endogenous SRp75 (right). DNA marker sizes are indicated between the panels. The exon 10, 18S and SRp75 PCR products are indicated on the left and right of the panels, respectively. (B, left panel) Quantitative RT-PCR of tau exon 10. (B, right panel) Quantitative RT-PCR of endogenous SRp75. The ratio of SRp75 to 18S is indicated. Primer pair: 75-PS/75RRRM-1N. The 18S:competimer ratio was 3:7. Exon ratio calculations, primers, graph conventions and p-value notations are as in Fig. 1C.
The result prompts the conclusion that SRp75 is an endogenous activator of tau exon 10 splicing, though it falls short of providing definitive proof.
3.5. SRp75 is enriched in the central nervous system
Given the effect of SRp75 on exon 10 splicing, we wanted to determine its expression profile. SR proteins are known to be ubiquitously expressed, although their relative amounts differ spatially and temporally (Hertel, 2008; Long and Cáceres, 2009). Quantitative PCR shows that SRp75 is enriched in the central nervous system (Fig. 7A), particularly in cortex and spinal cord (Fig. 7B, lanes 2 and 5). It is also elevated in fetal brain and adult cerebellum (Fig. 7B, lanes 1 and 3), whereas its levels in hippocampus (Fig, 7B, lane 4) are equivalent to those in non-neuronal tissues (skeletal muscle, heart and liver; Fig. 7B, lanes 6–8).
Fig. 7.
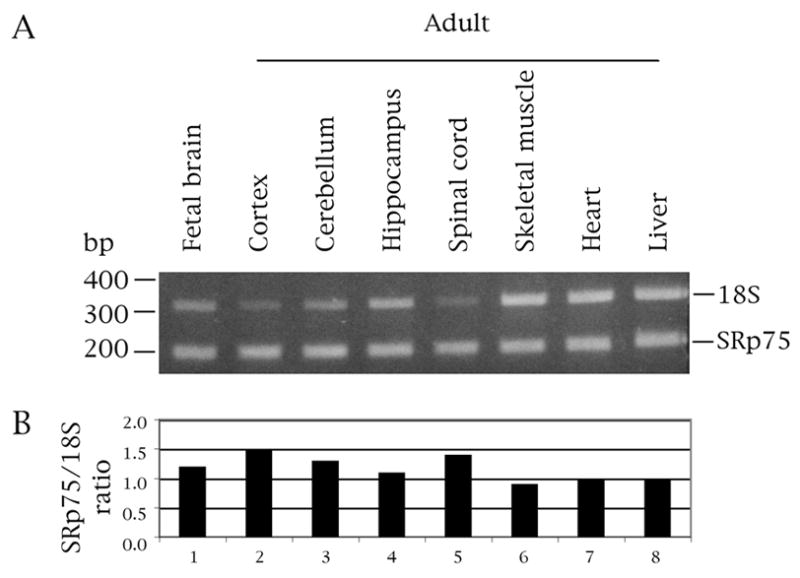
SRp75 is enriched in both the fetal and adult central nervous system, particularly in cortex and spinal cord. (A) Quantitative RT-PCR of polyA+ human RNA (Clontech). DNA marker sizes are indicated on the left, the 18S and SRp75 PCR products on the right of the panel. Primer pair: 75-PS/75RRRM-1N. 18S:competimer ratio was 3:7. (B) The graph shows the expression of SRp75 relative to 18S.
The SRp75 profile is totally different from that of hnRNPE2 and hnRNPE3 (Wang et al., 2010). The two hnRNP proteins are either highly enriched in (hnRNPE2) or restricted to (hnRNPE3) brain and the latter is adult-specific. Conversely, other studies (Heinrich et al., 2009) and Affymetrix arrays show that hnRNPG expression levels are similar in neuronal and non-neuronal tissues (http://www.genecards.org/cgi-bin/carddisp.pl?gene=gene=RBMX).
Our results agree with a previous study that reported SRp75 is enriched in brain (Zahler et al., 1993). The expression profile of SRp75 indicates that the factor is not the determinant of the developmental profile of exon 10, which is adult-specific (and thus would be compatible with an inhibitor with higher levels in fetal brain).
4. Discussion
4.1. SRp75 inhibits splicing of tau exon 10 by binding to the CA repeat in the intronic splicing silencer downstream of the exon
Previous work showed that tau exon 10 contains several splicing silencers and enhancers (reviewed in Andreadis, 2005; Liu and Gong 2008). A particularly important and interesting regulatory region of the exon is its proximal downstream intron, which diverges considerably between human and mouse (Fig. 1A). This contrasts with the near-total conservation of the exon itself and strongly suggests that the species-specific difference in the expression of exon 10 arises from regulation of this portion of the pre-mRNA. The region known to affect splicing of exon 10 consists of a silencer (3 to 16) followed by an enhancer (19 to 30; Fig. 8). This region is a hotspot for point mutations that cause tauopathies by either strongly increasing (M3, 11, 12, 13, 14, 16) or strongly decreasing (M19, M29) inclusion of exon 10 (reviewed in Andreadis, 2005).
Fig. 8.
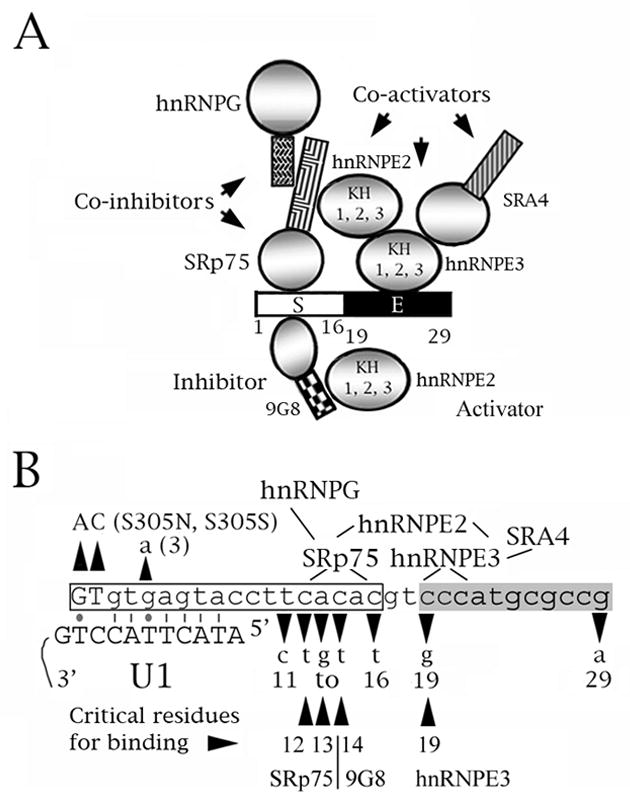
A speculative model of the role of SRp75 in the regulation of tau exon 10 splicing. The model comes from Wang et al. (2003), Broderick et al. (2004), Gao et al. (2007) and Wang et al (2009) and this study. (A, B) Diagrams of the proximal downstream intron of exon 10. The numbers below the sequence correspond to the residue number from the start of the intron. 9G8, SRp75 and hnRNPG inhibit exon 10 splicing whereas hnRNPE2, hnRNPE3 and SRA4 activate it. SRp75, 9G8 and hnRNPE3 bind to the tau pre-mRNA at sites around residues 12–13, 14 and 19, respectively. SRp75 interacts with hnRNPG and hnRNPE2, hnRNPE3 interacts with hnRNPE2 and SRA4. 9G8 also interacts with hnRNPE2 but with none of the other factors. (A) The proximal downstream intron of tau exon 10. S=intronic splicing silencer (white), E=intronic splicing enhancer (black). For the factors, circles represent RRM or KH domains, rectangles represent RS domains. (B) The boxed region is the intronic silencer, the shaded region the intronic enhancer. The exon is shown in uppercase, the intron in lowercase letters. The FTDP mutations are indicated. The lines show which factors interact and the regions of the tau pre-mRNA that interact with SRp75 and hnRNPE3. The residues critical for binding of SRp75 (12, 13), 9G8 (14) and hnRNPE3 (19) are indicated. Also shown is the base paring of the exon 10 5′ splice site with the U1 snRNA. Lines are Crick-Watson pairs, dots G-T base pairs.
The co-transfection and pull-down results indicate that SRp75 exerts its influence on exon 10 by directly binding to the CA repeat at position 12–16. In particular, the CA residues at position 12–13 appear critical to binding (Fig. 3). This, plus the inhibitory behavior of SRp75, is congruent with the finding that FTDP-17 mutants M11 to M16 strongly increase exon 10 inclusion. CA repeats are often found at alternative 5′ splice sites and can function as either splicing enhancers or silencers (Hui et al., 2005).
Interaction with the U1 snRNA itself would encompass residues −2 to 7 (Fig. 8B). Thus, although SRp75 would not mask the site necessary for base pair formation with U1, it and its interacting co-factors may sterically inhibit attachment of the U1 snRNP.
SRp75 has not been as widely studied as other SR proteins. As all SR proteins, it can either activate or inhibit splicing. A recent study demonstrated that SRp75 inhibits use of a 5′ splice site in HIV-1 RNA but the details of its action were not determined (Tranell et al., 2010).
4.2. SRp75 requires hnRNPG to exert its full regulatory effect on tau exon 10 and its action may be antagonized by hnRNPE2 and its interacting factors
The SR protein 9G8, which inhibits splicing of exon 10 more strongly than SRp75, binds to the same region as SRp75 and requires a wild-type residue at position 14 (Gao et al., 2007; Wang et al., 2004). However, SRp75 and 9G8 do not interact (Fig. 5C) nor act additively to enhance suppression of exon 10 (Fig. 5E). Instead, both inhibitors interact with hnRNPE2, a modest activator of exon 10 splicing (Broderick et al., 2004), as evidenced by co-IPs for SRp75 (Fig. 5C) and yeast two-hybrid assays for 9G8 (Funke et al., 1996).
These interactions imply that hnRNPE2 can antagonize the action of both SRp75 and 9G8. Our previous work indicates that it may do so by interacting with hnRNPE3, another modest activator of exon 10 splicing which binds to the C triplet in position 19–21 and also interacts with SR factor SRA4 (Fig. 8; Wang et al., 2010). The hnRNPE2/hnRNPE3 complex may prevent binding of SRp75 and 9G8 either sterically (by “covering” a region of the exon 10 RNA which would otherwise be available to either inhibitor) or kinetically (by titrating out the inhibitors).
If 9G8 and SRp75 act independently, their concentrations within a cell may determine which one becomes the crucial factor for exon 10 exclusion. This scenario may also explain the binding of SRp75 to deletion I10-Δ11/18 in vitro (Fig. 2). This deletion brings a CA motif, originally at position 21–22, to position 13–14 of the intron. The newly proximal CA motif may be sufficient to bind SRp75 (but not to suppress exon 10 splicing, which requires SRp75 cofactors), whereas 9G8 can no longer bind to this deletion (Gao et al., 2007).
Whereas 9G8 seems not to need partners to fully inhibit tau exon 10 splicing, SRp75 may need hnRNPG or other co-factors to exert its full effect. Co-IPs and co-transfections show that SRp75 and hnRNPG interact (Fig. 5D) and may act synergistically (Fig. 5E). Previous work (Wang et al., 2004) has shown that, unlike SRp75, the N-terminal region of hnRNPG (which contains both its RRM and a recently identified novel RNA binding domain; Heinrich et al., 2009) is not required to inhibit splicing of exon 10, suggesting it acts via protein interactions.
HnRNPG also interacts with htra2-beta1, antagonizing the action of the latter (Heinrich et al., 2009; Hoffman and Wirth, 2002; Nasim et al., 2003). Htra2-beta1 modestly activates exon 10 splicing by interacting with a purine-rich enhancer within the exon (defined by FTDP-17 mutations Δ280K and N279K). However, the inhibitory effect of hnRNPG persists when the purine-rich region is destroyed (Wang et al., 2005). The interaction with SRp75 pinpoint the additional entry point of the hnRNPG influence on exon 10.
Fig. 8 shows a speculative model that summarizes our cumulative knowledge of the splicing regulation at the proximal downstream intron of tau exon 10. The binding sites of 9G8, SRp75 and hnRNPE3 and the interactions between hnRNPE3, hnRNPE2, 9G8, SRp75 and hnRNPG have been defined (Broderick et al., 2004; Gao et al., 2007; Wang et al., 2004, 2010; this work). Future work will allow us to determine if additional elements and factors play a role in regulating exon 10 splicing via this region.
4.3. Connections of tau exon splicing to dementia
The accumulation of abnormal tau filaments into tangles is a hallmark of many neurodegenerative diseases, including AD and DS. The number of NFTs correlates with disease severity, although the emerging consensus is that the true toxic species are tau oligomers that form earlier in the process (Jellinger, 2009). In several neurodegenerative diseases, collectively termed tauopathies, tau pathology is solely and directly responsible for neuronal death and development of the clinical dementia manifestations (Goedert and Jakes, 2005).
Tau exon 10 is a developmentally regulated cassette. It codes for an additional microtubule binding domain which increases affinity of tau protein for microtubules (Andreadis, 2005; Liu and Gong, 2008). The tauopathy pedigrees analyzed thus far predominantly show mutations in tau exon 10, although several pedigrees carry mutations in tau exons 1, 9, 11, 12 and 13 which influence either microtubule binding or protein conformation (Goedert and Jakes, 2005).
The exon 10 mutations fall in two categories: half influence microtubule binding and half alter the ratio of exon 10 (Andreadis, 2005; Goedert and Jakes, 2005). The latter category is effectively a subtle version of a dosage disease, as the disease results from ratio alterations even though the gene produces wild-type protein. In addition to tangle-only tauopathies, the ratio of exon 10 is also disturbed in sporadic AD (Glatz et al., 2005; Conrad et al., 2007), DS (Mehta et al., 1999; Shi et al., 2008) and DM1 (Jiang et al., 2004; Hernández-Hernández et al., 2006).
Work by us and other researchers has linked tauopathies to the cognitive impairment in DS by showing that factors on chromosome 21 influence splicing of tau exon 10 (Shi et al., 2008; Wang et al., 2010). This work and related studies (Wang et al., 2004; Heinrich et al., 2009) link gender-specific modulation of neurodegeneration to tau splicing: hnRNPG, also known as RBMX, is X-linked and has a Y-linked paralogue, RBMY, which has a different set of co-factors and targets (Elliott, 2004).
It is becoming clear that many finely-tuned biological processes achieve their exquisite calibration by combinatorial methods. The overlap of SR protein functions and the regulation of alternative splicing both belong in this regulatory mode. This inherently complex regulation mode complicates the possibility of ameliorating or curing diseases caused through missplicing (such as FTDP-17) by tinkering with factor ratios. Nevertheless, work on the basic molecular biology of the tau molecule may give us the tools to comprehend and combat not only FTDP, but also other types of dementia, which is becoming increasingly prevalent as the human lifespan lengthens, as well as the common and pleiotropic DS and DM1. These diseases vary widely both in clinical phenotype and brain pathology, but they share tangles, neuronal death and cognitive impairment as invariable defining characteristics.
Acknowledgments
This work was supported by NIH grants R01 AG18486 to A. A. and R21 HD056195 to A. A. and S. S. We want to thank Dr. Alonso Ross for his generous subsidy of the shRNA clones via the UMMS SiRNA Core.
Abbreviations used
- AD
Alzheimer’s disease
- DM1
myotonic dystrophy type 1
- DS
Down syndrome, (k)bp, (kilo)base pairs
- FTDP-17
frontotemporal dementia with Parkinsonism
- hnRNP
heterogeneous nuclear ribonucleoprotein
- IP
immunoprecipitation
- ISS
intronic splicing silencer
- MAP
microtubule-associated protein
- nt
nucleotides
- NFT
neurofibrillary tangle
- RRM
RNA recognition motif
- R/S
arginine/serine
- snRNPs
small nuclear ribonucleoproteins
- SR proteins
serine/arginine-rich proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochem Biophys Acta. 2005;1739:91–103. doi: 10.1016/j.bbadis.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Broderick JA, Wang J, Andreadis A. Heterogeneous nuclear ribonucleoprotein E2 binds to tau exon 10 and moderately activates its splicing. Gene. 2004;331:107–114. doi: 10.1016/j.gene.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Conrad C, Zhu J, Conrad C, Schoenfeld D, Fang Z, Ingelsson M, Stamm S, Church G, Hyman BT. Single molecule profiling of tau gene expression in Alzheimer’s disease. J Neurochem. 2007;103:1228–1236. doi: 10.1111/j.1471-4159.2007.04857.x. [DOI] [PubMed] [Google Scholar]
- Elliott DJ. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int J Androl. 2004;27:328–334. doi: 10.1111/j.1365-2605.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- Funke B, Zuleger B, Benavente R, Schuster T, Goller M, Stevenin J, Horak I. The mouse polyC-binding protein exists in multiple isoforms and interacts with several RNA-binding proteins. Nucl Acids Res. 1996;24:3821–3828. doi: 10.1093/nar/24.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Wang J, Wang Y, Andreadis A. SR protein 9G8 modulates splicing of tau exon 10 via its proximal downstream intron, a clustering region for frontotemporal dementia mutations. Mol Cell Neurosci. 2007;34:48–58. doi: 10.1016/j.mcn.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, Rieder P, Moeller HJ, Andreadis A, Henkel K, Stamm S. The alternative splicing of tau exon 10 and its regulatory proteins clk2 and tra2-beta1 changes in sporadic Alzheimer’s disease. J Neurochem. 2005;96:635–644. doi: 10.1111/j.1471-4159.2005.03552.x. [DOI] [PubMed] [Google Scholar]
- Goedert M, Jakes R. Mutations causing neurodegenerative tauopathies. Biochim Biophys Acta. 2005;1739:240–250. doi: 10.1016/j.bbadis.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, Sperling J, Sperling R, Stamm S. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem. 2009;284:14303–14315. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Hernández O, Bermúdez-de-León M, Gómez P, Velázquez-Bernardino P, García-Sierra F, Cisneros B. Myotonic dystrophy expanded CUG repeats disturb the expression and phosphorylation of tau in PC12 cells. J Neurosci Res. 2006;84:841–851. doi: 10.1002/jnr.20989. [DOI] [PubMed] [Google Scholar]
- Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Wirth B. HnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- Hui J, Hung LH, Heiner M, Schreiner S, Neumuller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami S, Harada A, Hirokawa N. Muscle weakness, hyperactivity, and impairment in fear conditioning in tau-deficient mice. Neurosci Lett. 2000;279:129–132. doi: 10.1016/s0304-3940(99)00964-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Jellinger KA. Recent advances in our understanding of neurodegeneration. J Neural Transm. 2009;116:1111–1162. doi: 10.1007/s00702-009-0240-y. [DOI] [PubMed] [Google Scholar]
- Kirchhoff M, Bisgaard AM, Duno M, Hansen FJ, Schwartz M. A 17q21.31 microduplication, reciprocal to the newly described 17q21.31 microdeletion, in a girl with severe psychomotor developmental delay and dysmorphic craniofacial features. Eur J Med Genet. 2007;50:256–263. doi: 10.1016/j.ejmg.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Long JC, Cáceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. HnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Mehta PD, Patrick BA, Dalton AJ, Aisen PS, Emmerling ME, Sersen EA, Wisniewski HM. Increased levels of tau-like protein in patients with Down syndrome. Neurosci Lett. 1999;275:159–162. doi: 10.1016/s0304-3940(99)00754-5. [DOI] [PubMed] [Google Scholar]
- Nasim MT, Chernova TK, Chowdhury HM, Yue BG, Eperon IC. HnRNPG and Tra2beta: opposite effects on splicing matched by antagonism in RNA binding. Hum Mol Genet. 2003;12:1337–1348. doi: 10.1093/hmg/ddg136. [DOI] [PubMed] [Google Scholar]
- Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AH, Bollen M, Stamm S. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum Mol Genet. 2008;17:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, Gribble S, Curley R, Cumming S, Dunn C, Kalaitzopoulos D, Porter K, Prigmore E, Krepischi-Santos AC, Varela MC, Koiffmann CP, Lees AJ, Rosenberg C, Firth HV, de Silva R, Carter NP. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhang T, Zhou C, Chohan MO, Gu X, Wegiel J, Zhou J, Hwang YW, Iqbal K, Grundke-Iqbal I, Gong CX, Liu F. Increased dosage of Dyrk1A alters alternative splicing factor (ASF)-regulated alternative splicing of tau in Down syndrome. J Biol Chem. 2008;283:28660–28669. doi: 10.1074/jbc.M802645200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochem Biophys Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranell A, Fenyö EM, Schwartz S. Serine- and arginine-rich proteins 55 and 75 (SRp55 and SRp75) induce production of HIV-1 vpr mRNA by inhibiting the 5′-splice site of exon 3. J Biol Chem. 2010;285:31537–31547. doi: 10.1074/jbc.M109.077453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gao QS, Wang Y, Lafyatis R, Stamm S, Andreadis A. Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. J Neurochem. 2004;88:1078–1090. doi: 10.1046/j.1471-4159.2003.02232.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang J, Gao L, Lafyatis R, Stamm S, Andreadis A. Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c/SRp55 complex that either recruits or antagonizes htra2-beta1. J Biol Chem. 2005;280:14230–14239. doi: 10.1074/jbc.M413846200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao L, Tse SW, Andreadis A. Heterogeneous nuclear ribonucleoprotein E3 modestly activates splicing of tau exon 10 via its proximal downstream intron, a hotspot for frontotemporal dementia mutations. Gene. 2010;451:23–31. doi: 10.1016/j.gene.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Kar A, Kuo D, Yu B, Havlioglu N. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol Cell Biol. 2006;26:6739–6747. doi: 10.1128/MCB.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Stolk JA, Roth MB. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]


