Abstract
HNF-4α is a transcription factor of the nuclear hormone receptor family that is expressed in the hepatic diverticulum at the onset of liver development. Mouse embryos lacking HNF-4α fail to complete gastrulation due to dysfunction of the visceral endoderm. This early embryonic lethality has so far prevented any analyses of the contribution of HNF-4α toward liver development and hepatocyte differentiation. However, we have shown that complementation of HNF-4α−/− embryos with a tetraploid embryo-derived wild-type visceral endoderm rescues this early developmental arrest and allows HNF-4α−/− embryos to proceed normally through midgestation stages of development. Examination of these rescued embryos revealed that HNF-4α was dispensable for specification and early development of the liver. However, HNF-4α−/− fetal livers failed to express a large array of genes whose expression in differentiated hepatocytes is essential for a functional hepatic parenchyma, including genes encoding several apolipoproteins, metabolic proteins, and serum factors. In addition, we have demonstrated that HNF-4α is essential for expression of the transcription factors HNF-1α and PXR within the fetal liver. We therefore conclude that HNF-4α is both essential for hepatocyte differentiation during mammalian liver development and also crucial for metabolic regulation and liver function.
Keywords: HNF-4, liver development, hepatocyte, tetraploid, hepatogenesis, differentiation
Cell differentiation is a consequence of changes in gene expression that are primarily controlled at the level of transcription. This implies that regulated expression of specific transcription factors in response to extracellular signals is an important determinant of cell differentiation. This view is supported by the fact that mice lacking the transcription factor PDX-1 fail to form a pancreas (Jonsson et al. 1994; Offield et al. 1996). Transcription factors that control embryonic cell differentiation also are often required to maintain and regulate gene expression in the adult cell. Therefore, analyzing the promoters of genes expressed in adult tissues can identify factors that control embryonic development. Study of liver gene expression using hepatoma cells has identified several transcription factors acting in the adult liver. These include the hepatocyte nuclear factors (HNFs) -1α and -1β, -3α, β, and γ, -4α; and -6, as well as the c/EBP family (for review, see Cereghini 1996). Although many of these factors appear to regulate gene expression in mature hepatocytes, until now none have been shown to be essential for hepatic development.
Liver development in the mouse begins at ∼E8.25 days of gestation (approximately the four- to six-somite stage) following formation of the definitive endoderm (Gualdi et al. 1996). Although it has been proposed that the liver parenchyma may derive from the septum transversum mesenchyme, lineage tracing studies and tissue explant coculture experiments strongly support the alternative proposal that hepatocytes descend directly from the definitive endoderm (LeDouarin 1968; LeDouarin 1975; Gualdi et al. 1996). Signal(s) emanating from precardiac mesenchyme induce the ventral foregut endoderm to proliferate and subsequently adopt a hepatic fate (LeDouarin 1964; Houssaint 1980; Fukuda-Taira 1981; Gualdi et al. 1996). Recently, Jung et al. (1999) have demonstrated that fibroblast growth factors (FGFs) -1 and -2 can mediate this induction in endoderm explant cultures. One consequence of this induction is the expression of characteristic hepatocyte marker genes such as albumin and transthyretin within the developing liver (Cascio and Zaret 1991). In situ hybridization analyses have shown that like albumin, the transcription factor HNF-4α is expressed in the developing hepatic diverticulum at ∼E8.75 (10–12 somites) (Duncan et al. 1994; Taraviras et al. 1994). This expression of HNF-4α early in the genesis of the hepatic lineage precedes that of HNF-1α, which has also been implicated in regulation of liver gene expression (Cereghini et al. 1992; Cereghini 1996). Importantly, HNF-4α has been found to activate expression from the HNF-1α promoter, which contains an HNF-4α binding site (Tian and Schibler 1991; Kuo et al. 1992). These data suggest that HNF-4α may act upstream in a transcription factor cascade that could drive differentiation of the hepatocyte lineage during liver development.
HNF-4α was identified as a factor within hepatoma cell extracts that bound to the promoters of the transthyretin (TTR) and apolipoprotein CIII (apoCIII) genes (Sladek et al. 1990). It is a member of the nuclear hormone receptor family whose activity may be modulated by fatty-acyl thioesters (Hertz et al. 1998). Targeted disruption of the HNF-4α gene in mice resulted in an embryonic lethal phenotype caused by the failure of HNF-4α−/− embryos to complete gastrulation (Chen et al. 1994). This developmental disruption occurred prior to the onset of hepatogenesis and so prevented the use of these embryos to analyze the role of HNF-4α in liver development. A detailed study of these embryos revealed that their early developmental arrest was caused by a dysregulation of extraembryonic (visceral) endoderm function (Duncan et al. 1997). Furthermore, it was found that embryonic development could proceed normally through gastrulation if HNF-4α−/− fetuses were complemented with an HNF-4α+/+ visceral endoderm (Duncan et al. 1997). This complementation was achieved by aggregating HNF-4α−/− ES cells with HNF-4α+/+ tetraploid embryos. With this approach the resulting fetuses are derived solely from the diploid ES cells whereas the extraembryonic tissues are tetraploid embryo derived (Nagy et al. 1990, 1993; Nagy and Rossant 1993). We have now exploited this extraordinarily powerful procedure to determine the consequence of HNF-4α loss of function on liver development. Our studies demonstrate that although HNF-4α is dispensable for specification of the hepatic lineage, it is an essential and central regulator of liver gene expression and is required for hepatocyte differentiation.
Results
HNF-4α is dispensable for hepatic specification
At the onset of liver development FGFs secreted from the developing heart induce adjacent embryonic endoderm to commit to a hepatic fate. In situ hybridization studies reveal that the first expression of HNF-4α within the embryonic endoderm occurs at around this stage in development. This early developmental expression within the hepatic field raised the possibility that HNF-4α might have an integral role during specification of the hepatic lineage. Moreover, because HNF-4α is a transcription factor, it seemed likely that any such role would be cell autonomous. If HNF-4α was essential for specification of hepatic cells then it could be predicted that HNF-4α+/+ cells would have a strong selective advantage over HNF-4α−/− cells during hepatogenesis. Livers isolated from HNF-4α+/+: HNF-4α−/− chimeric embryos should, in this case, contain only HNF-4α+/+ hepatoblasts. To determine whether HNF-4α provided a selective advantage to developing hepatoblasts, we therefore produced chimeric embryos between ROSA26 HNF-4α+/+ morulae and HNF-4α−/− ES cells. ROSA26 embryos express a lacZ transgene in all fetal tissues including the developing liver (Fig. 1A). In contrast, the HNF-4α−/− ES cells used in these experiments do not contain lacZ. Staining for β-galactosidase expression could therefore be used to determine whether hepatoblasts present in livers isolated from chimeric embryos were of HNF-4α+/+ (lacZ +ve) or HNF-4α−/− (lacZ −ve) origin. Chimeric embryos were collected after 11.5 days of gestation (E11.5) and stained for expression of β-galactosidase. Figure 1A shows that β-galactosidase was expressed ubiquitously in fetal livers isolated from ROSA26 mice, whereas no expression was detected in livers isolated from wild-type (CD-1) mice. The majority of livers isolated from HNF-4α+/+ (ROSA26); HNF-4α−/− chimeric embryos contained a mixture of β-galactosidase +ve and −ve cells (Fig. 1A,B). Figure 1B shows that numerous β-galactosidase −ve hepatoblasts could be detected in semithin sections prepared from chimeric livers. These results demonstrate that HNF-4α offers no selective advantage to hepatoblasts during early liver development. This, in turn, implies that the ability of embryonic endoderm to embark upon a hepatic differentiation program does not require HNF-4α within the developing hepatic cells.
Figure 1.
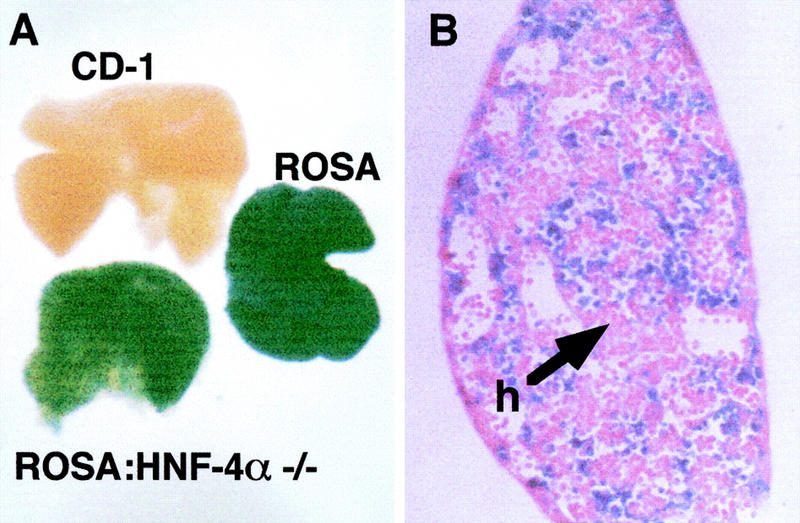
Hepatoblasts derived from HNF-4α−/− ES cells contribute toward the developing liver. (A) Fetal livers were isolated from wild-type (CD-1), ROSA26 (ROSA), or chimeric (ROSA: HNF-4α−/−) embryos at E11.5 and stained for β-galactosidase expression. (B) Photomicrograph showing a 5 μm section through the chimeric fetal liver shown in A. The section was counterstained with eosin to show HNF-4α−/− hepatoblasts (h).
HNF-4α−/− livers can be generated through aggregation of tetraploid embryos with HNF-4α−/− ES cells
Although the above data demonstrate that there is no genetic selection against the contribution of HNF-4α−/− cells to the developing liver, this assay does not determine the phenotypic consequence of loss of HNF-4α function on hepatocyte differentiation. We have shown previously that HNF-4α knockout embryos can proceed through gastrulation if they are provided with an HNF-4α+/+ visceral endoderm. As illustrated in Figure 2A, this was achieved by aggregating HNF-4α null ES cells with wild-type tetraploid embryos and allowing their development in utero (Duncan et al. 1997). Fetuses generated by the tetraploid aggregation procedure are entirely ES cell derived (Nagy et al. 1990, 1993; Nagy and Rossant 1993; Duncan et al. 1997). To directly determine the role of HNF-4α during hepatocyte differentiation we therefore attempted to generate HNF-4α null livers from HNF-4α−/− ES cells by tetraploid aggregation.
Figure 2.
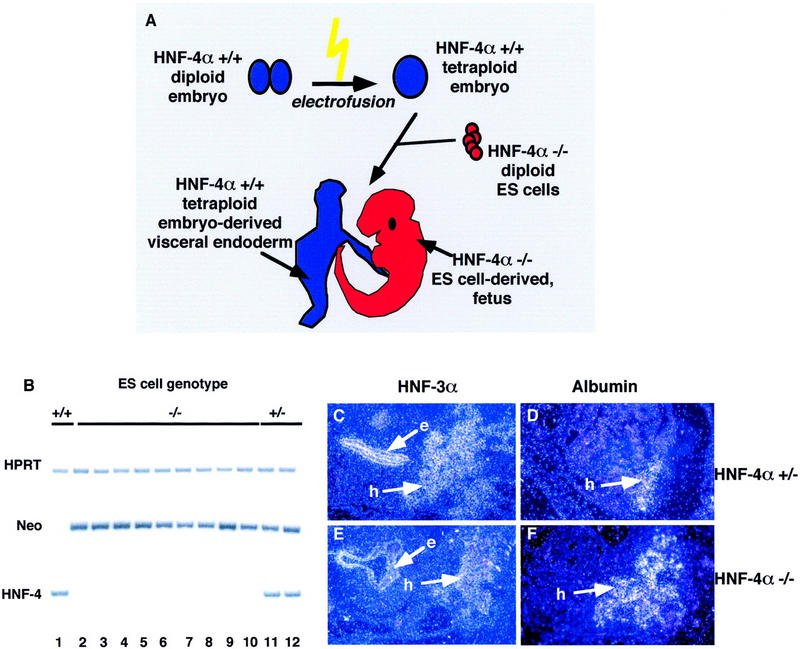
Hepatoblasts are present in embryos generated from HNF-4α−/− ES cells by tetraploid aggregation. (A) Embryos were generated from HNF-4α−/− ES cells (red) by aggregating the cells with HNF-4α+/+ embryos (blue) made tetraploid by electrofusion. Embryos continued their development in utero after transfer of the aggregates to psuedopregnant surrogate mothers. Fetal tissues generated by this procedure arise from HNF-4α−/− ES cells; the extraembryonic tissues, including the visceral endoderm, are tetraploid embryo derived. The presence of HNF-4α+/+ visceral endoderm allows HNF-4α−/− embryos to proceed through midgestation stages of development. (B) The genotype of embryos generated from HNF-4α+/+ (lane 1), HNF-4α+/− (lanes 11,12), and HNF-4α−/− ES cells (lanes 2–10) could be determined by PCR using primers which specifically amplified DNA fragments from the HPRT, bacterial neo, or HNF-4α (HNF-4) genes. (C–F) Dark-field micrographs showing in situ hybridization analyses that were performed on 5 μm paraffin sections of E10.5 embryos generated from HNF-4α+/− (C,D) and HNF-4α−/− (E,F) ES cells. In all embryos, HNF-3α mRNAs (C,E) could be detected in gut endoderm (e) and hepatoblasts (h), whereas albumin mRNAs (D,F) were detected only in hepatoblasts.
Figure 2B shows that the genotype of embryos derived from HNF-4α+/+, HNF-4α+/−, or HNF-4α−/− ES cells could be confirmed by PCR analysis of genomic DNA isolated from a rostral region of the embryos. Specific oligonucleotide primers were designed to amplify hypoxanthine phosphoribosyl transferase (HPRT), neomycin phosphotransferase (neo), and HNF-4α genomic sequences. All embryos amplified a 219-bp product from the HPRT gene, demonstrating the presence of DNA in each sample. HNF-4α+/− and HNF-4α−/− ES cell-derived embryos could be distinguished from wild-type ES cell-derived embryos by amplification of a 225-bp region of the neor transgene that was originally used to mutate HNF-4α in ES cells (Chen et al. 1994). Finally, amplification using HNF-4α-specific oligonucleotides demonstrated that embryos (n ≥ 20) derived from HNF-4α−/− ES cells were devoid of HNF-4α+/+ cells (Fig. 2B). We next determined whether E10.5 HNF-4α−/− ES cell-derived embryos were capable of producing hepatoblasts by identifying cells expressing albumin and HNF-3α mRNAs by in situ hybridization. Albumin expression within fetal tissue is restricted to the developing hepatoblasts, whereas HNF-3α is additionally expressed throughout the gut endoderm as well as in the notochord and floor plate of the neural tube (Cascio and Zaret 1991; Ang et al. 1993; Monaghan et al. 1993; Sasaki and Hogan 1993). Figure 2C–F shows that cells expressing both albumin and HNF-3α could readily be detected in both HNF-4α+/− and HNF-4α−/− ES cell-derived embryos. These results demonstrate conclusively that specification of the ventral foregut endoderm toward a hepatic fate does not require HNF-4α.
HNF-4α−/− hepatoblasts have a normal morphological appearance
Following specification of the hepatic primordium from the ventral foregut endoderm the hepatoblasts migrate from the ventral endoderm into the septum transversum mesenchyme. As they do so they lose their epithelial nature and adopt an irregular stellate morphology (Medlock and Haar 1983). At this stage the hepatoblasts continue to proliferate and the liver expands greatly in size due to increased hematopoiesis. At this time as many as 80% of cells within the developing liver are nucleated blood cells (Medlock and Haar 1983). Between E10.5 and E12.5 in the mouse, the primitive hepatoblasts begin to differentiate toward a hepatocyte phenotype and initiate the expression of a large array of genes associated with mature hepatocyte function. To determine whether HNF-4α was required for hepatocyte differentiation we therefore attempted to produce embryos that survived to 12 days postgestation from HNF-4α−/− ES cells by tetraploid aggregation.
Figure 3 shows that E12.0 embryos could be generated from both HNF-4α+/− and HNF-4α−/− ES cells. A comparison of wild-type embryos with all embryos produced from ES cells by tetraploid aggregation revealed that embryos generated from ES cells were generally slightly smaller (<10%). However, all ES cell-derived embryos examined had beating hearts and morphological features appropriate for this stage in development including limb buds, gut, neural structures, embryonic vasculature, and circulation (Kaufman and Bard 1999). As described previously, all of our ES cell-derived embryos, including those from wild-type J1 ES cells, were exencephalic to varying degrees. However, the presence of this defect in HNF-4α+/+ and HNF-4α+/− ES cell-derived embryos indicates that it is not related to an HNF-4α deficiency (Duncan et al. 1997).
Figure 3.
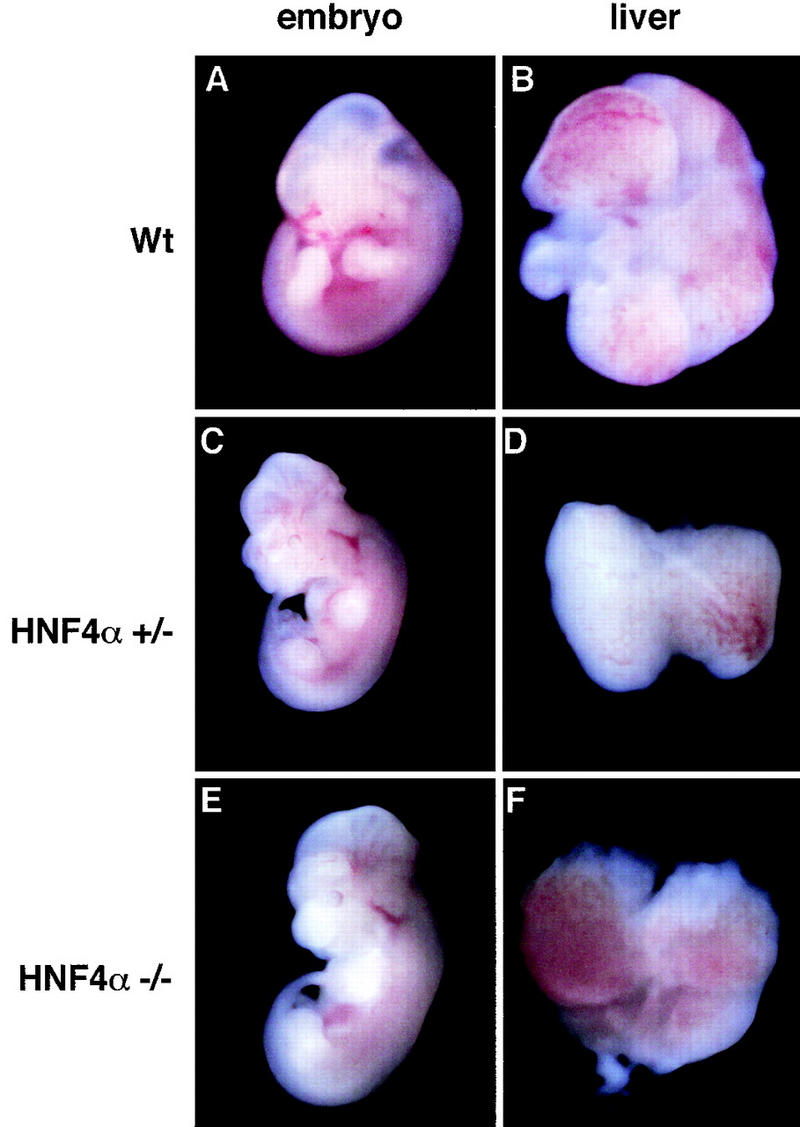
Whole mount micrographs showing wild-type CD-1, HNF-4α+/−, and HNF-4α−/− ES cell derived fetuses and livers after 12 days gestation. Embryos were removed and photographed under a dissecting microscope at 15× magnification. Livers were dissected and photographed at 70× magnification.
Livers were removed from these embryos and examined for morphological and histological characteristics of hepatogenesis. Figure 3 shows that all livers, including those lacking HNF-4α, were similar in size, had initiated hematopoiesis, and had formed distinct lobules that are indicative of normal morphogenesis. In addition, Figure 4A,B shows that sections through HNF-4α+/− and HNF-4α−/− ES cell-derived livers were indistinguishable and contained hepatoblasts, endothelial cells surrounding sinusoids, and numerous hematopoietic cells. Also, electron micrographs of HNF-4α+/− and HNF-4α−/− livers presented in Figure 4, C and D, show that both livers contained light and dark hepatoblasts typical of E12.0 livers (Medlock and Haar 1983). It has been suggested that dark hepatoblasts are more differentiated than light hepatoblasts. Consistent with this model the presence of rough endoplasmic reticulum and lipid vesicles could readily be detected in dark hepatoblasts of both genotypes. Both HNF-4α+/− and HNF-4α−/− hepatoblasts were also found to have a stellate shape. This cellular morphology is characteristic of midgestation stage hepatoblasts that do not exhibit the polarity associated with the mature cuboidal parenchyma (Medlock and Haar 1983). In sum, HNF-4α−/− fetal livers were morphologically identical to wild-type and HNF-4α+/− livers and included the presence of hepatoblasts.
Figure 4.
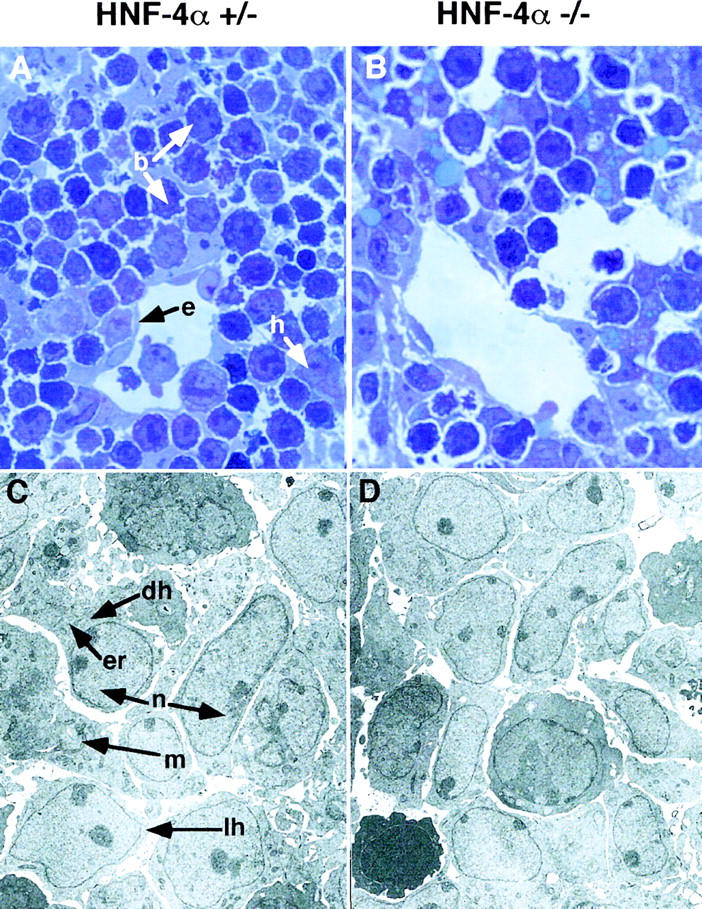
Light micrographs and electron micrographs comparing HNF-4α+/− and HNF-4α−/− fetal livers. (A,B) Sections (1 μm) through livers from HNF-4α−/− or HNF-4α+/− ES cell-derived mice at E12. Sections were stained with toluidine blue and photographed using a 100× objective. Both HNF-4α+/− and HNF-4α−/− livers contained hepatoblasts (h), nucleated fetal blood cells (b), and endothelial cells (e) surrounding sinusoidal spaces. (C,D) Electron micrographs of the same fetal livers at 3000× magnification. Both dark (dh) and light (lh) hepatoblasts can be identified in HNF-4α+/− and HNF-4α−/− livers. The light hepatoblasts are believed to be in a less differentiated state containing a high nuclear (n) to cytoplasmic ratio and few organelles. In contrast, the more differentiated dark hepatoblasts have a relatively small nucleus and contain many organelles including endoplasmic reticulum (er) and mitochondria (m).
HNF-4α is essential for hepatocyte differentiation
The liver has numerous functions including detoxification of metabolic waste products and synthesis of serum proteins, plasma lipoproteins, and clotting factors. It is also a center of several metabolic activities, including glycogen synthesis and gluconeogenesis. Although the hepatoblasts do not function as a parenchyma during midgestational stages of embryonic development, they do begin to express several gene products associated with mature liver function. Expression of these genes can therefore be used to measure the differentiated state of the maturing hepatocyte. To determine whether HNF-4α was necessary for differentiation of hepatoblast to hepatocyte we produced E11.5 fetal livers from HNF-4α+/+, HNF-4α+/−, or HNF-4α−/− ES cells by tetraploid aggregation and compared steady-state mRNA levels of several characteristic hepatocyte marker genes in these livers by RT–PCR (Fig. 5). RT–PCR using HPRT-specific primers detected equal amounts of product from all samples, demonstrating that a similar amount of starting material was used in each reaction. Also, the amplification of this product was dependent on the presence of reverse transcriptase, confirming the absence of genomic DNA in the starting material (not shown). We next compared mRNA levels for >60 genes that are known to define mature hepatocyte function. These included transcription factors, metabolic proteins, enzymes, and secreted serum factors. Figure 5 shows that loss of HNF-4α function had an extreme and dramatic effect on hepatocyte gene expression. In all HNF-4α−/− fetal livers examined, the expression of a large array of genes associated with mature hepatocyte function was virtually abolished. Steady-state mRNA levels of apoA1, apoAII, apoB, apoCIII, apoCII, as well as aldolase B, phenylalanine hydroxylase (pAH), L-type fatty acid-binding protein (LFABP), transferrin (TFN), retinol-binding protein (RBP), and erythropoietin (Epo) could readily be measured in HNF-4α+/+ or HNF-4α+/− ES cell-derived livers by RT–PCR. However, in the absence of HNF-4α the expression of these same genes was almost undetectable. These data show that HNF-4α is essential for the expression of genes that define a fully differentiated hepatocyte phenotype. Based on these results we propose that HNF-4α acts as a hepatocyte differentiation factor and is crucial for normal development of the liver.
Figure 5.
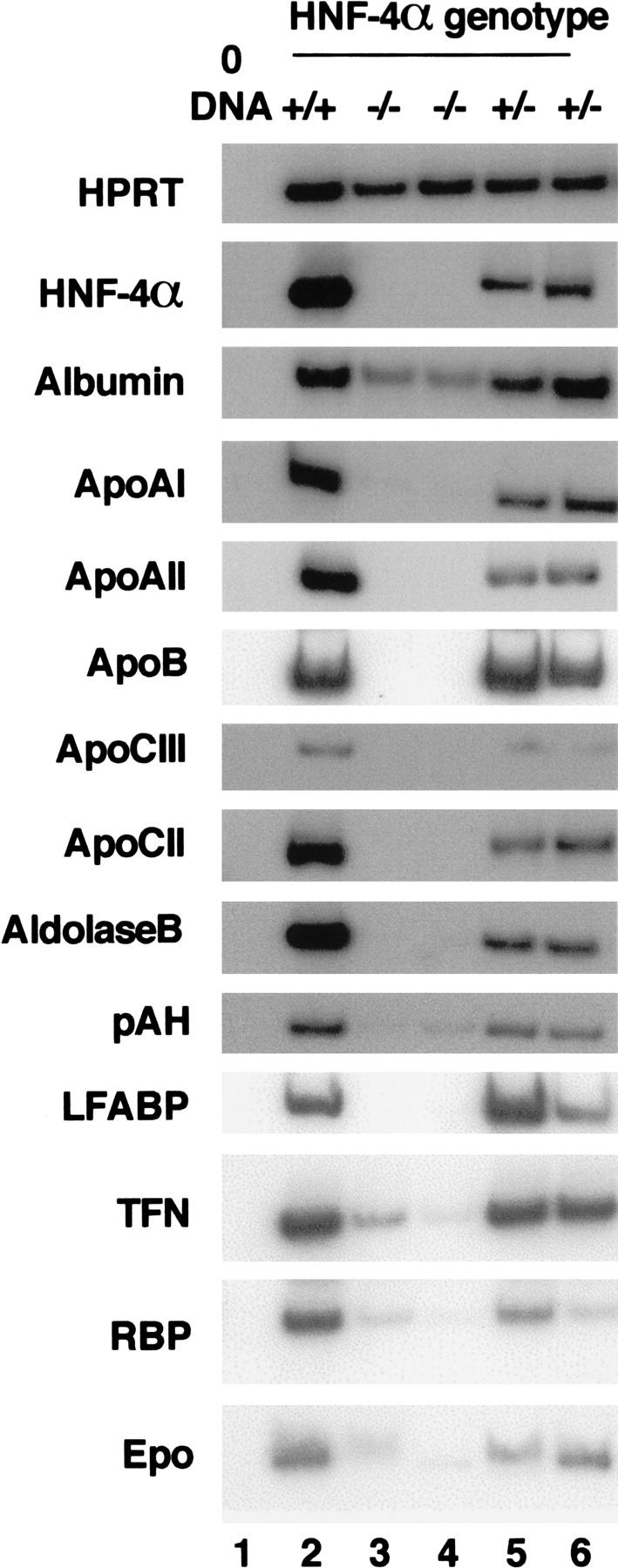
HNF-4α is essential for normal hepatocyte differentiation. Steady-state mRNA levels from several genes normally expressed in differentiated hepatocytes was measured in livers isolated from HNF-4α+/+ (lane 2), HNF-4α+/− (lanes 5,6), and HNF-4α−/− (lanes 3,4) ES-cell-derived embryos at day 12 of gestation by RT–PCR. All samples contained comparable amounts of cDNA, as HPRT primers amplified a similar level of product in all cases. HPRT primers did not amplify product in the absence of cDNA (lane 1). apoAI, apoAII, apoB, apoCIII, apoCII, and albumin, aldolase B, pAH, LFABP, TFN, RBP, and Epo mRNA levels were similar in HNF-4+/+ and HNF-4+/− livers. In HNF-4α−/− livers the relative level of the same mRNAs was decreased dramatically and in some cases undetectable.
HNF-4α acts upstream in a transcription factor cascade that drives hepatocyte differentiation
The regulatory regions of genes expressed in the liver usually bind several different liver-enriched transcription factors. On the basis of such observations, it has been proposed that hepatocyte gene expression is controlled by multiple transcription factors acting coordinately. Given that the expression of such a large array of genes was disrupted by loss of HNF-4α function, we wished to determine whether other liver transcription factors were still expressed in HNF-4α−/− livers. Steady-state mRNA levels of 15 transcription factors expressed in the fetal liver during midgestation stages of development were compared among HNF-4α+/+, HNF-4α+/−, and HNF-4α−/− E11.5 fetal livers generated by tetraploid aggregation. Figure 6 shows that all of these transcription factor mRNAs could be detected in HNF-4α+/+ and HNF-4α+/− livers by RT–PCR. HNF-3α, -β, and -γ, HNF-1β, GATA-4, HNF-6, c/EBP-β, c/EBP-γ, retinoic acid receptor-α (RAR-α), -β, and -γ, retinoid X-receptor-α (RXR-α), and COUP-TFII mRNAs could also be detected at similar levels in HNF-4α−/− fetal livers. However, in contrast to this, HNF-1α mRNA levels were significantly reduced and pregnane-x-receptor (PXR) mRNA was virtually undetectable. From these data we conclude that although the majority of liver-enriched transcription factors are still expressed in HNF-4α−/− fetal livers, HNF-4α acts upstream of at least two transcription factors with important roles in hepatocyte gene expression.
Figure 6.
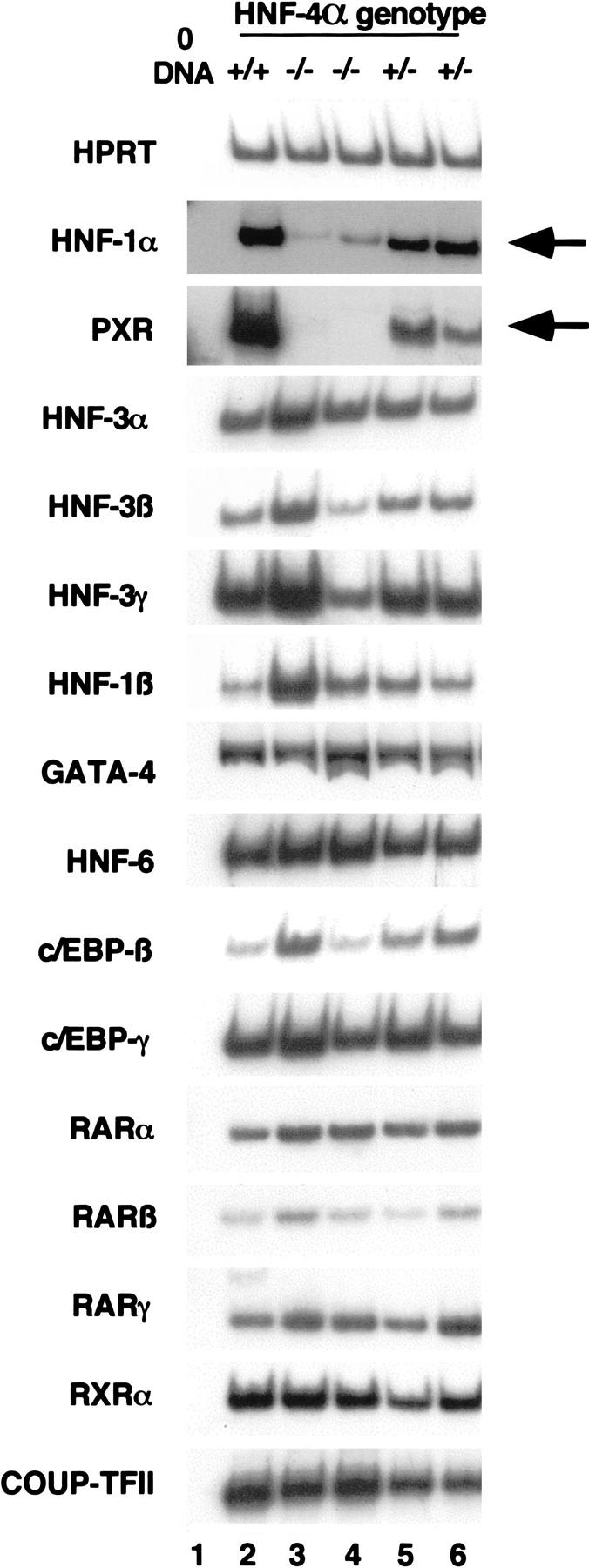
HNF-1α and PXR lie downstream of HNF-4α during hepatogenesis. Steady-state mRNA levels of several transcription factors acting in the fetal liver were measured in E12 HNF-4α+/+ (lane 2), HNF-4α+/− (lanes 5,6), and HNF-4α−/− (lanes 3,4) livers by RT–PCR. Primers recognizing HNF-3α, -3β, 3γ, HNF-1β, GATA-4, HNF-6, c/EBP-β and -γ, RAR-α, -β and -γ, RXR-α, and COUP-TFII all amplified similar levels of product in all fetal livers. However, HNF-1α and PXR steady-state mRNA levels (arrowed) were greatly decreased in HNF-4α−/− fetal livers.
Discussion
HNF-4α is a hepatocyte differentiation factor
Figure 7 shows a simplified model of early hepatic development that is considered in three stages: competency, specification, and differentiation. In this model transcription factors have defined roles in controlling these distinct steps of hepatic development. The first stage is described as competency, whereby the entire foregut endoderm has the potential to adopt a hepatic fate if given the correct developmental cues. Although the liver derives from a distinct portion of ventral endoderm called the hepatic field, Gualdi et al. (1996) have shown that dorsal regions of endoderm are also competent to enter a hepatocyte differentiation program when cultured as explants. It has been proposed that the winged-helix transcription factor HNF-3 acts as a competency factor within the endoderm by marking “liver” genes with the potential to be expressed in response to specific developmental cues (Gualdi et al. 1996; Cirillo et al. 1998; Shim et al. 1998). This may occur through chromatin remodeling, as HNF-3 has an inherent ability to reposition nucleosomes (Cirillo et al. 1998; Shim et al. 1998). In the second stage of liver development, FGF released from cardiac tissue induces the adjacent pluripotent endoderm to commit to a hepatic fate (Jung et al. 1999). The action of FGF is likely to result in the expression of specific transcription factors, called specification factors, which initiate a program of hepatic gene expression by binding to competent promoters. Because HNF-4α is first expressed in the hepatic field during this stage of development we considered the possibility that HNF-4α could act as a specification factor. According to this model, loss of HNF-4α function should lead to an ablation of the hepatic lineage, as is the case for PDX-1 during pancreatic development. However, analyses of HNF-4α−/− ES cell-derived embryos revealed that early stages of liver development were unaffected and morphologically normal HNF-4α−/− hepatoblasts could be detected. These data imply that HNF-4α acts downstream of the specification event or that other factors can compensate for loss of HNF-4α function during this period in liver development.
Figure 7.
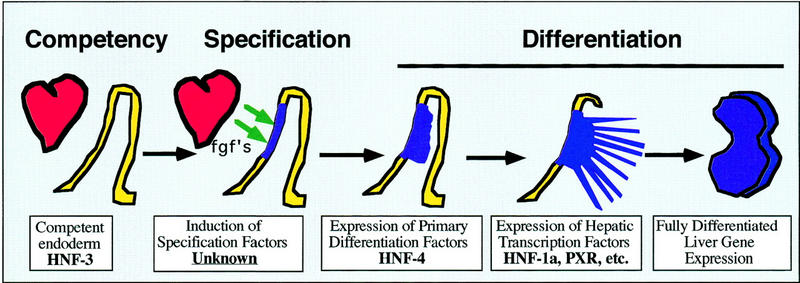
Schematic depicting key points during hepatic development and the stage at which specific transcription factors first act. During gastrulation, at ∼7.0 days of gestation in the mouse, the definitive endoderm (yellow) is generated from the primitive ectoderm. Both ventral and dorsal aspects of the definitive embryonic endoderm express HNF-3 and are competent to adopt a hepatic fate. By the four- to six-somite stage a ventral portion of the embryonic endoderm (blue) lies close to the developing heart. During hepatic specification, FGFs (fgf's) from the developing cardiac mesoderm (red) induce the underlying endoderm to commit to a hepatic fate. In response to this signal the endodermal cells proliferate, expand into the surrounding septum transversum, and adopt a stellate morphology. At this point these cells are considered hepatoblasts. The hepatoblasts continue to proliferate and begin to differentiate to become mature hepatocyte in a process that requires HNF-4α.
The final stage illustrated in Figure 7 describes the differentiation of a pluripotent endodermal cell to a fully functional hepatocyte. This process of differentiation is a transition accompanied by large changes in gene expression. Such a model predicts that hepatic differentiation is controlled by a gradual accumulation of multiple transcription factors over time. Moreover, it would imply the existence of a developmentally regulated transcription factor hierarchy with one factor regulating the expression of another. Until now, however, no transcription factor has been shown to be essential for hepatocyte differentiation in mammalian livers. In this report we have shown that loss of HNF-4α results in the formation of poorly differentiated hepatic cells due to a failure of these cells to express a large array of genes that define mature hepatocyte function. Moreover, our analyses of HNF-4α−/− livers show that two transcription factors, HNF-1α and PXR, require HNF-4α for their expression. This provides the first direct evidence for the existence of a transcription factor cascade acting during mammalian hepatogenesis and supports previous work showing that HNF-4α can activate expression from the HNF-1α promoter in transfected hepatoma cells (Tian and Schibler 1991; Kuo et al. 1992). It is worth noting that not all liver gene expression was down-regulated in the absence of HNF-4α. Genes whose expression was found to be unaffected by loss of HNF-4α function are shown in Table 1.
Table 1.
Genes whose expression was unaffected by loss of HNF-4α
| α1-antitrypsin | glucokinase |
| α-fetoprotein | glucose transporter (glut)-2 |
| apoAIV | indian hedgehog (iHH) |
| apoE | hepatocyte growth factor-like (HGFL) protein |
| bone morphogenic protein (BMP)2 | insulin growth factor (IGF)-1 |
| BMP4 | IGF-2 |
| cellular retinol protein (CRBP)1 | IGF-I receptor |
| CRBPII | insulin receptor |
| FGF-1 | leptin |
| FGF-7 | l-type pyruvate kinase |
| fructose2-6-biphosphate | signal transducer and activator of transcription (STAT)-3 |
| fetoprotein transcription factor (FTF) | TTR |
Although the expression of hepatocyte genes was grossly affected by loss of HNF-4α, liver morphology appeared normal. This seemed surprising given the significant dysregulation of hepatic gene expression seen in HNF-4α−/− livers. At E12, the developing hepatic cells appear morphologically undifferentiated, and have just begun their transition toward a polarized hepatic epithelium. Complete formation of the liver parenchyma does not occur until around birth and the hepatocytes continue to mature even in the neonate. This would suggest that differences in the morphology of hepatocytes lacking HNF-4α would not be observed until late developmental stages. In our experience, all embryos derived from J1 ES cells by tetraploid aggregation develop a severe exencephaly that results in embryonic lethality by E12.5. This defect, which is not associated with loss of HNF-4α function, has prevented us from generating HNF-4α−/− livers at later stages of development and directly investigating the role of HNF-4α in morphological differentiation (Duncan et al. 1997). However, Spath and Weiss have shown that expression of exogenous HNF-4α in stellate dedifferentiated hepatoma cells can reestablish an epithelial morphology (Spath and Weiss 1998). This strongly supports the proposal that in addition to regulating genes essential for mature hepatocyte function, HNF-4α has an integral role in generating a mature hepatocyte morphology.
In sum, our data show that HNF-4α acts as a hepatocyte differentiation factor during mammalian liver development. Although deletion of several factors, including c-Jun, RelA, N-myc, and hepatocyte growth factor, have resulted in increased hepatic apoptosis and cell death during liver development, this study is the first demonstration of a transcription factor required for hepatocyte differentiation (Hilberg et al. 1993; Johnson et al. 1993; Beg et al. 1995; Schmidt et al. 1995; Uehara et al. 1995; Giroux and Charron 1998; Li et al. 1999).
Regulation of gene expression by HNF-4α
Individual promoters and enhancers usually bind multiple different transcription factors that act coordinately to regulate gene expression. This, in turn, would suggest that loss of a single transcription factor could be compensated for by the presence of others. Support for such a model comes from analyses of mice harboring targeted mutations in genes encoding the transcription factors HNF-1α, HNF-3α, and HNF-3γ. These knockout mice exhibit only a moderate dysregulation of liver gene expression (Pontoglio et al. 1996; Kaestner et al. 1998, 1999; Shih et al. 1999). We have shown above that although HNF-1α and PXR require HNF-4α for expression, mRNAs for the majority of liver transcription factors are detected at normal levels. Why then is the expression of so many genes whose promoters can bind multiple transcription factors so drastically repressed in HNF-4α−/− fetal livers? The answer to this may lie in the mode of action of HNF-4α. HNF-4α is a nuclear hormone receptor, and many nuclear hormone receptors interact with transcriptional coactivators containing histone acetylase (HAT) activity (for review, see Torchia et al. 1998). It has been proposed that recruitment of coactivators results in transcriptional activation through chromatin remodeling. In essence, this could be achieved, at least in part, by increasing access of transcription factors to regulatory elements through the relaxation of the chromatin structure around enhancers and promoters. HNF-4α has been shown to interact with several coactivators including SRC-1, GRIP-1, and CBP/p300 (Yoshida et al. 1997; Wang et al. 1998; Dell and Hadzopoulou 1999). Moreover, the presence of CBP/p300 has been shown to enhance HNF-4α-dependent transactivation of the apoCIII promoter in cell-culture transfection assays (Dell and Hadzopoulou 1999). Therefore, we predict that the primary function of HNF-4α is to recruit coactivators to mediate chromatin remodeling, thereby facilitating transcriptional initiation. In this case, the presence of other transcription factors might well be unable to compensate for loss of HNF-4α.
HNF-4 is likely to have a crucial role in mature liver function and regulation of metabolism
Our finding that HNF-4α is required for expression of such a large array of genes in the fetal liver suggests that loss of HNF-4α activity in the adult liver would have severe physiological consequences. Support for this hypothesis comes from genetic studies in humans that have implicated HNF-4α in regulation of serum glucose levels. Yamagata et al. (1996) have shown that a HNF-4α haploinsufficiency results in maturity onset diabetes of the young (MODY). Although it is likely that dysregulation of glucose homeostasis in MODY1 patients is due, in a large part, to reduced insulin secretion by pancreatic islets it is also possible that loss of HNF-4α function in the liver contributes toward this disease. Analysis of MODY1 patients has shown that in addition to defects in insulin secretion, triglyceride metabolism is also significantly affected (Lehto et al. 1999). In this regard, the demonstration that HNF-4α is required for expression of several genes encoding secreted serum factors might have clinical implications. It is possible that decreased expression of serum factors might precede a clinical manifestation of MODY. Screening presymptomatic patients for reduced expression of these factors may therefore help in early diagnosis of MODY and possibly other forms of type II diabetes.
The fact that HNF-4α is so crucial for expression of several apolipoprotein genes implies that it has an important role in regulating serum cholesterol levels. A requirement for HNF-4α in apolipoprotein expression was predicted from analyses of apolipoprotein promoters. All of the apolipoprotein genes that depend on HNF-4α for expression in the fetal mouse liver have been shown to have promoters that interact with HNF-4α in tissue culture assays (Cardot et al. 1993; Metzger et al. 1993; Harnish et al. 1994, 1996; Kardassis et al. 1997, 1998; Hadzopoulou-Cladaras et al. 1998; Vorgia et al. 1998). We have also shown that HNF-4α is required for expression of the nuclear hormone receptor PXR. PXR is activated by pregnanes and regulates expression of the cytochrome P-450 gene CYP3A1 (Kliewer et al. 1998). This finding suggests that HNF-4α could indirectly influence cytochrome P-450 action, in processes such as hydroxylation of steroid hormones and xenobiotics, by controlling PXR expression. The exact role of HNF-4α in the adult liver will best be determined by the generation of mice harboring a hepatocyte-specific deletion of the HNF-4α gene. On the basis of our analyses of HNF-4α−/− fetal livers we believe it is likely that loss of HNF-4α activity in the adult liver will have pleiotropic consequences that grossly affect liver function and metabolism.
Materials and methods
Generation of ES cell-derived embryos and chimeras
Tetraploid embryos were produced from ROSA26 or CD-1 mice following the procedure described by Nagy and Rossant (1993). HNF-4α−/− ES cells were produced by selection of HNF-4α+/− ES cells in high concentrations of G418 as described previously (Duncan et al. 1997). Embryos were generated from ES cells by aggregating them with four-cell stage tetraploid embryos, allowing them to develop to blastocysts before transferring them to the uterus of a surrogate mother. Similarly, chimeric embryos were produced by aggregating ES cells with diploid CD-1 embryos. Embryos were allowed to develop in utero before harvesting for analysis. The exact methodology used in this procedure has been described in detail elsewhere (Nagy and Rossant 1993; Duncan et al. 1997).
In situ hybridization
Paraffin sections of 5 μm were processed and hybridized to 33P-labeled antisense RNA probes generated from albumin and HNF-3α partial cDNAs as described previously (Duncan et al. 1994; Rausa et al. 1998).
Electron micrscopy and histology
Fetal livers were dissected from embryos and fixed by overnight submersion in 2% gluteraldehyde in 0.1 m phosphate buffer. Sections were dehydrated and embedded in epon resin using standard procedures. Sections of 0.5 μm were stained with toluidine blue and observed under oil using a 100× objective. Sections for electron microscopy were 60–80 nm thick and contrasted with uranyl acetate and lead citrate.
RT–PCR
Conditions used for RT–PCR were those of Wilson and Melton (1994), with modifications described elsewhere (Duncan et al. 1997). Number of cycles and annealing temperature were optimized for each primer pair. The following forward and reverse primers were used for specific amplification: hprt; agcgcaagttgaatctgc, agcgacaatctaccagag, HNF-4α; cttccttcttcatgccag, acacgtccccatctgaag, albumin; cttaaaccgatgggcgatctcact, ccccactagcctctggcaaaat, apoAI; acacacgtagactctctg, ctgggctttggtcttaag, apoAII; gccatcatgaagctgctcgc, ccagactagttcctgctgac, apoB; cttcagggaacaaagcag, tcaagggtgagctgattg, apoCIII; ctcaatagctggagttgg, ctgaagaggtagagggat, apoCII; ttactggacctctgccaagg, cctaacttggggcttgcctg, aldolase b; tctccgtcaggaagcacctc, atggcctctctgaacgctgt, phenylalanine hydroxylase; tcgctatgacccctacactc, ggttgacctcctaagttctg, transferrin; tggcacaggaacactttg, tcctgctgattccgaatg, retinol-binding protein; atccagtggtcatcgtttcctcgct, gaacttcgacaaggctcgtttctctgg, hnf-1α; ttctaagctgagccagctgcagacg, gctgaggttctccggctctttcaga, pxr; gcgcggagaagacggcagcatc, cccaggttcccgtttccgtgtcg, hnf-3α; atgccagccacagcaccgggactc, ggcctactcctctgtccctgtcag, hnf-3β; actggagcagctactacg, cccacataggatgacatg, hnf-3γ; tcaagctggaggagaaggcaaaga, ctcgggctcagatggcgtagg, hnf-1β; gaaagcaacgggagatcctccgac, cctccactaaggcctccctctcttcc, gata-4; ctaagctgtccccacaaggctatgca, cagagctccacctggaaaggtgtttg, hnf-6; aactcccagcaaggacttccccact, ccatctgccctgaattacttccattgc, c/ebp-β; cgcccgcgcaccacgact, gaggcggcggcgggaagc, c/ebp-γ; gtcgcagccagccactactccag, caccgccatattgttccgctctc, rar-α; gccccatgccccgaggaaga, agagcgccgggtggggagag, rar-β; tgcgccgcggccacaagac, cagccagggcgggggttcc, rar-γ; gaagcggctgcctggttttaca, tcgtcgccgggcatagagc, rxr-α; cctgccgctcgacttctctaccc, taccagtcccgaagcccaatgtg, COUP-TFII; cccgcccagcacgccggccca, tacagcttcccgtctcatgccc, erythropoetin; aaggaggcagaaaatgtcacgatg, gtgagtgttcggagtggagcaggt, neo; gccaacgctatgtcctgatagcggt, agccggtcttgtcgatcaggatgat.
Acknowledgments
We thank Stephen Dorris for expert technical assistance. Drs. Sukov and Vassuer provided help with erythropoeitin and COUP-TFII primers, respectively and Dr. Paula Traktman critically read the manuscript. We also thank Drs. James E. Darnell, Jr., and Markus Stoffel for support and advice, and Dr. Andras Nagy for his help and guidance with the tetraploid aggregation procedure. Funding for this work was provided to S.A.D. by the March of Dimes and American Diabetes Association. S.A.D. was a recipient of a Sinsheimer Scholar Award and was a Naomi Judd American Liver Scholar. We particularly thank Ms. Judd for her continued support of research into liver disease.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL duncans@mcw.edu; WEB http://www.mcw.edu/cellbio/bios/duncan.html; FAX (414) 456-6517.
References
- Ang S-L, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF-3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- Cardot P, Chambaz J, Kardassis D, Cladaras C, Zannis VI. Factors participating in the liver-specific expression of the human apolipoprotein A-II gene and their significance for transcription. Biochemistry. 1993;32:9080–9093. doi: 10.1021/bi00086a013. [DOI] [PubMed] [Google Scholar]
- Cascio S, Zaret KS. Hepatocyte differentiation initiates during endodermal-mesenchymal interactions prior to liver formation. Development. 1991;113:217–225. doi: 10.1242/dev.113.1.217. [DOI] [PubMed] [Google Scholar]
- Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- Cereghini S, Ott M-O, Power S, Maury M. Expression patterns of vHNF1 and HNF1 homeoproteins in early postimplantation embryos suggest distinct and sequential developmental roles. Development. 1992;116:783–797. doi: 10.1242/dev.116.3.783. [DOI] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes & Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell H, Hadzopoulou CM. CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J Biol Chem. 1999;274:9013–9021. doi: 10.1074/jbc.274.13.9013. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE., Jr Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: Tetraploid rescue of HNF-4−/− embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- Fukuda-Taira S. Hepatic induction in the avian embryo: specificity of reactive endoderm and inductive mesoderm. J Embryol Exp Morphol. 1981;63:111–125. [PubMed] [Google Scholar]
- Giroux S, Charron J. Defective development of the embryonic liver in N-myc-deficient mice. Dev Biol. 1998;195:16–28. doi: 10.1006/dbio.1997.8823. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Yamada H, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes & Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Hadzopoulou-Cladaras M, Lavrentiadou SN, Zannis VI, Kardassis D. Transactivation of the ApoCIII promoter by ATF-2 and repression by members of the Jun family. Biochemistry. 1998;37:14078–14087. doi: 10.1021/bi9804176. [DOI] [PubMed] [Google Scholar]
- Harnish DC, Malik S, Karathanasis SK. Activation of apolipoprotein AI gene transcription by the liver-enriched factor HNF-3. J Biol Chem. 1994;269:28220–28226. [PubMed] [Google Scholar]
- Harnish DC, Malik S, Kilbourne E, Costa R, Karathanasis SK. Control of apolipoprotein AI gene expression through synergistic interactions between hepatocyte nuclear factors 3 and 4. J Biol Chem. 1996;271:13621–13628. doi: 10.1074/jbc.271.23.13621. [DOI] [PubMed] [Google Scholar]
- Hertz R, Magenheim J, Berman I, Bar TJ. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- Hilberg F, Aguzzi A, Howells N, Wagner EF. c-Jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- Houssaint E. Differentiation of the mouse hepatic primordiam. I. An analysis of tissue interactions in hepatocyte differentiation. Cell Differ. 1980;9:269–279. doi: 10.1016/0045-6039(80)90026-3. [DOI] [PubMed] [Google Scholar]
- Johnson RS, van LB, Papaioannou VE, Spiegelman BM. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes & Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Hiemisch H, Schutz G. Targeted disruption of the gene encoding hepatocyte nuclear factor 3γ results in reduced transcription of hepatocyte-specific genes. Mol Cell Biol. 1998;18:4245–4251. doi: 10.1128/mcb.18.7.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G. Inactivation of the winged helix transcription factor HNF3α affects glucose homeostasis and islet glucagon gene expression in vivo. Genes & Dev. 1999;13:2495–2504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardassis D, Tzameli I, Hadzopoulou-Cladaras M, Talianidis I, Zannis V. Distal apolipoprotein C-III regulatory elements F to J act as a general modular enhancer for proximal promoters that contain hormone response elements. Synergism between hepatic nuclear factor-4 molecules bound to the proximal promoter and distal enhancer sites. Arterioscler Thromb Vasc Biol. 1997;17:222–232. doi: 10.1161/01.atv.17.1.222. [DOI] [PubMed] [Google Scholar]
- Kardassis D, Sacharidou E, Zannis VI. Transactivation of the human apolipoprotein CII promoter by orphan and ligand-dependent nuclear receptors. The regulatory element CIIC is a thyroid hormone response element. J Biol Chem. 1998;273:17810–17816. doi: 10.1074/jbc.273.28.17810. [DOI] [PubMed] [Google Scholar]
- Kaufman MH, Bard JBL. The anatomical basis of mouse development. London, UK: Academic Press; 1999. [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE, Jr, Crabtree GR. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:458–460. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- LeDouarin N. Etude experimentale de l'organogenese du tube digestif et du foie chez l'embryon de poulet. Bull Biol Fr Belg. 1964;98:543–576. [Google Scholar]
- ————— Synthese du glycogene dans les hepatocytes en voie de differentiation: Role des mesenchymes homologue et heterologues. Dev Biol. 1968;17:101–114. doi: 10.1016/0012-1606(68)90092-4. [DOI] [PubMed] [Google Scholar]
- ————— An experimental analysis of liver development. Med Biol. 1975;53:427–455. [PubMed] [Google Scholar]
- Lehto M, Bitzen PO, Isomaa B, Wipemo C, Wessman Y, Forsblom C, Tuomi T, Taskinen MR, Groop L. Mutation in the HNF-4alpha gene affects insulin secretion and triglyceride metabolism. Diabetes. 1999;48:423–425. doi: 10.2337/diabetes.48.2.423. [DOI] [PubMed] [Google Scholar]
- Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- Medlock ES, Haar JL. The liver hemopoietic environment: I. Developing hepatocytes and their role in fetal hemopoiesis. Anat Rec. 1983;207:31–41. doi: 10.1002/ar.1092070105. [DOI] [PubMed] [Google Scholar]
- Metzger S, Halaas JL, Breslow JL, Sladek FM. Orphan receptor HNF-4 and bZip protein C/EBP alpha bind to overlapping regions of the apolipoprotein B gene promoter and synergistically activate transcription. J Biol Chem. 1993;268:16831–16838. [PubMed] [Google Scholar]
- Monaghan AP, Kaestner KH, Grau E, Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3α, β, and γ genes in determination of the definitive endoderm, chordamesoderm, and neurectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J. Production of completely ES cell-derived fetuses. In: Joyner A, editor. Gene targeting: A practical approach. Oxford, UK: Oxford University Press; 1993. pp. 147–179. [Google Scholar]
- Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early passage embryonic stem cells. Proc Natl Acad Sci. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Pontoglio M, Barra J, Hadchouei M, Doyen A, Kress C, Bach JP, Babinet C, Yaniv M. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- Rausa FM, Ye H, Lim L, Duncan SA, Costa RH. In situ hybridization with 33P-labeled RNA probes for determination of cellular expression patterns of liver transcription factors in mouse embryos. Methods. 1998;16:29–41. doi: 10.1006/meth.1998.0642. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BLM. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Navas MA, Kuwajima S, Duncan SA, Stoffel M. Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3alpha-deficient mice. Proc Natl Acad Sci. 1999;96:10152–10157. doi: 10.1073/pnas.96.18.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Woodcock C, Zaret KS. Nucleosome positioning by the winged helix transcription factor HNF3. Genes & Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek FM, Zhong W, Lai E, Darnell JE., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes & Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- Spath GF, Weiss MC. Hepatocyte nuclear factor 4 provokes expression of epithelial marker genes, acting as a morphogen in dedifferentiated hepatoma cells. J Cell Biol. 1998;140:935–946. doi: 10.1083/jcb.140.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraviras S, Monaghan AP, Schutz G, Kelsey G. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech Dev. 1994;48:67–79. doi: 10.1016/0925-4773(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Tian JM, Schibler U. Tissue-specific expression of the gene encoding hepatocyte nuclear factor 1 may involve hepatocyte nuclear factor 4. Genes & Dev. 1991;5:2225–2234. doi: 10.1101/gad.5.12a.2225. [DOI] [PubMed] [Google Scholar]
- Torchia J, Glass C, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- Vorgia P, Zannis VI, Kardassis D. A short proximal promoter and the distal hepatic control region-1 (HCR- 1) contribute to the liver specificity of the human apolipoprotein C-II gene. Hepatic enhancement by HCR-1 requires two proximal hormone response elements which have different binding specificities for orphan receptors HNF-4, ARP-1, and EAR-2. J Biol Chem. 1998;273:4188–4196. doi: 10.1074/jbc.273.7.4188. [DOI] [PubMed] [Google Scholar]
- Wang JC, Stafford JM, Granner DK. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem. 1998;273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PA, Melton DA. Mesodermal patterning by an inducer gradient depends on secondary cell-cell communication. Curr Biol. 1994;4:676–686. doi: 10.1016/s0960-9822(00)00152-4. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- Yoshida E, Aratani S, Itou H, Miyagishi M, Takiguchi M, Osumu T, Murakami K, Fukamizu A. Functional association between CBP and HNF4 in trans-activation. Biochem Biophys Res Commun. 1997;241:664–669. doi: 10.1006/bbrc.1997.7871. [DOI] [PubMed] [Google Scholar]


