Abstract
We investigated gene regulation at the IL-3/GM-CSF gene cluster. We found BRG1, a SWI/SNF remodeling ATPase, bound a distal element, CNSa. BRG1 binding was strongest in differentiated, stimulated T helper cells, paralleling IL-3 and GM-CSF expression. Depletion of BRG1 reduced IL-3 and GM-CSF transcription. BAF-specific SWI/SNF subunits bound to this locus and regulated IL-3 expression. CNSa was in closed chromatin in fibroblasts, open chromatin in differentiated T helper cells, and moderately open chromatin in naïve (undifferentiated) T helper cells; BRG1 was required for the most open state. CNSa increased transcription of a reporter in an episomal expression system, in a BRG1-dependent manner. The NF-κB subunit RelA/p65 bound CNSa in activated T helper cells. Inhibition of NF-κB blocked BRG1 binding to CNSa, chromatin opening at CNSa, and activation of IL-3 and GM-CSF. Together, these findings suggest CNSa is a distal enhancer that binds BRG1 and NF-κB.
Keywords: Gene Regulation, T cell regulation, Chromatin Remodeling, BRG1, SWI/SNF, Cytokine Transcription, Distal Regulatory Elements
1. Introduction
GM-CSF and IL-3 are two tightly linked and closely related proinflammatory cytokines that have arisen by a gene duplication event. Both cytokines are involved in the activation and survival of multiple myeloid lineages. Clinically, IL-3 and GM-CSF are used to replenish white blood counts in patients after chemotherapy. However, inappropriately elevated expression of IL-3 and GM-CSF is also associated with inflammatory disorders such as arthritis and psoriasis. The major biological source of GM-CSF and IL-3 appears to be activated, differentiated T cells, with little or no specificity with respect to T cell subsets. IL-3 is also produced by mast cells and eosinophils, while GM-CSF is also produced by monocytes and macrophages.
The transcriptional regulation and corresponding changes in the chromatin structure of the IL-3/GM-CSF gene cluster have been extensively studied (Cockerill, 2004). These are separated by only 13 kb in the mouse genome and located within one megabase of the Th2 cytokine cluster (IL-4, IL-13, and IL-5). The expression of IL-3 and GM-CSF in T cells is highly regulated; expression is limited to differentiated T cells, and requires T cell activation. Several enhancers, both upstream and intergenic, have been identified in the IL-3/GM-CSF locus. Transcription factors associated with T cell activation (NFAT and NF-kB) and otherwise (Sp1 and GATA) bind to and regulate the activity of these elements (Cakouros et al., 2001; Duncliffe et al., 1997; Holloway et al., 2003; Johnson et al., 2004). Changes in nucleosome mobility as detected by the generation of DNase I hypersensitive sites (DHS) following T cell activation, is associated with the promoters and enhancers of the IL-3/GM-CSF locus (Holloway et al., 2003; Johnson et al., 2004). Chromatin reorganization extends across a 3kb region around the intergenic GM-CSF enhancer region and is associated with gene activity (Bert et al., 2007). Although the tissue specific expression pattern of IL-3 and GM-CSF largely overlap they are not identical, as evidenced by the exclusive expression of GM-CSF in myeloid cells, indicating these genes can be regulated independently (Bert et al., 2007). The recent identification of an insulator element located between IL-3 and GM-CSF may provide a means to segregate the regulatory elements associated with this gene cluster (Bowers et al., 2009).
Although chromatin structure changes within the IL-3/GM-CSF locus have been well documented during both T cells development and after T cell activation, less is known about the enzymes that catalyze these changes. A recent study demonstrated that in early thymocyte development the IL-3/GM-CSF locus exists in an epigenetically silent state as defined both by nuclease accessibility and histone modifications (Mirabella et al., 2010). As the T cells develop into a mature T cells and T cell blasts the cytokine locus acquires a more active chromatin structure marked by DHS, active histone modifications and the expression of inducible noncoding RNA’s (Mirabella et al., 2010). BRG1, a remodeling enzyme, has been identified as a regulator functioning at the GM-CSF promoter. However in one study BRG1 recruitment to the promoter was reduced following T cell activation, while in another BRG1 was enriched (Brettingham-Moore et al., 2008; Holloway et al., 2003). A role for BRG1 in remodeling events outside of the proximal promoter regions has not been reported, thought distal BRG1 binding has been reported in a T cell line and primary T cells (Precht et al., 2010). ISWI, another type of remodeling enzyme, has also been found to regulate gene expression in T cells (Landry et al., 2011; Precht et al., 2010). In the T cell line EL4, ISWI activated expression of IL-3, while repressing expression of IL-2, IL-5, IL-13, and IL-17A (Precht et al., 2010). At these loci, remodeling enzyme binding is found at promoters and at distal regions (De et al., 2011; Precht et al., 2010; Wurster and Pazin, 2008).
Changes in chromatin structure are catalyzed by chromatin remodeling enzymes. ATP-dependent remodeling enzymes reposition, unfold, displace and assemble nucleosomes, while other classes of remodeling enzymes covalently modify histone proteins or DNA. ATP-dependent remodeling can directly alter gene expression in cell-free systems and in cells (Saha et al., 2006). Remodeling may alter binding of transcription factors and RNA polymerase, transcription factor and RNA polymerase function, and alter higher-order or long-range chromatin structure. Remodeling enzymes are thought to bind DNA non-specifically, and are recruited by interactions with transcription factors, modified histones and non-coding RNA (Biddie and Hager, 2009; Clapier and Cairns, 2009; Hassan et al., 2001; Ho and Crabtree, 2010; Neely et al., 1999; Tarakhovsky, 2010; Wysocka et al., 2006; Yudkovsky et al., 1999). ATP-dependent remodeling enzymes are often multi-protein complexes, classified by their ATPase subunit into subfamilies such as SWI/SNF, ISWI and Mi2 (Saha et al., 2006).
BRG1 is an ATPase in the SWI/SNF subfamily, and is essential for embryonic development (Bultman et al., 2000). In cell-free systems SWI/SNF enzymes can displace, unfold and slide nucleosomes (Lorch et al., 2006; Schnitzler et al., 1998; Whitehouse et al., 1999). Mammalian SWI/SNF has been found in BAF and PBAF complexes containing a small number of distinct subunits and many common subunits (Lemon et al., 2001; Nie et al., 2000; Yan et al., 2005), as well as complexes specific to ES cells and neurons (Ho et al., 2009; Lessard et al., 2007).
BRG1 has been found to play an important role in T cell development (Chi, 2004). BRG1 also plays an important role in macrophages (Ramirez-Carrozzi et al., 2006) and differentiated T helper cells, including T helper 1 (Th1), T helper 2 (Th2) and T helper 17 cells (De et al., 2011; Letimier et al., 2007; Wurster and Pazin, 2008; Zhang and Boothby, 2006). Genome-wide analysis of BRG1 binding during Th differentiation suggested BRG1 activated many genes in each fate, in response to activation-specific and lineage-specific signals (De et al., 2011). Distal regulatory elements are frequently involved in Th gene regulation, and may be sites for remodeling enzyme function (Agarwal and Rao, 1998a; De et al., 2011; Jones and Flavell, 2005; Lee et al., 2003; Placek et al., 2009; Wurster and Pazin, 2008). Distal regulatory elements may play a widespread, if underappreciated, role in gene regulation, and distal chromatin structure may be better correlated with gene activity than promoter chromatin structure (Heintzman et al., 2009; Visel et al., 2009).
Here, we asked whether the SWI/SNF subunit BRG1 is required for IL-3/GM-CSF gene expression and remodeling of the cytokine locus. We found that knockdown of BRG1 expression in primary effector T cells impaired the expression of both cytokines. BRG1-containing BAF complexes bound to multiple known regulatory elements in the IL-3/GM-CSF cytokine cluster, in an inducible manner; little if any binding was found in naïve cells. Comparative sequence analysis revealed the existence of additional conserved noncoding sequence (CNS) regions 25 to 40 kb downstream of the cytokine cluster; one in particular, CNSa, binds BRG1 to an especially high degree. We detected changes in chromatin accessibility at CNSa in when BRG1 expression was reduced, suggesting a dependence on BRG1 for establishing an active chromatin conformation at this site. Activation-induced recruitment of BRG1 to CNSa appears to depend at least in part on NFKb pathway. Finally, CNSa appears to possess BRG1-dependent enhancer activity in a chromatin-based reporter assay. BRG1 appears to be an important regulator of chromatin structure and gene expression in IL-3/GM-CSF locus and was a useful marker in the identification of a novel, distal regulatory element.
2. Materials and methods
2.1 Lymphocyte isolation from mice and culture
Animal approval was from the NIA ACUC, protocol ASP-365-MJP-Mi, and all experiments conform to the relevant regulatory standards. T cells were isolated and cultured essentially as described previously (De et al., 2011; Precht et al., 2010; Wurster and Pazin, 2008). CD4+ T cells were purified from the lymph nodes of 4–6 week old Balb/c mice (Taconic) by CD4 Macs per manufacturer’s instructions (Miltenyi). Naïve Thp (T helper precursor) cells were purified from lymph node and spleens by using CD4+CD62+ T cell isolation kit (Miltenyi) to 95% purity (unpublished). Lymphocytes were cultured in RPMI 1640 supplemented with 10% FCS, 100U/ml penicillin, 100 μg/ml streptomycin, 1mM Sodium Pyruvate, 2 mM L-glutamine, 25mM Hepes, 50 μM β-mercaptoethanol.
2.2 In Vitro T Helper Cell Culture
Murine primary T cells were cultured essentially as described previously (De et al., 2011; Precht et al., 2010; Wurster and Pazin, 2008). Purified CD4+ LN T cells or naïve Thp cells were plated onto anti-CD3 (1 μg/ml), anti-CD28 (2 μg/ml) coated plates at 1–2 × 106/ml in the presence of IL-2. In some experiments cells were also cultured under Th2 skewing conditions (10ng/ml IL-4, 10 μg/ml anti-IFNγ). Cultures were expanded in IL-2 (100U/ml) three days after initial culture.
2.3 Transduction of Th cells
Th cells were transduced essentially as described (Wurster and Pazin, 2008). 2.5–4×106 Th cells were resuspended in 100 μl “Mouse T cell Nucleofector Solution” (Amaxa) and mixed with indicated siRNA oligo (500nM). Electroporation was performed as described by manufacturer’s protocol using the program x-001 and the cells were transferred to prewarmed “Mouse T cell Growth Medium” (Amaxa) supplemented with hIL-2 (100U/ml). Efficiency of transcfection by GFP plasmid ~75% in effector cells (data not shown). siRNA oligos (BRGm sc-29830) and negative control siRNA (sc-37007) used in this study were obtained from Santa Cruz Biotechnology. Similar results were obtained using independent BRG1-specific siRNA oligos obtained from Santa Cruz Biotechnology (sc-44289) and Dharmacon (m-041135-00) ((Wurster and Pazin, 2008) and data not shown). BAF250a siRNA oligos were from Dharmacon (Arid1a SMARTpool M-040694-00) while BAF180 siRNA oligos were from Invitrogen (pool PB1M55227873-5).
2.4 mRNA Quantitation
Total RNA was purified using RNeasy columns (Qiagen). cDNA was made using iScript (BioRad) according to the manufacturer’s instructions. The mRNA levels were determined by real time PCR using SYBR green (Qiagen) on an ABI 7500. Expression levels were normalized to mTBP or m-actin as indicated. Primer sequences for steady state mRNA are in Table 1, while primer sequences for unspliced (nascent) RNA are in Table 3.
Table 1.
Primers for steady-state RNA
| Gene | Primer name | Primer 1 | Primer 2 |
|---|---|---|---|
| IL-3 | MP 771 | GGAAGCTCCCAGAACCTGAAC | TCAGAGAGGGTCCTTCATCATCA |
| GMCSF | MP 773 | GCCATCAAAGAAGCCCTGAA | GCGGGTCTGCACACATGTTA |
| IFNγ | MP 592 | GGATGCATTCATGAGTATTGC | CCTTTTCCGCTTCCTGAGG |
| TBP | MP 935 | CTTCGTGCAAGAAATGCTGAATAT | TGTCCGTGGCTCTCTTATTCTCA |
| actin | MP 598 | AGAGGGAAATCGTGCGTGAC | CAATAGTGATGACCTGGCCGT |
Table 3.
Primers for nascent (unspliced) RNA
| Gene | Primer name | Primer 1 | Primer 2 |
|---|---|---|---|
| GM-CSF | MP 1077 | CCGCATAGGTGGTAACTTGTGTT | TTACCTGGACTCAAGTGCTGTTTATTT |
| IL-3 | MP 1078 | CGAAAGTCATCCAGATCTCGAA | TGTTCACATCTAATGCCTTCTTTTCT |
2.5 Immunoblot Analysis
Whole cell extracts were prepared by lysing cells in 50mM Tris 7.4, 1% NP40, 150mM NaCl, 0.5% Deoxycholate, 0.1% SDS and clearing the lysates by centrifugation. Protein extracts were separated on a 6% polyacrylamide gel and transferred to a PVDF membrane (BioRad). The immunoblots were blocked for 1 hour at room temperature in 5% milk in TBST (50mM Tris pH7.5, 100mM NaCl, 0.03% Tween 20) and incubated with the indicated antibody overnight at 4°C. The blots were washed with TBST and incubated with anti-rabbit HRP-conjugated antibody (Zymed) at room temperature. After washing the blots with TBST, detection was carried out using enhanced chemiluminescence (Amersham) according to manufacturer’s instructions.
2.6 ChIP Assay
Chromatin immunoprecipitation was performed using methods similar to those described previously (De et al., 2011; Precht et al., 2010; Wurster and Pazin, 2008); details are available on request. Approximately 20 million cells (for 3–5 immunoprecipitations) were crosslinked with 1% formaldehyde and quenched with glycine. Cells were lysed with buffer containing 1% SDS, treated with micrococcal nuclease, sonicated until the average DNA size was approximately 500 bp, and adjusted to 0.1% SDS, 1% Triton X-100 and 150 mM NaCl at 5 ml. Sonicates were precleared with protein A Sepharose (Upstate) and IP was performed with the following antibodies: 1 ug H3K9,14 (Upstate 06-599), 0.5 ul BRG1 (J1), 1 ug Brm (Abcam 15597-100), 2 ug BAF250a/Arid1a (Bethyl A301-041A), 2 ug BAF180/PB1 (Bethyl A301-591A), or rabbit IgG (Santa Cruz sc-2027). Chromatin was collected with protein A, washed, eluted with sodium bicarbonate/SDS and crosslinks were reversed, followed by protease treatment. Chromatin was quantified by real-time PCR (Q-PCR) using an Applied Biosystems 7500 with Sybr Green detection (Qiagen). Graphs indicate immunoprecipitated chromatin amounts relative to input DNA (% input). Primer sequences are in Table 2; identical results were obtained using CNSa primer sets MP849 and MP1310 (data not shown).
Table 2.
Primers for ChIP DNase I and MNase
| Locus | Primer name | Primer 1 | Primer 2 |
|---|---|---|---|
| IL3 CNSb | MP 851 | TGCAGCATCTCTAGCCGTCTT | AGATACCAGAGGCACTCAAGGAA |
| IL3 CNSa | MP 849 | TTTTTGGTGTGCAGCACTAATACA | GTCACCGCTCTGCCTGCTA |
| IL3 CNSc | MP 853 | GGCTTCAGCTCCCCACTTCT | GGTTTTCTAGCTTATATCCCCAGACA |
| GMCSF pro | MP 843 | AAGGCCGGGTGACAGTGAT | TTCCTGGGCATTGTGGTCTAC |
| GMCSF Enh | MP 847 | GTGGAGTGACCCCTCTTTGG | GAAACTCCTTCCAGAGGGTTCTC |
| IL3 pro | MP 841 | CCCGGCCACTGATTGAAG | CCAGCATCCACACCATGCT |
| IL3 Enh | MP 845 | CACACCTGCTTCTTGTCATCATC | TCCCTGCCAGTGGTGGAT |
| Nfm | MP 855 | CCACGGCGCTGAAGGA | CTGGTGCATGTTCTGGTCTGA |
| IL3 CNSa | MP 1310 | GTGGAAAGCGAAGCTGGTTT | GAGCCCAAGGCCCAAAA |
2.7 Nuclease Hypersensitivity Analysis
DNase I hypersensitivity analysis was performed essentially as described (Precht et al., 2010; Wurster and Pazin, 2008), with the following modifications. Nuclei were purified from differentiated or effector Th2 cells as described previously (Agarwal and Rao, 1998b). Briefly, nuclei were released by hypotonic lysis in the presence of 0.5% NP40 and digested with the indicated amounts of DNase I (Worthington) for three minutes at room temperature. The samples were then treated with proteinase K, RNase A and the DNA was recovered after phenol/chloroform extraction and ethanol precipitation. Stimulated effector Th2 cells were enriched for viable cells prior to nuclear isolation by ficoll-hypaque separation. MNase I hypersensitivity analysis was performed essentially as described (Weinmann et al., 1999; Wurster and Pazin, 2008) with the following modifications. Cells were washed twice with 10 mM Tris pH 7.5 containing 10 mM NaCl, 3 mM MgCl2, 0.5 % NP40) resuspended in 10 mM Tris pH 7.5 containing 10 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 4% NP40 and treated with the indicated amounts of micrococcal nuclease (Sigma). Nuclease accessibility was assessed by real-time PCR of DNA samples (McArthur et al., 2001) with the following modifications. Briefly, nuclease-treated DNA was subjected to real-time PCR using primers to indicated regulatory elements in the IL-3/GM-CSF locus. PCR reactions were performed using QuantiTect SYBR green PCR kit (Qiagen) per manufacturer’s instructions on an ABI 7500. The DNase I sensitivity is indicated by % DNA remaining compared to undigested sample and DNA content is normalized to a known DNase I resistant locus (Nfm). Primer sequences are in Table 2.
2.8 Episomal reporter assays
Episomal reporter assays were performed essentially as described (De et al., 2011). A 450bp region including CNSa was amplified using primers MP1616c and MP1617d using Phusion (Finnzymes) from mouse genomic DNA, primer sequences in Table 4. The BamHI fragment was cloned upstream (BglII site) or 2.2kb downstream (BamHI) of the SV40 promoter (200bp promoter region from pGL3 promoter) in the episomal luciferase reporter pREP4-luc (Liu et al., 2001). The reporter constructs were transfected into the human lymphoblastoid T cell line CEM using Amaxa Nucleofection Kit V (C-16) (Lonza). After 72 hours, the cells were either left resting or stimulated with PMA/Ionomycin for 5 hours. Protein extracts were harvested and assayed for luciferase activity using Dual-Luciferase Reporter Assay System (Promega). Results were normalized to a cotransfected renilla luciferase control plasmid.
Table 4.
Primers for amplification of CNSa
| Primer name | Primer |
|---|---|
| MP 1617c | CCGGATCCTGCACGTACATGTTTGTACC |
| MP 1617d | CAGGATCCCAATGATTCAGGCAGCTCGG |
2.9 Genome-wide Analysis of BRG1 binding and NF-kB motifs
BRG1 binding data from (De et al., 2011), were used to create BRG1 binding tracks (Fig 1) using CisGenome (Ji et al., 2008). The BRG1 binding tracks are normalized to the number of reads in each track; thus, the y axis for each track covers the identical tag frequency range. BRG1 binding regions that were enriched relative to control (input) DNA were identified from resting cells (Naïve, Th1, Th2) and stimulated cells (Th1, Th2, and Th17); quality scores for “tags per position” and “Diff strand fold enrichment” were 1.74 and 2.7 for Naïve, 1.42 and 3.2 for resting Th1, 1.59 and 3.2 for resting Th2, 1.14 and 2.8 for stimulated Th1, 1.15 and 2.9 for stimulated Th2, and 1.04 and 1.7 for stimulated Th17 using Homer (van de Wetering et al.). BRG1 binding regions were analyzed for NF-kB motifs using Homer (van de Wetering et al.) using the command “findMotifsGenome” with the options of motif lengths 8 and 10 bp, testing the central 50 bp of the binding regions. Sequence motifs in CNSa were identified using TESS (Transcription Element Search System) (http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=WELCOME).
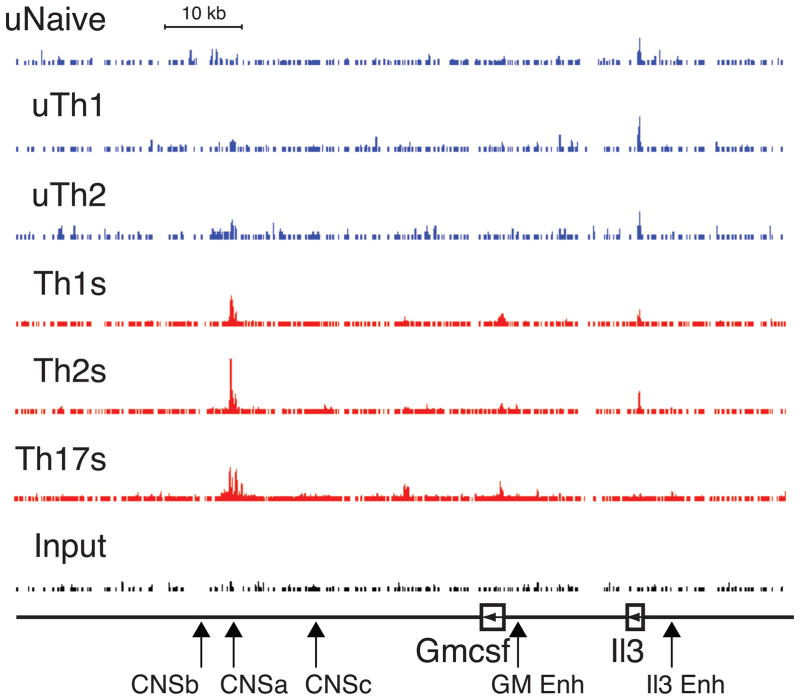
Figure 1. BRG1 binding to IL-3 and GM-CSF locus.
Data from unstimulated Naïve, Th1 and Th2 cells is in blue; stimulated Th1, Th2, and Th17 cells in red; Input (control) is in black. IL-3 and GM-CSF genes are indicated below, with horizontal arrowheads indicating direction of transcription. CNSa-c, IL-3 enhancer and GM-CSF enhancer are also labeled, and a scale bar indicates 10,000 bp. BRG1 occupancy (y axis) for all graphs is identical to allow direct comparison (minimum tag frequency of 0, maximum tag frequency of 1.14×10–5).
3. Results
3.1 Developmental and Activation Specific Recruitment of BRG1 to IL-3/GM-CSF Locus
We recently performed a genome-wide survey of BRG1 binding in a variety of Th subsets using mouse primary cells (De et al., 2011). Global analysis revealed BRG1 binding was highly dynamic; BRG1 binding was responsive to T cell activation signals and lineage-specific signals, resulting in enrichment at active genes at both promoter and distal elements. At the IL-3/GM-CSF locus we found strong, inducible BRG1 peak located 34kb downstream of the GM-CSF start site (Fig 1) corresponding with an element we recently identified as CNSa, a DNase hypersensitive site (DHS) that binds the remodeling enzyme SNF2H (Precht et al., 2010). We found weaker BRG1 binding at the GM-CSF and IL-3 promoters.
3.2 BRG1 Recruitment to CNSa Corresponds with IL-3/GM-CSF transcription
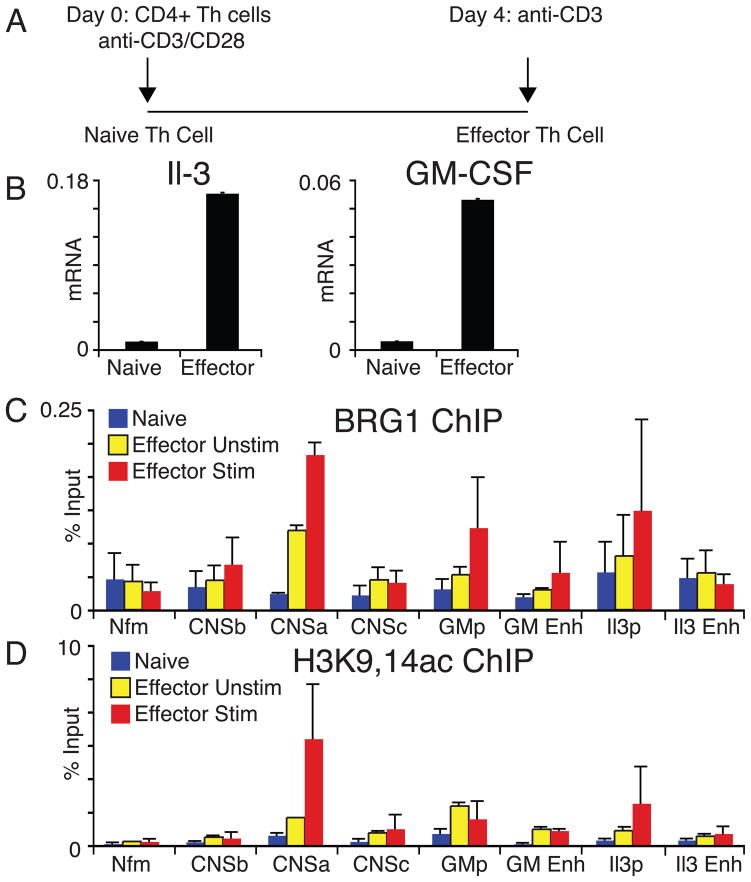
The expression of both IL-3 and GM-CSF has been suggested to be a marker for effector T cells. It has been recently demonstrated that freshly isolated splenocytes do not produce as much of these cytokines as previously activated T cells blasts (Mirabella et al., 2010). We confirmed these findings by examining IL-3 and GM-CSF mRNA expression in stimulated naïve Th cells or effector Th cells (Fig 2A and B). We found that differentiated effector Th cells were capable of producing 10 to 25 fold more GM-CSF and IL-3 message, respectively, when compared to undifferentiated precursors. We characterized the changes in chromatin modifications at the locus and found that both BRG1 binding as well as histone H3 acetylation at lysines 9 and 14 correlated with cytokine expression, particularly at the distal CNSa element (Fig 2C,D). Enhanced BRG1 binding was also detected at both promoters. Our findings are in agreement with a recent report on histone modifications associated with more proximal elements surrounding Il-3/GM-CSF (Mirabella et al., 2010). Two adjacent conserved non-coding sequences CNSb and CNSc do not appear to bind BRG1 strongly. We verified our ChIP-seq results that BRG1 binding is highly enriched at the CNSa element and responsive to both differentiation from naïve to effector Th cells as well as to T cell stimulation.
Figure 2. BRG1 binding to IL-3/GM-CSF locus correlates with gene expression.
A) Schematic of cell culture conditions. B) IL-3/GM-CSF mRNA expression, normalized to actin, was compared between CD3/CD28 stimulated Thp or CD3 stimulated effector Th cells. Results represent average and standard deviation of four independent experiments. C) BRG1 binding and D) H3K9,14 acetylation at the IL-3/GM-CSF locus. Nfm is a neuron-specific gene, a negative control locus in T cells (De et al., 2011). Results represent average and standard deviation of two independent experiments.
3.3 BRG1-containing BAF Complexes Regulate Expression Of IL-3/GM-CSF In Effector Th Cells
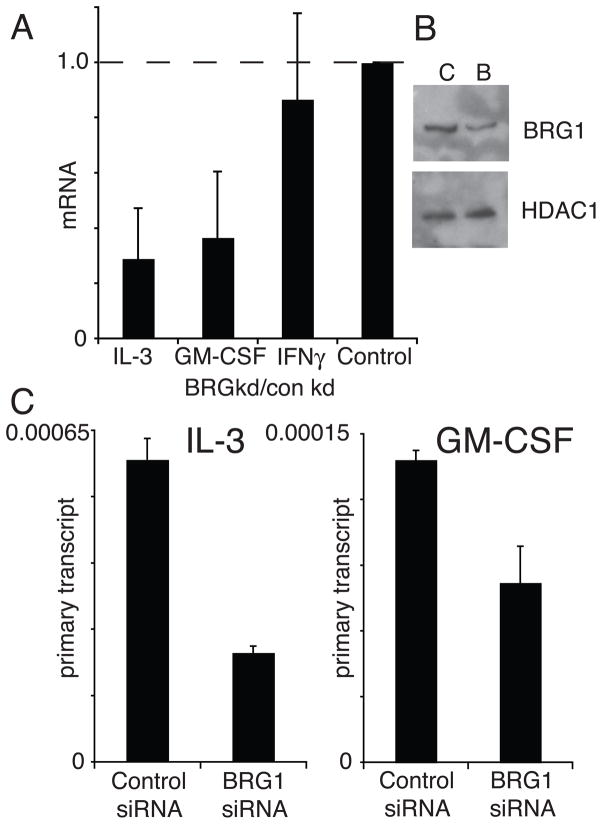
Previously we had found that BRG1 was required for Th2 cytokine gene expression and chromatin remodeling (Wurster and Pazin, 2008). Given the highly regulated BRG1 recruitment to the IL-3/GM-CSF locus and the correspondence of BRG1 binding with gene expression, we asked whether BRG1 modulates IL-3/GM-CSF gene expression. We found that the expression of both IL-3 and GM-CSF mRNA was reduced in cells following depletion of BRG1 using siRNA (Fig 3A). Treatment of primary effector Th cells with siRNA reagents targeted to BRG1 led to a modest reduction of BRG1 protein (Fig 3B). The amount of primary transcripts for IL-3 and GM-CSF were also reduced following BRG1 depletion, suggesting this effect was at the level of transcription, rather than RNA stability (Fig 3C). These results suggest that BRG1 plays a positive role in the regulation of activation-induced expression of the IL-3/GM-CSF locus.
Figure 3. BRG1 is required for IL-3/GM-CSF transcription in effector Th cells.
A) Impaired IL-3 and GM-CSF mRNA induction in BRG1 knockdown cells. mRNA was quantified by real-time RT-PCR, and the ratio of mRNA in stimulated cells treated with BRG1 siRNA was normalized to control siRNA-treated cells. Each bar is the average and standard deviation of at least 3 independent experiments. B) siRNA knockdown of BRG1 in effector Th cells. Lysates were analyzed by SDS PAGE, immunoblotted with BRG1 and HDAC1 (loading control). C) Impaired transcription of IL-3 and GM-CSF genes in BRG1 knockdown cells. RNA from stimulated cells was quantified by real-time PCR using primers specific to the primary transcripts including intronic sequences of the IL-3 and GM-CSF genes (Precht et al., 2010; Wurster and Pazin, 2008). The results shown are the average and standard deviation of two independent experiments.
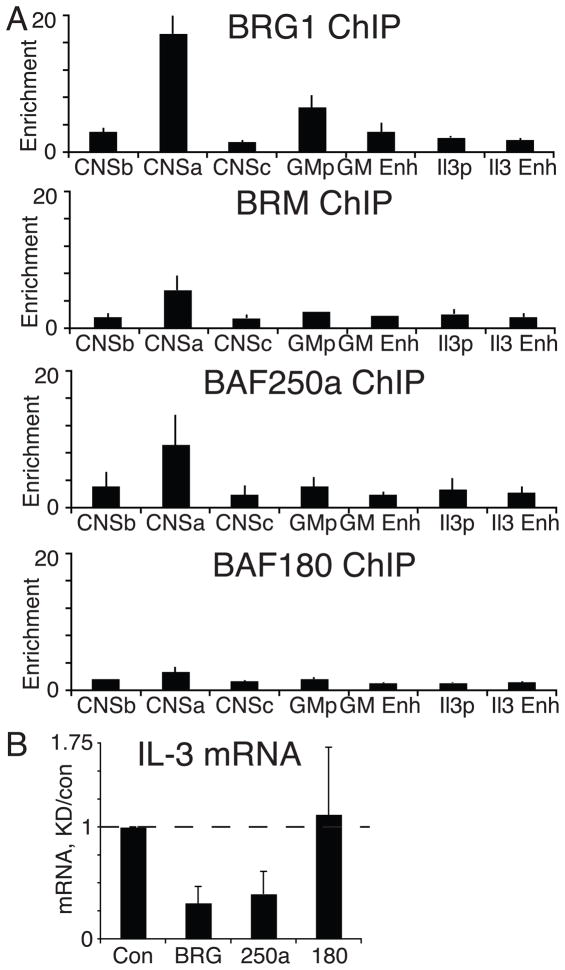
A number of structural variants have been described for SWI/SNF remodeling complexes that are identified by signature subunits (Boon et al., 2002). For example, BAF complexes contain BRG1 or Brm, and either BAF250a or BAF250b. PBAF complexes contain BAF180, BAF200 and BRG1 but not Brm. Importantly, BAF and PBAF complexes appear to regulate different target genes (Yan et al., 2005). Using ChIP, we found BAF-specific components (BAF250a and BRM) associated with elements in the IL-3/GM-CSF locus, including CNSa (Fig 4A). Additionally, siRNA knockdown of BRG1 and the BAF-specific BAF250a resulted in decreased IL-3 expression (Fig 4B). We did not detect a role for the PBAF complex (BAF180/polybromo-1) either in binding to the IL-3/GM-CSF locus or functionally by siRNA knockdown (Fig 4). These results suggest that BAF-specific BRG complexes are directly regulating the IL-3/GM-CSF locus.
Figure 4. BAF complexes bind to and regulate IL-3/GM-CSF expression.
A) BRG1, BAF250a, Brm and BAF180 binding to IL-3/GM-CSF locus. Results are the average and standard deviation of three independent experiments. Results are normalized to binding to the Nfm promoter, a silent and DNase I-resistant site in T cells. B) siRNA knockdown of BRG1, BAF250a and BAF180 in effector Th cells. IL-3 mRNA was harvested from stimulated cells, quantified by real-time PCR. The results are the average and standard deviation of at least three independent experiments.
3.4 BRG1-dependent Remodeling of CNSa
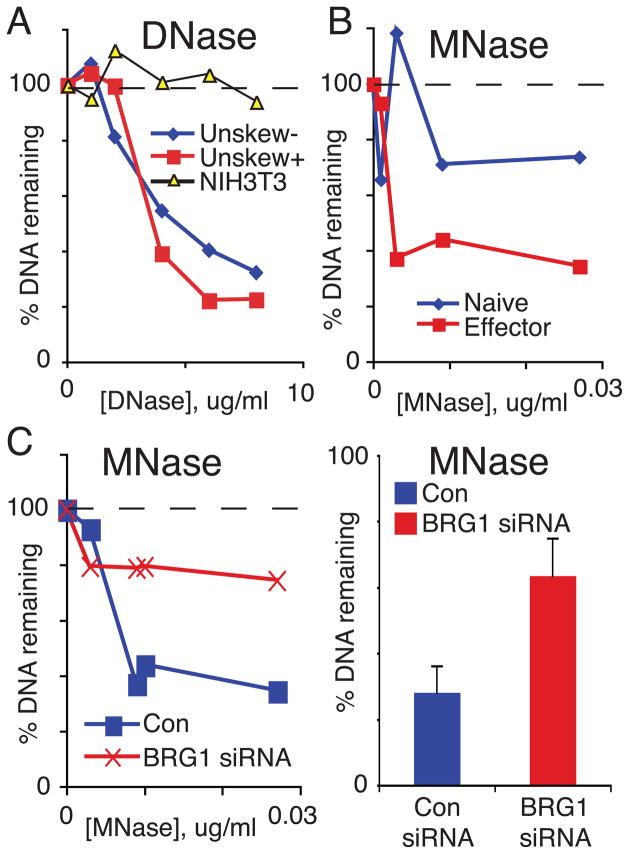
Many of the enhancer elements already characterized within the IL-3/GM-CSF locus are associated with increased chromatin accessibility as assayed by nuclease hypersensitivity (Cockerill, 2004). Given the enrichment of BAF complexes at CNSa and the role of BRG1 complexes in nucleosome remodeling, we asked whether CNSa was associated with nuclease hypersensitivity (HS). In fibroblast cells we found that CNSa was largely inaccessible to nucleases, however in effector T cells CNSa was acutely hypersensitive to two different nucleases, DNase and MNase digestion, as we previously found for another enhancer (Wurster and Pazin, 2008) (Fig 5A,B). Thus, CNSa is not hypersensitive in all cell types. The extent of digestion was increased following T cell effector differentiation and modestly by T cell activation (Fig 5A,B). The remodeling that occurred at CNSa was determined to be dependent on BRG1 as CNSa was significantly more resistant to nuclease digestion in BRG1 knockdown cells (Fig 5C). This suggests that BRG1 directly regulates expression of IL-3 and GM-CSF, as BRG1 is required for expression, binds to the locus, and programs chromatin structure.
Figure 5. BRG1 is required for DHS at CNSa.
A) DHS at CNSa was compared between unstimulated (−) and stimulated (+) effector Th cells and NIH3T3 fibroblasts B) Micrococcal nuclease accessibility was compared between naïve (blue) and effector (red) Th cells C) BRG1 is required for HS in effector Th cells. A representative dose response to MNase I in control (blue) and BRG1 knockdown (red) cells is shown (left panel). The average and standard deviation of accessibility of three independent experiments at one MNase concentration is shown (right panel).
3.5 CNSa has BRG1-dependent Enhancer Activity in Chromatinized Reporters
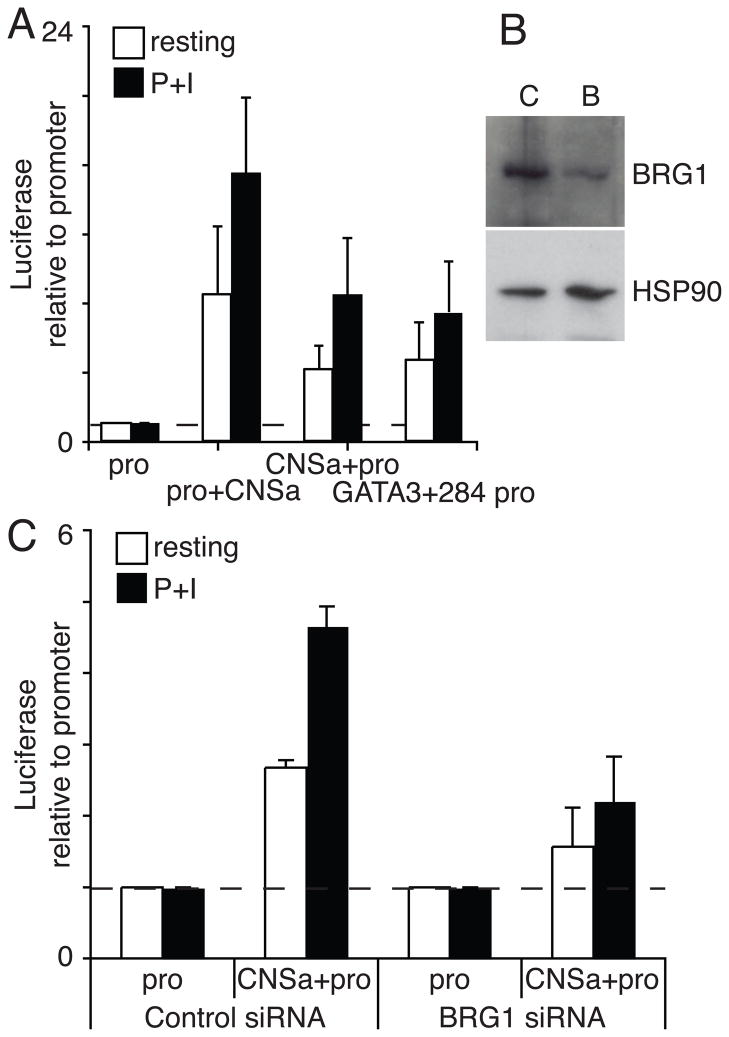
As CNSa sequence is conserved across species, highly nuclease accessible in T cells and marked with activate histone signatures, we tested whether CNSa is a novel enhancer element in the IL-3/GM-CSF locus. We cloned a 440bp element encompassing CNSa upstream and downstream of a reporter in an episomal vector. Traditional reporter constructs often do not form physiological chromatin, while episomal reporters contain an origin of replication that allows for assembly of a native chromatin structure (Liu et al., 2001; Smith and Hager, 1997). We transfected these constructs along with one carrying the promoter alone into CEM (human T lymphoblastoma) cells. We found that CNSa was able to consistently enhance reporter activity over promoter alone similar to activity observed for a bona fide enhancer element in the GATA3 locus (GATA3 +284), previously shown to function in both episomal reporters and transgenic mice (De et al., 2011; Hosoya-Ohmura et al., 2011) (Fig 6A). This element functioned both upstream and downstream of the reporter, suggesting it was functioning as an enhancer rather than a promoter or regulator of RNA stability. Depletion of BRG1 reduced the enhancer activity of the CNSa element, suggested the CNSa enhancer activity is dependent on BRG1 (Fig 6B,C). CNSa did not function in a standard transient reporter assay (data not shown), suggesting CNSa function might be chromatin-dependent, similar to other recently described elements (De et al., 2011). Definitive proof of CNSa enhancer activity requires testing in mouse knockout experiments.
Figure 6. CNSa has BRG1-dependent enhancer activity.
A) Episomal reporter constructs with CNSa either upstream or downstream of reporter transcription unit can augment promoter activity in CEM cells. Cells were either left untreated for stimulated with P+I for the last 5 hours of culture. The SV40 promoter activity along was unaffected by cell stimulation. The results are the average and standard deviation of six independent experiments. B) Depletion of BRG1 using siRNA in CEM cells. C) BRG1 knockdown in CEM cells results in reduced reporter activity. The results are the average and standard deviation of two independent experiments.
3.6 BRG-1 Recruitment to CNSa and IL-3/GM-CSF activation are Dependent on NF-κB
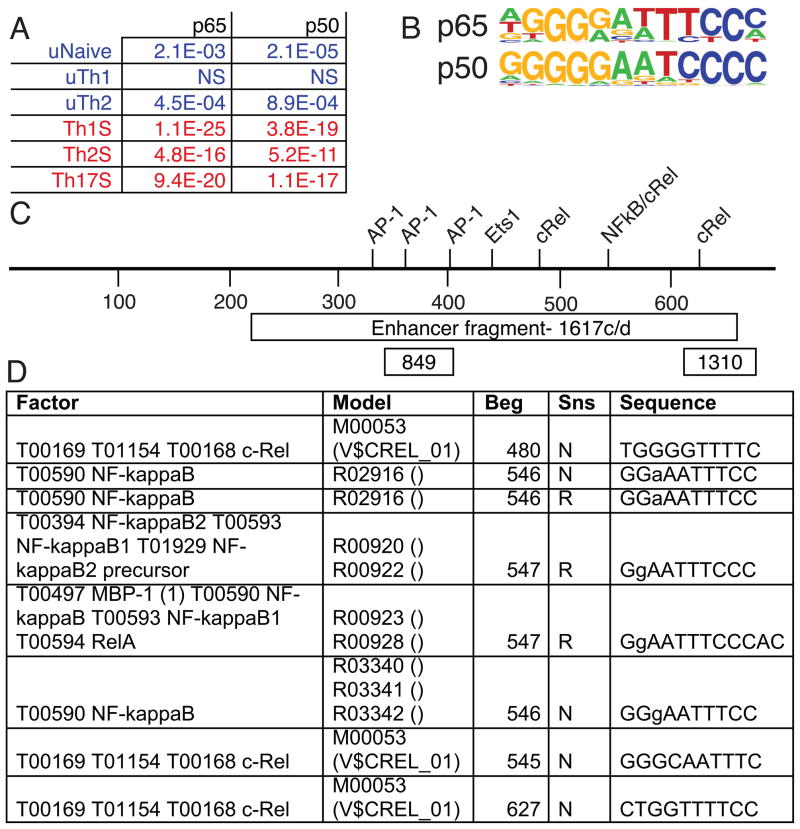
Previous reports demonstrated that regulatory elements in the IL-3 and GM-CSF locus are regulated by the NF-κB pathway (Brettingham-Moore et al., 2005; Holloway et al., 2003). Additionally, c-rel-deficient mice have significantly reduced IL-3 and GM-CSF gene expression (Gerondakis et al., 1996). T cell activation is known to induce NF-κB activity (Gebuhr et al., 2003). Genome-wide analysis of BRG1 binding regions in Th cells revealed that NF-κB sequence motifs were enriched in stimulated Th cells relative to unstimulated cells (Figure 7A,B). In particular, the CNSa element contained NF-κB sequence motifs (Fig 7C).
Figure 7. NF-κB motifs are enriched in BRG1 binding regions in activated T cells, and are found in CNSa.
A) Significance of enrichment of NF-κB sequence motifs in BRG1 binding regions from resting (blue) and stimulated (red) Th cells was calculated as described in the methods. B) NF-κB motifs from Homer. C) Sequence motifs in CNSa; motif names detected by TESS on top axis, scale indicated in bp beneath axis, box labeled Enhancer fragment indicates enhancer fragment amplified using primers MP1617c and MP1617d for enhancer assays, and boxes labeled 849 and 1310 indicate PCR products from primer set MP849 and MP1310, respectively. D) Rel sequence motifs predicted by TESS in CNSa; Factor is the name of the protein/family, Model is the motif matrix, Beg is the beginning of the match, Sns is strand (N- forward, R- reverse), and Sequence indicates the motif searched (mismatches shown in lower case).
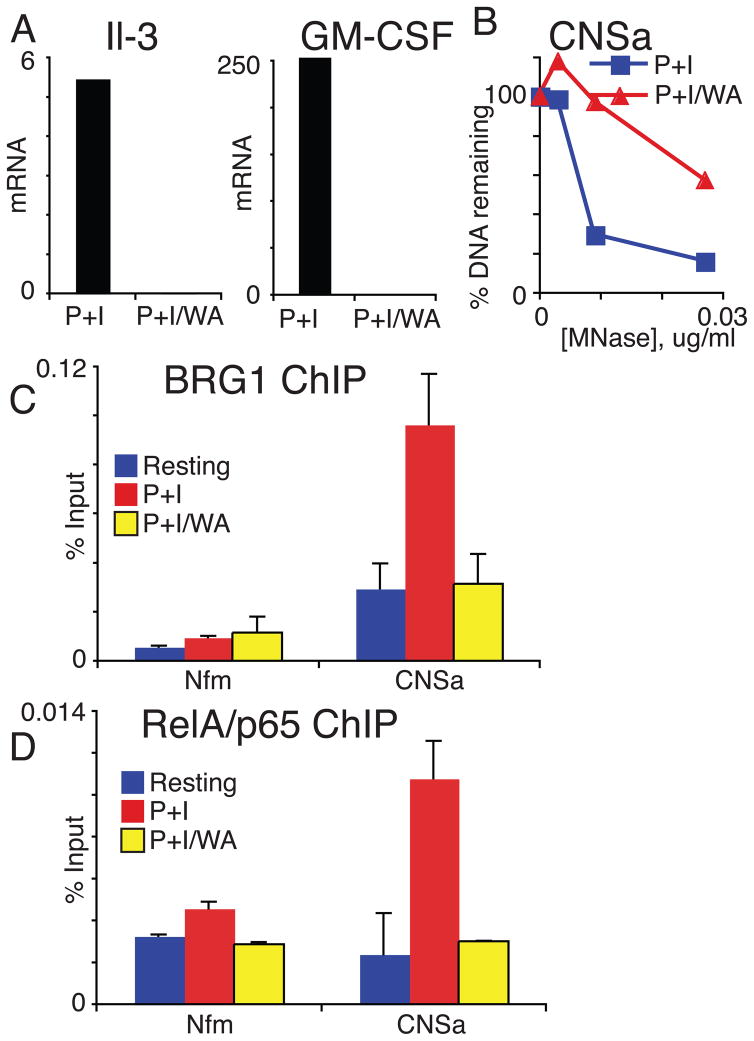
We tested the function of NF-κB using a pharmacological inhibitor. Withaferin A (WA) specifically inhibits NF-κB by reducing I-kappa-B phosphorylation and subsequent degradation (Kaileh et al., 2007). Pretreatment of effector T cells with Withaferin A reduced T cell activation-induced expression of both IL-3 and GM-CSF (Fig 8A). Withaferin A reduced the accessibility of CNSa, similar to what we observed when BRG1 was depleted (compare Fig 8B and 5B). Withaferin A pretreatment also reduced binding of BRG1 and the NF-κB subunit p65 to CNSa (Fig 8C, D). By contrast, pretreatment with Cyclosporine A (CSA), an inhibitor of NFAT activation, had no effect on BRG1 recruitment to CNSa (data not shown). Together, these results suggest that NF-κB activation after T cell stimulation results in the direct binding of p65 to CNSa, which in turn facilitates the recruitment of BRG1 and subsequent chromatin remodeling at CNSa. However, a significant amount of BRG1 is associated with CNSa in resting effector cells suggesting that additional mechanisms exist for BRG1 recruitment to CNSa independent of the NF-κB pathway.
Figure 8. IL-3/GM-CSF transcription and BRG1 recruitment to CNSa are dependent on NF-κB signals.
A) IL-3 and GM-CSF expression are reduced following Withaferin A pretreatment. The results are representative of three independent experiments B) HS at CNSa is reduced with pretreatment of Withaferin A. C) BRG1 and D) p65 binding to CNSa are blocked by withaferin A. The results are the average and standard deviation of 3 independent experiments.
4. Discussion
We examined the role of a distal, conserved element in the IL-3/GM-CSF locus. We found CNSa bound BRG1, and binding was induced by differentiation and stimulation. BRG1 contributed to an open chromatin structure at CNSa, and was required for maximal expression of IL-3 and GM-CSF. The BAF-specific cofactors Brm and BAF250a also bound CNSa; BAF250a was required for maximal IL-3 expression. CNSa activated expression of a reporter when placed upstream or downstream of the reporter. Activation was BRG1-dependent and limited to episomal vectors, suggesting chromatin and chromatin remodeling were required. Finally, we found RelA/p65 bound CNSa and IL-3/GM-CSF expression were abolished by an NF-κB inhibitor. NF-κB inhibition prevented BRG1 binding and chromatin opening at CNSa. Together, these findings suggest CNSa is a distal, chromatin-specific enhancer requiring NF-κB and BRG1 for function. However, definitive proof of enhancer activity requires genetic analysis in mice.
An outstanding question in chromatin biology is why there are so many different ATP-dependent remodeling enzymes and whether specificity exists in their activity. We recently reported that SNF2H, an ATP-dependent remodeling enzyme in the ISWI family, activates expression of IL-3 in a T cell line (Precht et al., 2010). SNF2H bound to a number of sites in the IL-3/GM-CSF locus, including CNSa. BRG1 binding to this locus was activation-dependent, while SNF2H binding is largely independent of stimulation. Subsequent to BRG1 binding, we observed increases in histone acetylation at CNSa and elsewhere in this locus. Together these results suggest the possibility of a cooperative relationship between SWI/SNF and ISWI complexes at the distal CNSa element, where SNF2H might bind constitutively in the T cell lineage, while BRG1 binding is induced following differentiation and stimulation. However, it remains to be determined experimentally whether ISWI and SWI/SNF complexes bind to the same allele of CNSa simultaneously and what the functional outcome of that interaction would be.
Many previous studies on BRG1 have focused on the role of ATP-dependent remodeling in the regulation of accessibility and activity of promoter proximal elements. However, there are reports of remodeling-independent function of BRG1 (Jani et al., 2008; Trotter and Archer, 2004). Moreover, an increasing number of studies have also pointed to a prominent role for SWI/SNF proteins in the function of distal enhancer elements. We previously reported that BRG1 regulates expression of the TH2 cytokines, and is recruited in a Th2-specific manner to the distal locus control region (LCR) in the IL-4/IL-13/IL5 locus (Wurster and Pazin, 2008). We also found that BRG1 binding to numerous distal elements in T helper cells correlated with lineage- and activation- specific gene expression (De et al., 2011). We identified novel distal regulatory elements at the GATA3 locus using BRG1 binding as a marker for active enhancers. BRG1 was found to localize to both promoter proximal and distal LCR’s of both the β- and γ-globin genes in developing erythrocytes (Kim et al., 2009a; Kim et al., 2009b). BRG1 also required for the interferon-stimulated induction of the CIITA gene through both proximal and remote regulatory elements (Ni et al., 2008). For both the β-globin and CIITA loci, BRG1 appears to play a role in higher order chromatin structure, facilitating distal enhancer and promoter contacts through looping. For the CIITA locus in particular, it was suggested that BRG1 did not directly trigger the looping itself but was required to facilitate cytokine-induced loop formation and cooperative interaction of multiple regulatory elements. It remains to be determined whether the distal enhancer element, CNSa, located over 30 kb downstream of the GM-CSF promoter, is in direct contact through a looping mechanism with the IL-3/GM-CSF promoters, enhancers and/or insulator. However, it is intriguing to consider a role for BRG1 in coordinating higher-order chromatin structure through these elements, as suggested for the MHC locus (Christova et al., 2007).
Highlights.
BRG1, an ATP-dependent chromatin remodeling enzyme, activates expression of the cytokines IL-3 and GM-CSF.
BRG1 binds the target genes, and opens the chromatin at a distal element, suggesting the regulation is direct.
NF-kB is required for activation of IL-3 and GM-CSF, BRG1 binding to a distal regulatory element, and chromatin opening at that element.
BRG1 binds to distal regulatory sites as well as promoters.
This work shows that chromatin remodeling plays a causal role in regulation of these genes, rather than a consequence of transcription.
Acknowledgments
We thank Weidong Wang for BRG1 antibody, Keji Zhao for the pREP reporter vectors. We also thank Sebastian Fugmann, Peter Cockerill, Nan-Ping Weng, Rebecca Potts and Mary Kaileh for helpful discussions.
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging, 1 Z01 AG000524. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Author contributions
The authors have made the following declaration about their contributions: Conceived and designed the experiments: ALW, MJP; Performed the experiments: ALW, PP; Analyzed the data: ALW, PP, MJP; Wrote the paper ALW, MJP.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal S, Rao A. Long-range transcriptional regulation of cytokine gene expression. Curr Opin Immunol. 1998a;10:345–52. doi: 10.1016/s0952-7915(98)80174-x. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998b;9:765–75. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Bert AG, Johnson BV, Baxter EW, Cockerill PN. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol Cell Biol. 2007;27:2870–85. doi: 10.1128/MCB.02323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, Hager GL. Glucocorticoid receptor dynamics and gene regulation. Stress. 2009;12:193–205. doi: 10.1080/10253890802506409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon EM, van der Neut R, van de Wetering M, Clevers H, Pals ST. Wnt signaling regulates expression of the receptor tyrosine kinase met in colorectal cancer. Cancer Res. 2002;62:5126–8. [PubMed] [Google Scholar]
- Bowers SR, Mirabella F, Calero-Nieto FJ, Valeaux S, Hadjur S, Baxter EW, Merkenschlager M, Cockerill PN. A conserved insulator that recruits CTCF and cohesin exists between the closely related but divergently regulated interleukin-3 and granulocyte-macrophage colony-stimulating factor genes. Mol Cell Biol. 2009;29:1682–93. doi: 10.1128/MCB.01411-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettingham-Moore KH, Rao S, Juelich T, Shannon MF, Holloway AF. GM-CSF promoter chromatin remodelling and gene transcription display distinct signal and transcription factor requirements. Nucleic Acids Res. 2005;33:225–34. doi: 10.1093/nar/gki161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettingham-Moore KH, Sprod OR, Chen X, Oakford P, Shannon MF, Holloway AF. Determinants of a transcriptionally competent environment at the GM-CSF promoter. Nucleic Acids Res. 2008;36:2639–53. doi: 10.1093/nar/gkn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Cakouros D, Cockerill PN, Bert AG, Mital R, Roberts DC, Shannon MF. A NF-kappa B/Sp1 region is essential for chromatin remodeling and correct transcription of a human granulocyte-macrophage colony-stimulating factor transgene. J Immunol. 2001;167:302–10. doi: 10.4049/jimmunol.167.1.302. [DOI] [PubMed] [Google Scholar]
- Chi T. A BAF-centred view of the immune system. Nat Rev Immunol. 2004;4:965–77. doi: 10.1038/nri1501. [DOI] [PubMed] [Google Scholar]
- Christova R, Jones T, Wu PJ, Bolzer A, Costa-Pereira AP, Watling D, Kerr IM, Sheer D. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNgamma. J Cell Sci. 2007;120:3262–70. doi: 10.1242/jcs.012328. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Cockerill PN. Mechanisms of transcriptional regulation of the human IL-3/GM-CSF locus by inducible tissue-specific promoters and enhancers. Crit Rev Immunol. 2004;24:385–408. doi: 10.1615/critrevimmunol.v24.i6.10. [DOI] [PubMed] [Google Scholar]
- De S, Wurster AL, Precht P, Wood WH, 3rd, Becker KG, Pazin MJ. Dynamic BRG1 Recruitment during T Helper Differentiation and Activation Reveals Distal Regulatory Elements. Mol Cell Biol. 2011;31:1512–27. doi: 10.1128/MCB.00920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncliffe KN, Bert AG, Vadas MA, Cockerill PN. A T cell-specific enhancer in the interleukin-3 locus is activated cooperatively by Oct and NFAT elements within a DNase I-hypersensitive site. Immunity. 1997;6:175–85. doi: 10.1016/s1074-7613(00)80424-0. [DOI] [PubMed] [Google Scholar]
- Gebuhr TC, Kovalev GI, Bultman S, Godfrey V, Su L, Magnuson T. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J Exp Med. 2003;198:1937–49. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondakis S, Strasser A, Metcalf D, Grigoriadis G, Scheerlinck JY, Grumont RJ. Rel-deficient T cells exhibit defects in production of interleukin 3 and granulocyte-macrophage colony-stimulating factor. Proc Natl Acad Sci U S A. 1996;93:3405–9. doi: 10.1073/pnas.93.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–27. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–6. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway AF, Rao S, Chen X, Shannon MF. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor kappaB proteins. J Exp Med. 2003;197:413–23. doi: 10.1084/jem.20021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya-Ohmura S, Lin YH, Herrmann M, Kuroha T, Rao A, Moriguchi T, Lim KC, Hosoya T, Engel JD. An NK and T cell enhancer lies 280 kilobase pairs 3′ to the gata3 structural gene. Mol Cell Biol. 2011;31:1894–904. doi: 10.1128/MCB.05065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani A, Wan M, Zhang J, Cui K, Wu J, Preston-Hurlburt P, Khatri R, Zhao K, Chi T. A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. J Exp Med. 2008;205:2813–25. doi: 10.1084/jem.20080938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BV, Bert AG, Ryan GR, Condina A, Cockerill PN. Granulocyte-macrophage colony-stimulating factor enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization. Mol Cell Biol. 2004;24:7914–30. doi: 10.1128/MCB.24.18.7914-7930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EA, Flavell RA. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J Immunol. 2005;175:7437–46. doi: 10.4049/jimmunol.175.11.7437. [DOI] [PubMed] [Google Scholar]
- Kaileh M, Vanden Berghe W, Heyerick A, Horion J, Piette J, Libert C, De Keukeleire D, Essawi T, Haegeman G. Withaferin a strongly elicits IkappaB kinase beta hyperphosphorylation concomitant with potent inhibition of its kinase activity. J Biol Chem. 2007;282:4253–64. doi: 10.1074/jbc.M606728200. [DOI] [PubMed] [Google Scholar]
- Kim SI, Bresnick EH, Bultman SJ. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 2009a;37:6019–27. doi: 10.1093/nar/gkp677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci U S A. 2009b;106:2259–64. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C. Chromatin remodeling complex NURF regulates thymocyte maturation. Genes Dev. 2011;25:275–86. doi: 10.1101/gad.2007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–53. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–8. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–15. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L. Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation. Embo J. 2007;26:1292–302. doi: 10.1038/sj.emboj.7601586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–18. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur M, Gerum S, Stamatoyannopoulos G. Quantification of DNaseI-sensitivity by real-time PCR: quantitative analysis of DNaseI-hypersensitivity of the mouse beta-globin LCR. J Mol Biol. 2001;313:27–34. doi: 10.1006/jmbi.2001.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabella F, Baxter EW, Boissinot M, James SR, Cockerill PN. The human IL-3/granulocyte-macrophage colony-stimulating factor locus is epigenetically silent in immature thymocytes and is progressively activated during T cell development. J Immunol. 2010;184:3043–54. doi: 10.4049/jimmunol.0901364. [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–55. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- Ni Z, Abou El Hassan M, Xu Z, Yu T, Bremner R. The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat Immunol. 2008;9:785–93. doi: 10.1038/ni.1619. [DOI] [PubMed] [Google Scholar]
- Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–88. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placek K, Gasparian S, Coffre M, Maiella S, Sechet E, Bianchi E, Rogge L. Integration of distinct intracellular signaling pathways at distal regulatory elements directs T-bet expression in human CD4+ T cells. J Immunol. 2009;183:7743–51. doi: 10.4049/jimmunol.0803812. [DOI] [PubMed] [Google Scholar]
- Precht P, Wurster AL, Pazin MJ. The SNF2H chromatin remodeling enzyme has opposing effects on cytokine gene expression. Mol Immunol. 2010;47:2038–2046. doi: 10.1016/j.molimm.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–96. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–47. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- Smith CL, Hager GL. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J Biol Chem. 1997;272:27493–6. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- Tarakhovsky A. Tools and landscapes of epigenetics. Nat Immunol. 2010;11:565–8. doi: 10.1038/ni0710-565. [DOI] [PubMed] [Google Scholar]
- Trotter KW, Archer TK. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Molecular and cellular biology. 2004;24:3347–58. doi: 10.1128/MCB.24.8.3347-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–75. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–7. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- Wurster AL, Pazin MJ. BRG1-mediated chromatin remodeling regulates differentiation and gene expression of T helper cells. Mol Cell Biol. 2008;28:7274–85. doi: 10.1128/MCB.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–7. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–74. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med. 2006;203:1493–505. doi: 10.1084/jem.20060066. [DOI] [PMC free article] [PubMed] [Google Scholar]