Abstract
Background
In Germany, lung cancer causes more deaths than any other malignant disease. Its main etiology is smoking, but other risk factors need to be considered as well. The morphological, molecular and biological phenotype is complex and should no longer be just categorized as either small-cell or non–small cell lung cancer.
Methods
This review article is based on the authors’ longstanding involvement in the scientific investigation and diagnostic evaluation of lung cancer, including contributions to the current WHO classification and collaboration in the new interdisciplinary classification of adenocarcinoma. The relevant literature was selectively reviewed.
Results
Lung cancer is morphologically classified into four main subtypes—small-cell carcinoma, squamous-cell carcinoma, adenocarcinoma, and large-cell carcinoma. Genetic and molecular analyses have revealed distinct differences within subtypes; in particular, adenocarcinomas can be further subdivided. Complex techniques of genomic analysis are now available, but clinicopathological data are still the most important determinants of prognosis and are clearly better for this purpose than molecular classification alone. Nonetheless, the assessment of specific molecular markers is becoming increasingly important.
Conclusion
The morphological and molecular classification of lung cancer is undergoing a re-evaluation which will lead to more accurate assessment of individual prognoses and to improved prediction of the response to specific treatment regimens.
Lung cancer is responsible for 14.2% of neoplasms in men and 7.4% in women; it is the third most common cancer in Germany. In terms of mortality, however, it is in first place: 25.7% of male cancer patients (1st place) and 12.1% of female cancer patients (3rd place) die due to lung cancer. The reasons include the aggressiveness of the tumor and its strong tendency to metastasize. The current 5-year survival rates for men are 15% and for women, 18%; these rates have not really improved in recent years (1).
Further risk factors in addition to smoking include environmental and occupational factors. In Germany, lung cancer may be accepted as an occupational disease, which is the case especially for exposures to asbestos and radon; more rarely, polycyclic aromatic hydrocarbons, chromates, crystalline silicium dioxide, arsenic, nickel, and chloromethyl methylether (2). Viruses also play a part in the genesis of lung cancer. Large cell lymphoepithelial lung carcinoma, a rare variant of large cell carcinoma, is associated with the Epstein-Barr virus (3). Human papillomaviruses (HPV) have also been associated with the development of lung cancer. There are notable geographical differences, however. In Germany, maximum HPV detection rates of 4.2% have been reported, whereas in certain regions of Asia these were as high as 80% (4). Smoking is, however, by some margin the most common cause for the development of lung cancer (2).
Morphological classification
The 2004 classification from the World Health Organization (WHO) is currently the standard system for the morphological classification of lung cancer (etable). The WHO classification was the first to consider genetic parameters in the characterization of subtypes (3). The practicing clinician should at least be aware of the four main types of lung cancer: squamous cell carcinoma, adenocarcinoma, and large cell carcinoma—which as a group are known as the non–small cell carcinomas—as well as the small cell carcinoma (figure 1).
eTable. WHO classification of malignant epithelial lung tumors.
| Squamous cell carcinoma | 8070/3 |
| Papillary | 8052/3 |
| Clear cell | 8084/3 |
| Small cell | 8073/3 |
| Basaloid | 8083/3 |
| Small-cell carcinoma | 8041/3 |
| Combined small-cell carcinoma | 8045/3 |
| Adenocarcinoma | 8140/3 |
| Mixed subtype | 8255/3 |
| Acinar | 8550/3 |
| Papillary | 8260/3 |
| Bronchioloalveolar | 8250/3 |
| – Non-mucinous | 8252/3 |
| – Mucinous | 8253/3 |
| – Mixed or undetermined | 8254/3 |
| Solid (with mucus formation) | 8230/3 |
| Variants | |
| – Fetal | 8333/3 |
| – Mucinous (colloidal) | 8480/3 |
| – Mucinous cystadenocarcinoma | 8470/3 |
| – Signet ring cell adenocarcinoma | 8490/3 |
| – Clear cell | 8310/3 |
| Large-cell carcinoma | 8012/3 |
| Large-cell neuroendocrine carcinoma | 8013/3 |
| – Combined subtype | 8013/3 |
| Basaloid carcinoma | 8123/3 |
| Lymphoepithelioma-like carcinoma | 8082/3 |
| Clear cell carcinoma | 8310/3 |
| Carcinoma with rhabdoid phenotype | 8014/3 |
| Adenosquamous carcinoma | 8560/3 |
| Sarcomatoid carcinoma | 8033/3 |
| – Pleomorphic carcinoma | 8022/3 |
| – Spindle cell carcinoma | 8032/3 |
| – Giant cell carcinoma | 8031/3 |
| – Carcinosarcoma | 8980/3 |
| – Pulmonary blastoma | 8972/3 |
| Carcinoid tumor | 8040/3 |
| – Typical carcinoid | 8240/3 |
| – Atypical carcinoid | 8249/3 |
| Salivary gland tumors | |
| – Mucoepidermoid carcinoma | 8030/3 |
| – Adenoid cystic carcinoma | 8200/3 |
| – Epithelial-mesenchymal carcinoma | 8562/3 |
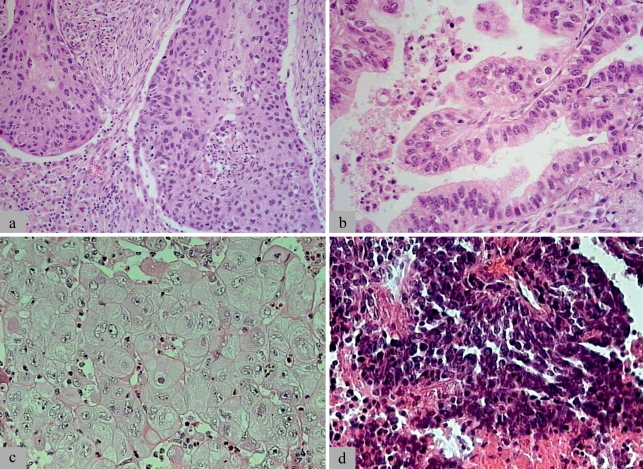
Figure 1.
Examples of the histomorphology of the four main types of lung cancer:
a) squamous cell carcinoma (p63, CK5/6); b) adenocarcinoma (TTF1, CK7); c) large cell carcinoma.
These three main types constitute the group of non–small cell lung cancers.
d) Small cell carcinoma (synaptophysin, chromogranin, CD56/NCAM).
Typical immunochemical marker proteins of each individual entity are listed in parentheses. These may, however, be lacking or expressed in other entities, and the immunophenotype should therefore always be interpreted in the morphological context
Squamous cell carcinoma is defined by the identification of keratinization or intercellular bridges. Adenocarcinoma is either characterized by mucus formation, which may be discrete or intracellular, or by distinct growth patterns such as glandular/acinar growth, papillar differentiation, or a single-layer, wallpaper-like spread along the alveolar septum and bronchioles; the latter is characteristic for bronchioloalveolar carcinoma. Large cell carcinoma is an exclusion diagnosis; the term refers to a barely differentiated, non–small cell cancer with a poor prognosis, in which neither the characteristics of squamous cell carcinoma nor those of adenocarcinoma are detectable. Small cell carcinoma represents the other extreme of a poorly differentiated lung cancer with a poor prognosis. On the one hand it is a tumor with a high proliferative activity and small tumor cells, which cannot be larger than three lymphocytes; and on the other hand, neuroendocrine differentiation has been identified (3).
When looking more closely at the classification, several mixed entities become obvious—such as the combined small cell carcinoma, which has a proportion of non-small cells; the adenosquamous carcinoma (adenocarcinoma and squamous cell carcinoma); or the carcinosarcoma. The most common form of adenocarcinoma is also a mixed type, which consists of a combination of the growth patterns described above. The diversity of lung cancer may cause problems with the diagnostic evaluation. The mixed entities, the heterogeneity of the tumors, and the observed phenotypic transitions between subtypes reflect the high genetic instability, which is also responsible for the high malignancy of and mortality due to lung cancer.
Genotype of lung cancer—ploidy and chromosomal changes
From the perspective of tumor genetics, lung cancers should be differentiated not so much on the basis of their cell size but rather on the basis of the size of their cell nuclei, because the nucleus is the location of the DNA and thus the primary information for the tumor genotype. Since the small cell carcinoma has hardly any cytoplasm, the denomination “small cell” actually means “small nucleus.” The situation is different for non–small cell carcinomas, where cell size and nucleus size may differ substantially. This problem is now well known, and newer classifications take this into account (5, 6).
Of importance is the fact that the nucleus size correlates to the DNA content of the tumor cells, and both variables significantly differ between small cell and non–small cell cancers. Small cell lung cancer typically has a reduced chromosome set—it is hypodiploid—whereas non–small cell cancers are usually hyperdiploid and often have chromosome numbers in the triploid range and higher. However, wide variation exists between the nucleus size and the ploidy level within the individual entities as well as within individual tumors. Atypical small cell carcinomas exist that contain large nuclei and hyperdiploid DNA content, whereas individual non–small cell carcinomas have been observed that have small nuclei and hypodiploid chromosome sets (5, 7). It is currently not known whether such tumors behave in clinically atypical ways.
Aneuploidy—the chromosomal changes in the tumor genome that are associated with the gain or loss of individual chromosomes or chromosome sections (DNA imbalances)—are of major importance in the context of lung cancer. It is found in all carcinomas; aneuploidy and the frequency of certain chromosomal imbalances clearly exceed the rate of specific gene mutations (3, 8).
DNA imbalances have been shown in archived tumor tissue by means of genomic screening procedures, such as comparative genomic hybridization (CGH) or array-CGH (a-CGH). These analyses show up characteristic changes that are associated with differences in tumor differentiation relating to adenocarcinomas, squamous cell carcinomas, large cell carcinomas, and small cell carcinomas. Small cell lung cancers show deletions of the short arm of chromosome 3 (3p deletions) in more than 90% of cases. These often affect the entire chromosomal arm and are often associated with a gain of the long chromosome arm, forming a so-called 3q isochromosome. In more than 80% of cases, deletions were seen on chromosomes 17p13 in 50% and 13q14 in 15% to 30% of cases for non–small cell carcinomas. Changes associated with tumor progression and metastasis have also been observed at the chromosome level (3).
The biological importance of the chromosomal imbalances lies in the change of the number of copies of the genes localized in the respective chromosomal regions. If these are transcribed into RNA and translated into proteins—that is, expressed—the result of the loss in DNA is a reduced expression of the respective genes. Deletions on chromosomes 17p13 and 13q14 are often associated with a reduced expression or inactivation of the tumor suppressor gene p53 and RB that are localized there. Accordingly, DNA gain may produce gene overexpression. The extreme variant of DNA gain is gene amplification. This is rare but may be crucial for the biology of the tumor in question, in case certain oncogenes are amplified (8).
Specific gene mutations, concept of oncogene addiction
In addition to amplifications, point mutations are often observed in lung cancer. KRAS mutations were described as one of the first alterations in 1987 (8). They are present in 10% to 15% of non–small cell carcinomas, most often in adenocarcinomas (20% to 30%) (3, 8). In the meantime, identifying this mutation has gained diagnostic relevance, because it is associated with primary resistance to treatment with small molecular antagonists of the epidermal growth factor receptor (EGFR). In 2004, an association was observed between activating mutations of the EGFR gene and successful treatment with EGFR inhibitors. The mutations are present in a maximum of 10% to 15% of lung carcinomas, primarily adenocarcinomas. Since July 2009, their identification has been the necessary requirement for first-line treatment with the EGFR inhibitor gefitinib (2, 8).
Activating mutations of the EGFR gene are an example of the so-called oncogene addiction of a tumor. This means that a specific oncogene is crucial for the tumor’s proliferation and growth; the tumor is dependent on the effects of the oncogene. If the oncogene is switched off then growth stops or the tumor may even regress. This is the reason for the success of targeted therapy with EGFR antagonists in non–small cell carcinomas. Within the tumor subgroups with activating EGFR mutations, response rates have been observed that are substantially higher than those associated with conventional chemotherapy (8). A similar association has been observed in the meantime for the detection of the so-called EML4-ALK translocation, which is present in 3% of all adenocarcinomas, and treatment with the ALK inhibitor crizotinib.
In small cell carcinomas, oncogene amplifications have been confirmed, especially of the MYC gene. Activating point mutations such as in the EGFR gene do not occur as such. This may explain why approaches using targeted molecular therapy have thus far not been successful in this tumor entity (3).
Molecular markers in differential diagnosis
Molecular markers, especially immunohistologically detectable antigens, have gained relevance for the diagnostic evaluation of lung cancer (3, 8). The immunohistological makers that are most often used in lung cancer are given in Figure 1. They include neuroendocrine markers such as synaptophysin, chromogranin, or CD56/NCAM, cytokeratines (CK5/6, CK7), or transcription factors (p63, TTF1), which, as lineage-specific antigens, may indicate a line of differentiation.
Since the lungs are often the location of cancer metastases, differential diagnosis uses further biomarkers. In the case of adenocarcinomas, these are in particular molecules that indicate a particular line of differentiation of the tumor cells and thus the origin from another organ—such as CDX-2 and CK20 as markers for colorectal cancer or prostate specific antigen in prostate cancer (9, 10). In squamous cell carcinomas, such markers do not exist, but the molecular genetic confirmation or exclusion of infection with human papillomavirus (HPV) may be helpful in deciding whether a squamous cell carcinoma of the lung is a primary tumor or a metastasis (11).
Genomic approaches to classification
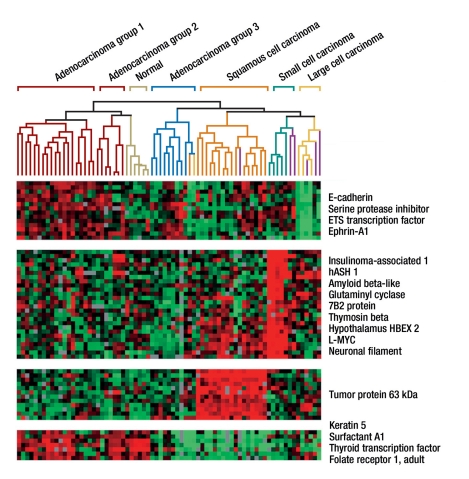
Gene expression studies have made a crucial contribution to identifying new molecular markers in lung cancer. Molecular classification recapitulated morphological subtyping and, for adenocarcinomas, showed a further subclassification into three groups that proved to be of prognostic importance (12, 13). Genes and classes of genes were identified whose overexpression or underexpression was characteristic for the individual tumor groups (figure 2). Large cell lung cancers have a reduced expression of the gene E-cadherin, which may be interpreted as a sign of epithelial-mesenchymal transition. The loss of E-cadherin has generally been associated with a poorer survival in patients with non–small cell lung cancers (12, 14). Reduced expression of TTF1 in adenocarcinomas has also been associated with a poorer prognosis; the gene may not be identifiable in less differentiated cancers, which should be considered in molecular diagnosis.
Figure 2.
Classification of lung cancer by means of hierarchical clustering (adapted from [12]). The columns correspond to individual tumor specimens, the rows to individual genes, which are grouped according to their similarity by means of cluster analysis. The analysis identified the four main types of lung cancer and the three subtypes of adenocarcinoma. Characteristic genes that are responsible for the grouping of the tumor types are listed to the right. In case of green coloring the respective genes of the tumor specimens were subject to reduced expression at the mRNA level, red signifies overexpression.
Molecular versus morphological classification
Further gene signatures are also of prognostic relevance. Over the years, the number of genes with relevance for prognostic assessment fell from 835 (12) to 50 (15), 25 (16), and finally only 5 genes (17). The number of genes to be analyzed is important because the analytic technique is selected on this basis. Several 100 or several dozens of genes can be analyzed using complex methods such as chip analysis, whereas five or a dozen genes can be analyzed using simpler techniques, such as immunohistochemistry or polymerase chain reaction.
In general, global gene expression analysis is currently not relevant in the diagnosis of lung cancer. An important study from 2008 showed that classifiers that were established solely on the basis of gene expression yielded poorer results than those that also considered clinical data, such as age, sex, and stage (18).
These results may seem to call into question the relevance of complex molecular analytic techniques in the classification of lung cancer. But it can be stated without any doubt that comprehensive genomic analytic methods for characterizing lung cancers have resulted in a new quality in the understanding of disease mechanisms and possible therapies (19, 20). Furthermore, the molecular, radiological, histomorphological, and clinical insights have helped to develop a new interdisciplinary classification for adenocarcinoma of the lung (8) under the aegis of the International Agency for the Study of Lung Cancer (IASLC) and the American Thoracic Society (ATS) in collaboration with the European Respiratory Society (ERS).
New classification of adenocarcinoma
The new classification of the adenocarcinoma of the lung is shown in the Box. It is based on the understanding that histomorphologically, distinction can be made not only between subtypes with a distinct prognosis but that the pathology can also give an idea of different genetic defects and the therapeutic response (8). Preinvasive lesions (atypical adenomatous hyperplasia, adenocarcinoma in situ [AIS]), and minimally invasive adenocarcinoma (MIA) have an excellent prognosis.
Adenocarcinoma in situ corresponds to the entity formerly known as pure bronchioloalveolar carcinoma without invasive growth. The term bronchioloalveolar carcinoma had caused confusion in the old WHO classification, because it was associated with the named tumor entity as well as with the characteristic growth pattern. The new classification dropped the term and replaced it with “adenocarcinoma in situ” and “lepidic tumor pattern.”
Minimally invasive adenocarcinoma is defined as a tumor of less than 3 cm in diameter with an invasive part of less than 5 mm. Such tumors can present a characteristic image in computed tomography scans, i.e. ground glass opacity with central consolidation. Ultimately, the final diagnosis of MIA requires full pathological work-up of the resected tumor.
Invasive adenocarcinomas are classified according to their predominant growth pattern; micropapillary adenocarcinoma was added as an individual subtype. It is recommended that growth patterns that are present in the tumor be documented and quantified and a decision be reached about the predominant growth type. This means that no mixed subtypes will exist in future. This distinction is also of prognostic relevance. The predominantly lepidic (G1) adenocarcinoma has the best prognosis, followed by the predominantly papillary subtype and acinar subtype (G2), whereas the predominantly micropapillary adenocarcinoma and solid adenocarcinoma are classified as G3 tumors and have the worst survival rates.
The growth pattern can be reliably classified only on the basis of histological analysis of the resected tumor. However, most lung cancers are diagnosed on the basis of small biopsy samples or cytology samples. For the first time, the classification acknowledges this problem and provides recommendations for the terminology and how to use such limited specimens. To simplify, analyzing a biopsy or cytology specimen should suffice to not only differentiate between small cell cancers and non–small cell cancers, but also to decide on whether the tumor is an adenocarcinoma or a squamous cell carcinoma. If that is not possible using molecular markers then making a diagnosis of a non–small cell cancer that is not otherwise specified (NSCLC-NOS) is justifiable.
In conclusion it may be stated that the classification of lung cancer is undergoing a phase of transition. In addition to subtle morphological analysis, targeted use of molecular markers and close interdisciplinary collaboration are required in order to decide on the best possible therapy for a patient. The hope is that all this will yield an improved prognosis for this disease.
Box. New classification of adenocarcinoma*.
-
Pre-invasive lesions
Atypical adenomatous hyperplasia (AAH)
-
Adenocarcinoma in situ (AIS)
(≤ 3 cm, formerly: “pure” bronchioloalveolar carcinoma)
Non-mucinous
Mucinous
Mixed non-mucinous/mucinous
-
Minimally invasive adenocarcinoma (MIA)
-
Predominantly lepidic
(adenocarcinoma up to ≤ 3 cm in size and ≤ 5 mm invasion)
Non-mucinous
Mucinous
Mixed non-mucinous/mucinous
-
-
Invasive adenocarcinoma
-
Predominantly lepidic
(formerly: non-mucinous bronchioloalveolar growth patterns with >5 mm invasion)
Predominantly acinar
Predominantly papillary
Predominantly micropapillary
Predominantly solid and mucinous
-
-
Variants of invasive adenocarcinoma
-
Invasive mucinous adenocarcinoma
(formerly: mucinous bronchioloalveolar carcinoma)
Colloidal adenocarcinoma
Fetal adenocarcinoma (of low of high malignancy)
Enteric adenocarcinoma
-
* under the aegis of the International Agency for the Study of Lung Cancer (IASLC) and the American Thoracic Society (ATS,) in collaboration with the European Respiratory Society (ERS), please also note details in the original publication (8)
Key Messages.
Lung cancer is classified into four main types: small cell carcinoma, squamous cell carcinoma, adenocarcinoma, and large cell carcinoma.
Using molecular analytic techniques has resulted in more precise typing, especially in adenocarcinoma.
The new international consensus classification of adenocarcinoma enables better prognostic assessment.
Some lung cancers are particularly dependent on the activating mutation of an oncogene.
Identifying such mutations is the prerequisite for successful targeted therapy.
Acknowledgments
Translated from the original German by Dr Birte Twisselmann.
Footnotes
Conflict of interest statement
Professor Petersen has received honoraria for speaking at continuing medical educational events and expert meetings from Lilly, Roche, AstraZeneca, Novartis, and Menarini. Furthermore he has acted as an adviser to Lilly and Boehringer-Ingelheim.
References
- 1.Robert Koch-Institut und die Gesellschaft der epidemiologischen Krebsregister in Deutschland (GEKID) e. V. (eds) Krebs in Deutschland 2005/2006. www.gekid.de. 7th revised edition. Berlin: 2010. Häufigkeiten und Trends. [Google Scholar]
- 2.Goeckenjan G, Sitter H, Thomas M. Prävention, Diagnose, Therapie und Nachsorge des Lungenkarzinoms Interdisziplinäre S3-Leitlinie der Deutschen Gesellschaft für Pneumologie und Beatmungsmedizin und der Deutschen Krebsgesellschaft. Pneumologie. 2010;64(Suppl 2):e1–e164. [Google Scholar]
- 3.Travis WD, Brambilla E, Müller-Hermeling K. In: World Health Organization Classification of tumours. Pathology and genetics of tumours of the lung, pleura, thymus and heart. Harris CC, editor. Lyon: IARCPress; 2004. [Google Scholar]
- 4.Klein F, Amin Kotb WFM, Petersen I. Incidence of human papilloma virus in lung cancer. Lung Cancer. 2009;65:13–18. doi: 10.1016/j.lungcan.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Petersen I, Amin Kotb WFM, Friedrich KH, Schlüns K, Böcking A, Dietel M. Core classification of lung cancer: Correlating nuclear size and mitoses with ploidy and clinicopathological parameters. Lung Cancer. 2009;65:312–318. doi: 10.1016/j.lungcan.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Nakazato Y, Minami Y, Kobayashi H. Nuclear grading of primary pulmonary adenocarcinomas: correlation between nuclear size and prognosis. Cancer. 2010;116:2011–2019. doi: 10.1002/cncr.24948. [DOI] [PubMed] [Google Scholar]
- 7.Junker K, Petersen I. Kleinzelliges Lungenkarzinom: Pathologie und Molekularpathologie. Onkologe. 2008;14:762–773. doi: 10.1007/s00292-008-1115-y. [DOI] [PubMed] [Google Scholar]
- 8.Travis WD, Brambilla E, Noguchi M. IASLC/ATS/ERS international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller KM, Wiethege T. Pathology, classification, and staging of malignant lung tumors. Radiologe. 2004;44:415–426. doi: 10.1007/s00117-004-1052-6. [DOI] [PubMed] [Google Scholar]
- 10.Neben K, Hübner G, Folprecht G, Jäger D, Krämer A. Metastases in the absence of a primary tumor: Advances in the diagnosis and treatment of CUP syndrome. Dtsch Arztebl Int. 2008;105:733–740. doi: 10.3238/arztebl.2008.0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weichert W, Schewe C, Denkert C, Morawietz L, Dietel M, Petersen I. Molecular HPV typing as a diagnostic tool to discriminate primary from metastatic squamous cell carcinoma of the lung. Am J Surg Pathol. 2009;33:513–520. doi: 10.1097/PAS.0b013e3181938319. [DOI] [PubMed] [Google Scholar]
- 12.Garber ME, Troyanskaya OG, Schluens K. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyerson M, Franklin WA, Kelley MJ. Molecular classification and molecular genetics of human lung cancers. Semin Oncol. 2004;31(1) Suppl 1:4–19. doi: 10.1053/j.seminoncol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Bremnes RM, Veve R, Hirsch FR, Franklin WA. The E-cadherin cell-cell adhesion complex and lung cancer invasion, metastasis, and prognosis. Lung Cancer. 2002;36:115–124. doi: 10.1016/s0169-5002(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 15.Beer DG, Kardia SL, Huang CC. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 16.Yanagisawa K, Tomida S, Shimada Y, Yatabe Y, Mitsudomi T, Takahashi T. A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J Natl Cancer Inst. 2007;99:858–867. doi: 10.1093/jnci/djk197. [DOI] [PubMed] [Google Scholar]
- 17.Chen HY, Yu SL, Chen CH. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 18.Shedden K, Taylor JM Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motoi N, Szoke J, Riely GJ. Lung adenocarcinoma: modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. Am J Surg Pathol. 2008;32:810–827. doi: 10.1097/PAS.0b013e31815cb162. [DOI] [PubMed] [Google Scholar]
- 20.Thomas RK, Weir B, Meyerson M. Genomic approaches to lung cancer. Clin Cancer Res. 2006;12:4384s–4391s. doi: 10.1158/1078-0432.CCR-06-0098. [DOI] [PubMed] [Google Scholar]