Abstract
Collapsin response mediator proteins (CRMPs) are cytosolic phosphoproteins that are functionally important during vertebrate development. We have generated a zebrafish genetrap line that produces fluorescently tagged Crmp1 protein, which can be dynamically tracked in living fish at subcellular resolution. The results show that Crmp1 is expressed in numerous sites in the developing nervous system. Early expression is apparent in the forebrain, epiphysis, optic tectum and the developing spinal cord. In the larval brain, Crmp1 is expressed in several distinct brain regions, such as the telencephalon, habenula and cerebellum. In addition, it is expressed in the spinal cord in a manner that persists in the larva. The results suggest that this Crmp1 protein trap line offers a powerful tool to track selected neuronal populations at high resolution.
Keywords: Crmp1, zebrafish, nervous system, development
Collapsin response mediator proteins (CRMPs) are a family of cytosolic phosphoproteins proteins thought to play important roles during neural development (CRMP 1–5; previously called TUC, Drp, Ulip and TOAD-64; reviewed by (Charrier et al., 2003; Quinn et al., 1999). CRMP-1 and CRMP-2 are key downstream mediators of the Semaphorin-3A signal transduction pathway involved in axon guidance and neuronal migration (Goshima et al., 1995; Uchida et al., 2005; Yamashita et al., 2007). Analysis of mouse CRMP1 suggests a function during maintenance of long term potentiation and spatial learning and memory (Su et al., 2007).
Recent advances in transgenesis have made the zebrafish a powerful model organism for examining expression and function of developmentally important genes. Here, we describe a zebrafish transgenic line for Crmp1. Generated by a Tol2 transposase mediated recombination (Trinh et al, submitted), we report visualization and dynamic tracking of fluorescently tagged endogenous Crmp1 fusion protein expression in living embryos at subcellular resolution. Previous in situ hybridization analysis has described the distribution of Crmp1-5 mRNA at 16 and 24 hours post fertilization (hpf) (Schweitzer et al., 2005). We extend these findings and report the high-resolution expression pattern of Crmp1 protein in live zebrafish during embryonic and larval development.
1. Results and discussion
From a gene and protein trap screen, in which a Tol2 transposase based vector containing Citrine (a variant of the yellow fluorescent protein) was randomly inserted into the genome, we discovered Gt(crmp1:Citrine)ct130a based on its interesting expression pattern in the developing nervous system. Molecular analysis by 3′ Rapid amplification of cDNA ends using Citrine primers determined genomic integration within the locus of Zebrafish crmp1. The crmp1 gene is comprised of 14 exons based on previous published data (Schweitzer et al., 2005). Our 3′ Rapid amplification of cDNA ends (RACE) results matched the previously published sequence downstream from exon 2 (Schweitzer et al., 2005) (data not shown), with the Citrine insertion occurring in the intronic region located between exons 1 and 2 (Fig. 1). Citrine is flanked by splice acceptor and splice donor sites and thus becomes integrated into the Crmp1 protein as an artificial exon (Fig. 1). Gt(crmp1:Citrine)ct130a homozygotes are viable suggesting that the Citrine integration does not impair the function of full length Crmp1.
Figure 1.
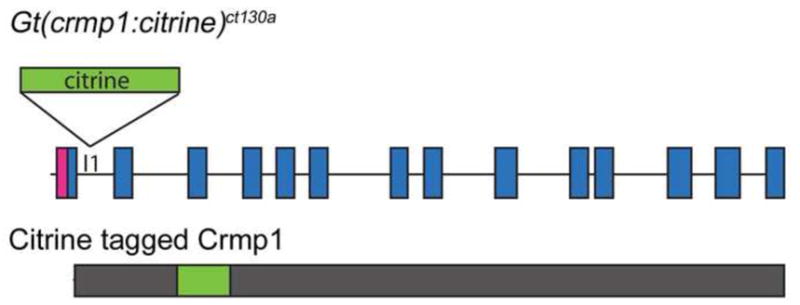
The Citrine insertion (green) occurred within the first intron (I1) in crmp1 in the Gt(crmp1:citrine)ct130a line and produces a fluorescently tagged full-length Crmp1 protein. The exons (blue boxes) and introns are not drawn to scale. Pink, 5′ Untranslated region.
1.1. Live Citrine expression in the Gt(crmp1:Citrine)ct130a line
As Crmps are highly enriched in the nervous system, we focused on its expression and localization between 1 to 4 days of development, as early born neurons were differentiating and eliciting axons and different regions of the brain were forming.
At 24–31 hours post-fertilization (hpf), high levels of expression were noted in the forebrain regions: the telen- and diencephalons (Fig. 2A, 5C). This protein expression is consistent with previous in situ hybridization data at this time point (Schweitzer, J et al 2005). Crmp1 expression persists and heightens in the forebrain at 31 hpf (Fig. 2A) and also becomes apparent in the epiphysis. Faint signal is also observed in developing spinal cord neurons (asterisks, Fig. 2B).
Figure 2.
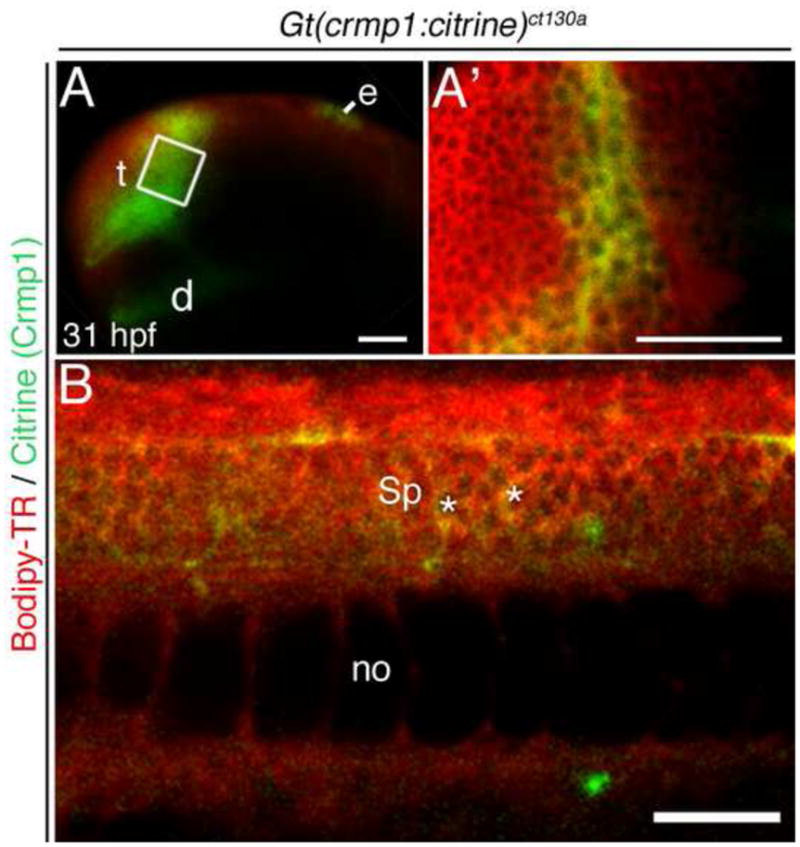
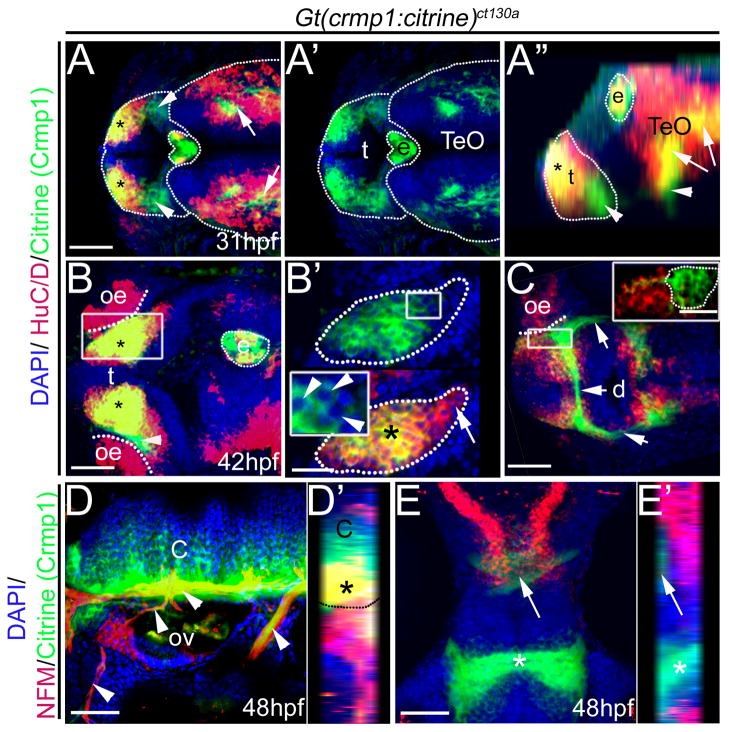
Crmp1 (green) is expressed in the forebrain at 31 hpf. All embryos were treated with the vital dye Bodipy-TR (red) to act as a contrast against the citrine signal. (A–B) Live confocal images. (A) Confocal z-stack showing Citrine/ Crmp1 (green) expression in the forebrain regions: the telencephalon (t) and diencephalon (d). Also, expression in epiphysis (e) is apparent. Boxed area shown in (A′). (A′) Z-section through the telencephalon. (B) Expression also found in spinal cord neurons (Sp) (asterisks). no, notochord. Scale bar: 50 μm.
Figure 5.
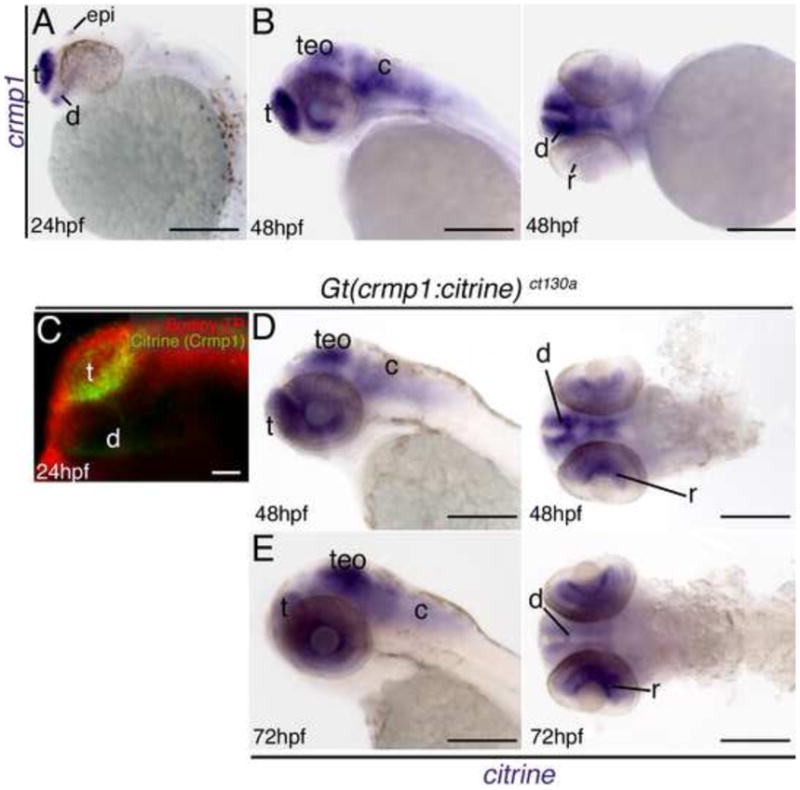
In situ hybridization results showing expression of crmp1 (A–B) and citrine (D–E). In wildtype fish crmp1 expression is localized to the nervous system at 24 hpf (A) and 48 hpf (B). (B) Lateral and frontal views at 48 hpf. (C) Confocal z-stack at 24 hours post fertilization (hpf) showing Citrine/ Crmp1 (green) expression in the forebrain regions. Red: Bodipy-TR vital dye. (D–E) Lateral and ventral views at 48 hpf and 72 hpf. Transcripts for citrine are expressed in selected neural regions at both stages consistent with wildtype expression. Abbreviations: epi, epiphysis; t, telencephalon; teO, optic tectum/ midbrain; c, cerebellum/ hindbrain; d, diencephalon; r, retina. Scale: 200 μM, 50 μM (C).
Tracking protein expression from the embryo to early larval revealed that, once expressed, Crmp1 appears to be maintained in selected sites, including the epiphysis, forebrain regions and spinal cord (Fig. 3A,B) at 42 hpf as well as in the larva (Fig. 3C). As the brain enlarged and compartmentalized, Crmp1 was observed in selected brain regions, including a portion of the telencephalon, the habenula, the optic tectum, and the cerebellum (Fig. 3C–E). Crmp1 expressing cells are observed in closely packed linear arrays in the cerebellum (Fig. 3E).
Figure 3.
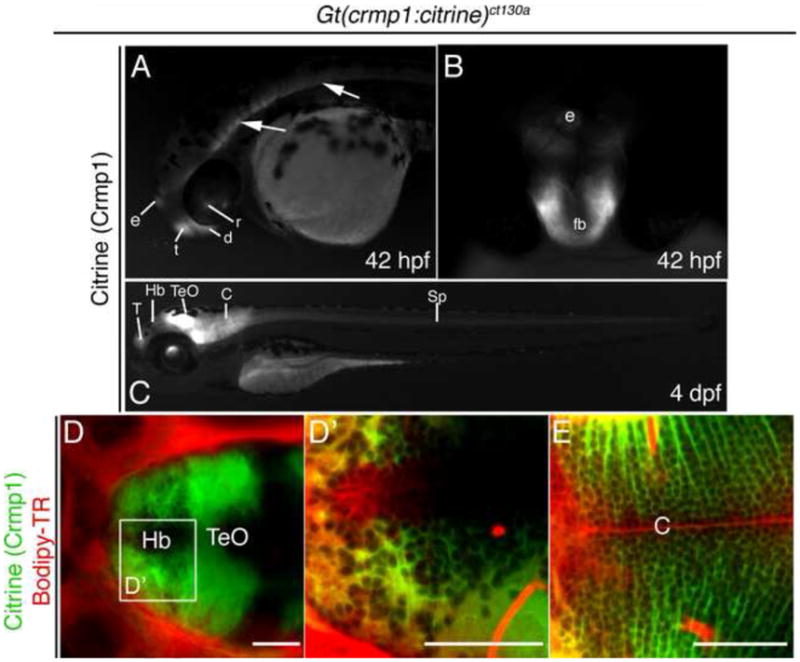
Crmp1 expression in the developing brain in live 42 hpf- 4 day fish. (A) Lateral view at 42 hpf. Crmp1 is expressed in the epiphysis (e) and forebrain regions: telencephalon (t) and diencephalons (d). Expression is also detected in the retina (r). Arrows show expression in brain and spinal cord. (B) Frontal view at 42 hpf of the forebrain (fb) and epiphysis. (C) Expression in the 4 day live larva showing expression in the T, telecephalon, Hb, habenula, TeO, optic tectum and Cb, cerebellum. Expression in spinal cord (Sp) also persists. (D–E) Larvae were treated with the vital dye Bodipy-TR (red) to act as a contrast against the citrine signal. (D) Confocal z-stack at 4 dpf, dorsal view showing expression in in Hb and TeO. (D′) Shows zoomed view but z-section. (E) Z-section at the level of cerebellum. Scale bar: 50 μm.
1.2. Further analysis of Crmp1 expression during zebrafish development compared to neuronal markers
We examined Crmp1 expression and its relationship to neuronal markers HuC/D and Neurofilament (NFM) (Fig. 4). HuC/D is expressed by newly born post-mitotic neurons and marks RNA binding proteins of the Elav family (Kim et al., 1996). There are clearly two populations of cells within the telencephalon that express Crmp1 (Fig. 4A–B). One population of Crmp1 expressing cells co-localizes with HuC/D (asterisk, Fig. 4A–B) whereas the second population of cells is HuC/D negative (arrowheads, Fig. 2D and Fig. 4A). For example, in the developing epiphysis, a proportion of Crmp1 positive cells do not express HuC/D (Fig. 4A–B). In addition, a substantial proportion of HuC/D positive neurons fail to express Crmp1 in the optic tectum at 31 hpf, whereas the majority of neurons in the telencephalon are Crmp1+ (Fig. 4A). By 42 hpf, the population of cells co-expressing HuC/D and Crmp1 has expanded in the telencephalon and the epiphysis (Fig. 4B). A ventral view at 42 hpf reveals that unlike in the telencephalon, in the diencephalon there is little to no overlap in Crmp1 and HuC/D expression (Fig. 4C and inset). Within the diencephalon, cell tracts or cell extensions can be visualized (arrows, Fig. 4C).
Figure 4.
Crmp1 expression (green) compared to neuronal markers HuC/D (A–C) and NFM (D–E) (red). Gt(crmp1:citrine)ct130a heterozygotes were stained with anti-green fluorescent protein to enhance the citrine signal and the nuclear stain (DAPI; blue) to visualize nuclei. (A–B) Dorsal view z-stack projection at the level of telencephalon (t) and optic tectum (TeO) at 31 hpf (A) and 42 hpf (B). Asterisk: co-expression of HuC/D and Crmp1. e; epiphysis. (A) Arrowheads: Neurons positive for Crmp1 in the telencephalon. Arrows: Crmp1 and HuC/D positive cells in the optic tectum (TeO). (A′) Crmp1 expression alone. (A″) Lateral reconstructed projection of the z-stack from (A). Arrowhead: Crmp1 expression devoid of HuC/D expression. Arrows: Crmp1 and HuC/D positive cells. Asterisk: HuC/D and Crmp1 positive cells in the telencephalon. (B–C) oe, olfactory epithelium. (B′) Magnified z-section from boxed region in (B) showing that majority of HuC/D cells are also positive for Crmp1 (highlighted dotted region; asterisk). Arrow: HuC/D positive and Crmp1 negative cells. Inset in (B′) shows that Crmp1 is localized to the cytoplasm (arrowheads) and excluded from the nucleus. (C) Ventral view z-stack projection of the diencephalon (d) at 42 hpf. Arrows: cell projections positive for Crmp1. Inset: magnified view of boxed region. Highlighted Crmp1 cells are negative for HuC/D. (D) Lateral view z-projection at the level of the otic vesicle (ov) and cerebellum (c). Arrowheads: axon projections that are NFM and Crmp1 positive. (E) Crmp1 cells at the level for the forebrain (asterisk) and optic tectum (arrow) are negative for NFM. (D′, E′) Ninty-degree rotated z-stack reconstructed projection of (D) and (E) respectively. (D′) Dotted outline delineates cerebellum (C) from the peripheral tissue. Asterisk: NFM and Crmp1 positive axon tracks. Scale: 50 μM (A–B, C–D), 25 μM (B′ and inset C).
By 48 hpf, NFM positive axon fascicles expressing Crmp1 can be seen emanating from the cerebellum (arrowheads, Fig. 4D), although the Crmp1 expressing domain is greater than the NFM expression within the cerebellum (Fig. 4D). This is consistent with the proposed role for Sema3a signaling in axon guidance and fasciculation (Kawasaki et al., 2002; Yamashita et al., 2007). In contrast, within the forebrain (asterisk) and optic tectum region (arrow), Crmp1 positive cells do not co-express NFM (Fig. 4E). At 48hpf, Crmp1 expression in the forebrain is remarkably similar to the radial glial marker Glial Fibrillary Acidic Protein (GFAP) (Barresi et al., 2005). It is known that glial bridge formation mediated by Slit-Robo signaling facilitates the correct formation of commissures in the zebrafish forebrain (Barresi et al., 2005). Intriguingly, there is evidence to suggest that Robo signaling modulates Semaphorin signaling during interneuron migration in the forebrain (Hernandez-Miranda et al., 2011). Thus, in a similar manner, Robo-expressing glia in the forebrain may also integrate Semaphorin signaling during commissure formation.
A significant advantage of the Crmp1 transgenic line is that it reflects expression of the endogenous protein that can be visualized at high magnification, revealing the subcellular distribution of the protein. The results reveal that Crmp1 expression is clearly restricted to cell bodies and excluded from the nucleus (arrowheads, Fig. 4B′ inset).
1.3. In situ expression of citrine in Gt(crmp1:Citrine)ct130a line during development
The above results demonstrate that Crmp1 protein is expressed as early as 24 hpf and maintained at later stages. To compare this distribution with that of the crmp1 transcript and of the citrine fusion transcript, we analyzed the expression of crmp1 in wildtype fish at 24 and 48 hpf. The 24 hpf crmp1 expression is consistent with previously published data (Schweitzer, J et al 2005) and overlaps with that of both Citrine protein and transcript expression in the Gt(crmp1:Citrine)ct130a line (Fig. 5). At 48hpf, crmp1 and citrine expression correlates well with Crmp1 protein expression observed in the genetrap line (Fig. 5B and 5D). However, crmp1 expression in the retina in wildtype fish is not as obvious as citrine expression in the genetrap line. At 72 hpf, Citrine mRNA is expressed and matches well with Citrine protein expression (Fig. 5E).
1.3. Conclusion
Gt(crmp1:Citrine)ct130a is a useful tool to study the nervous system. This transgenic line reflects the endogenous pattern of Crmp1 expression and the tagged version of the Crmp1 can be used in proteomic studies to identify binding partners in an in vivo relevant manner. By revealing expression of the endogenous protein at subcellular resolution, we show that Crmp1 is present throughout the cytoplasm and axon projections, consistent with its proposed role as a mediator of signal transduction downstream of Semaphorins and possibly other signaling pathways (Goshima et al., 1995; Uchida et al., 2005; Yamashita et al., 2007). Furthermore, the protein distribution largely correlates with that of its mRNA transcripts. The distribution of Crmp1 in subpopulations of neurons in the developing central nervous system of zebrafish is consistent with important functions in neuronal development, synaptic plasticity, and behavior.
2. Experimental Procedures
2.1. Zebrafish embryos
Wildtype and transgenic zebrafish lines were maintained according to Institutional Animal Care and Use protocols. Embryos were collected, maintained and staged according to (Kimmel et al., 1995). Gt(crmp1:citrine)ct130a AB/TL F1 were generated by crossing AB founder to TL wildtype.
2.2. 3′RACE
RNA was extracted from 3 dpf citrine positive fish and a complementary DNA (cDNA) library was generated using random hexamers. 3′ RACE was performed with the following primers: Citrine primer: 5′-GACAACCACTACCTGAGCTACC-3′ and dT15 primer: 5′-GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGGTTTTTTTTTTTTTTT-3′. A second round of amplification was performed using dT15 primer and nested Citrine primer-5′: ACATGGTCCTGCTGGAGTTC-3′. A 2.1 kb product was amplified, cloned into pGEMTeasy (Promega) and sequenced.
2.3. In situ hybridization
To generate crmp1 probe the following primers were used: crmp1 forward primer-5′-TGCATGCAGAGAATGGAGACCTGA-3′ and reverse primer containing a T7 promoter site T7-crmp1-R-5′-TAATACGACTCACTATAGGGACTGGACCATCATACATGCCACGA-3′. The 3′ RACE product was used to generate crmp1 probe. To generate citrine anti-sense probe the following primers were used with the reverse primer containing a T7 promoter site; Sp6-citrine-F-5′-GATTTAGGTGACACTATAGATGGTGAGCAAGGGCGAGGA-3′ and T7-citrine-R-5′-TAATACGACTCACTATAGGGCTTGTACAGCTCGTCCATGC-3′. In situ hybridization was carried out according to standard protocols (Jayasena et al., 2008) and detailed protocols are available upon request.
2.4. Imaging and immunofluorescent staining
Live Gt (crmp1:citrine)ct130a homozygous fish were soaked in BODIPY® TR methyl ester vital dye (Invitrogen) for 30–60 min at 28 °C to label all cells. Fish of appropriate stage were anesthetized using 0.1% Tricaine (pH 7.2) in egg water (Sigma) and mounted laterally onto imaging moulds (1% agarose in egg water). Confocal z-stack images ranging from 40–100 μM were collected using Zeiss LSM 510 microscope and Zeiss 25x LD LCI Plan-Apochromat NA 0.80 DIC 1mm Korr or Zeiss LD C-Apochromat 40X/ 1.1W Korr UV-VIS-IR objectives. Wholemount live images were collected using an Olympus MVX10 microscope and Zeiss software LSM 510 version 4. Images were processed using ImageJ 1.38x (NIH) and Adobe Photoshop 7. Z-stack projections were generated using LSM Image Examiner version 4.
Immunofluorescent staining was performed as follows. Embryos were manually deyolked with a pulled glass Pasteur pipette and fixed in 4% paraformaldehyde in 0.1M Phosphate buffer at room temperature for 1–2 hours. After several rinses in Phosphate buffer saline containing 0.1% Tween-20 (PBSTw), the embryos/ larvae were Trypsin treated (1mg/ml) at room temperature. Following a 20 minute post-fix in 4% paraformaldehyde in 0.1M Phosphate buffer at room temperature, the embryos/ larvae were blocked in 5% goat serum containing 0.5% Triton-X in PBS (5% GS+ PBSTx). Primary and secondary antibody incubations were performed at 4 °C overnight in 5% GS+ PBSTx. Post antibody washes were performed in PBS containing 0.1% Triton-X. The embryos were mounted as stated above for live confocal imaging. The following antibodies were used in this study: mouse anti-HuC/D (Molecular probes), mouse anti-NFM (Zymed), rabbit-anti-GFP (Molecular probes), goat anti-rabbit Alexa-488 (Molecular probes) and donkey anti-mouse Alexa-594 (Molecular probes).
Acknowledgments
We wish to thank the following people: Dr. Le. A. Trinh for helpful discussions and technical help, Ho-yin Leung for performing 3′ RACE and Leigh-Anne Fletcher for technical help in the Caltech Centers of Excellence in Genomic Science fish facility. This work is supported by USPHS P50HG004071.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barresi MJF, Hutson LD, Chien CB, Karlstrom RO. Hedgehog regulated Slit expression determines commissure and glial cell position in the zebrafish forebrain. Development. 2005;132:3643–3656. doi: 10.1242/dev.01929. [DOI] [PubMed] [Google Scholar]
- Charrier E, Reibel S, Rogemond V, Aguera M, Thomasset N, Honnorat J. Collapsin response mediator proteins (CRMPs) Molecular Neurobiology. 2003;28:51–63. doi: 10.1385/MN:28:1:51. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- Hernandez-Miranda LR, Cariboni A, Faux C, Ruhrberg C, Cho JH, Cloutier JF, Eickholt BJ, Parnavelas JG, Andrews WD. Robo1 Regulates Semaphorin Signaling to Guide the Migration of Cortical Interneurons through the Ventral Forebrain. The Journal of Neuroscience. 2011;31:6174–6187. doi: 10.1523/JNEUROSCI.5464-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena CS, Ohyama T, Segil N, Groves AK. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 2008;135:2251–2261. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Bekku Y, Suto F, Kitsukawa T, Taniguchi M, Nagatsu I, Nagatsu T, Itoh K, Yagi T, Fujisawa H. Requirement of neuropilin 1-mediated Sema3A signals in patterning of the sympathetic nervous system. Development. 2002;129:671–680. doi: 10.1242/dev.129.3.671. [DOI] [PubMed] [Google Scholar]
- Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL, Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neuroscience Letters. 1996;216:109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Quinn CC, Gray GE, Hockfield S. A family of proteins implicated in axon guidance and outgrowth. Journal of Neurobiology. 1999;41:158–164. [PubMed] [Google Scholar]
- Schweitzer J, Becker CG, Schachner M, Becker T. Expression of collapsin response mediator proteins in the nervous system of embryonic zebrafish. Gene Expression Patterns. 2005;5:809–816. doi: 10.1016/j.modgep.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Su K-Y, Chien W-L, Fu W-M, Yu IS, Huang H-P, Huang P-H, Lin S-R, Shih J-Y, Lin Y-L, Hsueh Y-P, et al. Mice Deficient in Collapsin Response Mediator Protein-1 Exhibit Impaired Long-Term Potentiation and Impaired Spatial Learning and Memory. J Neurosci. 2007;27:2513–2524. doi: 10.1523/JNEUROSCI.4497-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Ohshima T, Sasaki Y, Suzuki H, Yanai S, Yamashita N, Nakamura F, Takei K, Ihara Y, Mikoshiba K, et al. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3β phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer’s disease. Genes to Cells. 2005;10:165–179. doi: 10.1111/j.1365-2443.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Morita A, Uchida Y, Nakamura F, Usui H, Ohshima T, Taniguchi M, Honnorat J, Thomasset N, Takei K, et al. Regulation of Spine Development by Semaphorin3A through Cyclin-Dependent Kinase 5 Phosphorylation of Collapsin Response Mediator Protein 1. J Neurosci. 2007;27:12546–12554. doi: 10.1523/JNEUROSCI.3463-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]