Abstract
BACKGROUND & AIMS
Adenomatous polyps are precursors to colorectal cancer (CRC), whereas hyperplastic polyps (HPPs) have a small risk of progression to CRC. Mutations in KRAS are found in ~40% of CRCs and large adenomas and a subset of HPPs. We investigated the reasons that HPPs with KRAS mutations lack malignant potential; we compared the effects of Kras/KRAS activation to those of Adenomatous polyposis coli (Apc)/APC inactivation, which promotes adenoma formation.
METHODS
We activated a KrasG12D mutant allele or inactivated Apc alleles in mouse colon epithelium and analyzed phenotypes and expression of selected genes and proteins. The mouse data were validated using samples of human HPPs and adenomas. Signaling pathways and factors that contribute to Kras/KRAS-induced phenotypes were studied in intestinal epithelial cells.
RESULTS
Activation of Kras led to hyperplasia and serrated crypt architecture akin to that observed in human HPPs. We also observed loss of Paneth cells and increases in goblet cell numbers. Abnormalities in Kras-mediated differentiation and proliferation required mitogen-activated protein kinase (MAPK) signaling and were linked to activation of the Hes1 transcription factor. Human HPPs also had activation of HES1. In contrast to Apc/APC inactivation, Kras/KRAS activation did not increase expression of crypt stem cell markers in colon epithelium or colony formation in vitro. Kras/KRAS activation was not associated with substantial induction of p16INK4a protein expression in mouse colon epithelium or human HPPs.
CONCLUSIONS
Although Kras/KRAS mutation promotes serrated and hyperplastic morphological features in colon epithelium, it is not able to initiate adenoma development, perhaps in part because activated Kras/KRAS signaling does not increase the number of presumptive stem cells in affected crypts.
Keywords: Colon cancer, oncogene, tumor suppressor, transgenic mice
Introduction
While it has long been recognized that many colorectal cancers (CRCs) arise from adenomatous polyps, it is now appreciated that about 15% of CRCs likely arise via an alternative pathway – the so-called serrated adenoma pathway. 1, 2 Hyperplastic polyps (HPPs) are common lesions in the colorectum, and HPPs also display a serrated (sawtooth) gland morphology. However, unlike serrated adenomas, the epithelium in HPPs does not show dysplasia, and most HPPs, particularly small lesions in the distal colon, are presumed to have nominal if any risk of progression to CRC.2, 3 Regardless of the progression pathway, CRCs arise due to accumulated defects in genes regulating cell metabolism, proliferation, differentiation, and death.4 The gene lesions lead to novel or increased oncogene function or loss of tumor suppressor gene (TSG) function.
Inactivation of the APC (adenomatous polyposis coli) TSG is viewed as an initiating event in most adenomas and CRCs.5 The APC protein functions as a regulator of the β-catenin protein in the “canonical” (β-catenin-dependent) Wnt signaling pathway.6 In cells where both APC alleles are inactivated, coordinated phosphorylation and destruction of β-catenin is disrupted, mimicking β-catenin stabilization by Wnt ligands. β-catenin can complex with TCF (T cell factor) DNA binding proteins to co-activate transcription. The TCF-dependent transcription program induced by β-catenin stabilization in CRC appears to resemble that normally present in presumptive stem cells at the intestinal/colonic crypt base.7
Other somatic defects, such as KRAS and p53 mutations, collaborate with APC inactivation to promote progression of some adenomatous lesions to CRC.4, 5 KRAS somatic mutations are found in about 40% of CRCs.1 Mutant KRAS alleles have a key role in CRC cells, when present, as inactivation of mutant KRAS abrogated tumorigenic properties of CRC cells in in vitro and animal studies.8 Of note, KRAS mutations are present in some HPPs, including about 50% of so-called goblet cell-rich HPPs arising usually in the left colon and rectum.2 Prior studies of the phenotypic effect of Kras mutant alleles in mouse small intestine and/or colon have yielded highly variable results, ranging from no effect at all,9, 10 to increased goblet cell numbers,11 reduced Paneth cell numbers,12 epithelial hyperplasia,11, 12 epithelial architectural changes,12 and even the appearance of grossly visible tumors.13
In light of the data indicating KRAS mutations are functionally significant in human CRCs but also present in colon tumors with negligible malignant potential (e.g., HPPs), we sought to explore the functional consequences of Kras mutations in mouse colon epithelium and to compare the results to human HPPs. Using a CDX2P-G22Cre transgene, which expresses Cre in crypts of the terminal ileum, cecum, and colon14 to activate a KrasLSL-G12D mutant allele,15 we found dramatic epithelial hyperplasia and crypt architecture changes in the colon, reminiscent of those seen in human HPPs. We also observed Paneth cell loss and increased goblet cells. We established Kras mediated the differentiation and proliferation abnormalities via the mitogen activated protein kinase (MAPK) signaling pathway. We also implicated the transcriptional regulator Hes1 in altered cell fate. Notably, our studies revealed that in contrast to Apc/APC inactivation, Kras activation does not enhance expression of markers of the presumptive crypt stem cell pool or enhance colony formation in culture, leading us to propose that Kras/KRAS mutation does not initiate colorectal adenoma-carcinoma progression perhaps in part because activated Kras/KRAS signaling by itself cannot enhance formation of crypt stem cells.
Results
Activation of KrasG12D Mutant Allele in Mouse Colon Epithelium Promotes Hyperplastic Polyp-like Features
We used the CDX2P-G22Cre transgenic mouse line (referred to as G22Cre),14 which expresses Cre in epithelial cells in terminal ileum, cecum, colon and rectum, to activate a latent oncogenic KrasLSL-G12D allele.15 Cre-mediated recombination removes transcriptional stop elements, resulting in expression of the mutant KrasLSL-G12D allele under control of its endogenous regulatory elements. Using mice carrying a gene for enhanced yellow fluorescence protein (EYFP) at the Gt(ROSA)26Sor locus16 to report on Cre expression, we found G22Cre activated EYFP in epithelial cells of 40–50% crypts in proximal colon and 10–20% crypts in terminal ileum (Supplementary Figure 1B and 1E), consistent with prior data on G22Cre activity.14 In G22Cre mice carrying the EYFP reporter and the KrasLSL-G12D gene, more than 90% of crypts in proximal colon and terminal ileum had Cre-activated EYFP expression in epithelial cells (Supplementary Figure 1C and F). These findings imply KrasLSL-G12D activation may enhance proliferation and/or survival advantage of intestinal epithelial cells.
In G22Cre KrasLSL-G12D mice of one to two months of age, we observed marked hyperplasia of more than 90% of crypts in the terminal ileum, cecum, and colon compared to that of control mice (i.e. wild type mice and KrasLSL-G12D mice lacking the G22Cre transgene) (Figure 1A and B). No evidence of dysplasia, like that seen in adenomatous epithelium following Cre-mediated inactivation of both Apc alleles,14 was observed in G22Cre KrasLSL-G12D mice. Besides hyperplasia, the G22Cre KrasLSL-G12D colon epithelium showed serrated glandular morphology and increased goblet cells, reminiscent of epithelium in goblet cell-rich type human HPPs with activating somatic mutations in the KRAS or BRAF genes (Figure 1C and Supplementary Table 1). 2, 3 Increases in proliferating cells and decreases in apoptotic cells per crypt were seen in G22Cre KrasLSL-G12D epithelium (Figure 1, D–F and Supplementary Figure 2). Because the G22Cre KrasLSL-G12D mice developed one or two progressive papillary squamous lesions at the anal verge (due to stochastic Cre activation in caudal-derived epithelial cells), we could not study G22Cre KrasLSL-G12D mice beyond two months age. We used a tamoxifen-regulated Cre transgene (EGFP-IRES-CreERT2) integrated into the locus encoding leucine-rich-repeat-containing G-protein coupled receptor 5 (Lgr5), a presumptive intestinal crypt stem cell marker,17 as another approach to activate KrasG12D. In four Lgr5-EGFP-IRES-CreERT2 Kras LSL-G12D mice studied between 2 and 8 months after tamoxifen treatment, mutant Kras activation led to hyperplastic effects in intestine and colon epithelium (Supplementary Figure 3), without evidence of dysplasia or adenoma formation. For both drivers, the effects of Kras activation on Paneth cell loss, increased Goblet cells, increased proliferation, decreased cell death, and crypt hyperplasia all appeared restricted to the crypts where Kras was activated, without evidence of effects on adjacent non-targeted crypts (Supplementary Figure 4 and data not shown).
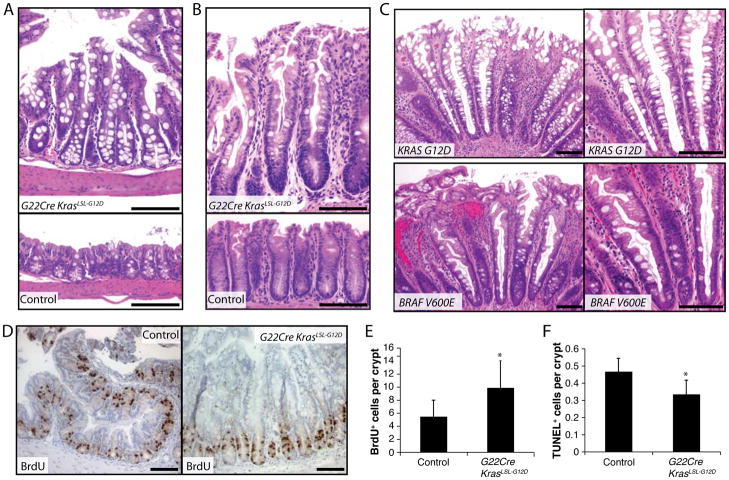
Figure 1.
Hyperplastic and hyperproliferation changes in mouse and human colon epithelium following Kras/KRAS pathway activation. (A and B) Hematoxylin and eosin (H&E) staining of proximal (A) and distal (B) colon tissues from a G22Cre KrasLSL-G12D mouse (A and B, upper) or a wild type mouse (A and B, lower). (C) H&E staining of a human HPP with KRAS G12D mutation (upper) and a HPP with BRAF V600E mutation (lower). Proliferating cells per crypt in mouse proximal colon were assessed by BrdU staining (D); BrdU incorporation was quantified (panel E; n = 4 mice and 160 crypts counted; *P < .001). (F) Apoptotic cells in proximal colon were assessed by TUNEL assay (n=4 mice; *P < .02). Scale bars, 100 μm.
Kras Activation Alters Expression of Cell Cycle Regulatory Proteins
Phospho-Erk1/2 levels were increased in Kras-activated epithelium (Supplementary Figure 5A). We found increased expression of the DNA synthesis (S)-phase cyclin E2 protein and decreased expression of cyclin-dependent kinase (Cdk) inhibitory proteins p27Kip1 and p57Kip2 in Kras-activated epithelium (Supplementary 5A). Expression of the p19ARF protein was decreased, and no change in p16INK4a protein expression was seen (Supplementary 5A and B). Transcript and protein levels were concordantly changed only for cyclin E2. Other genes showed either no change (e.g., p19ARF, p27Kip1) or divergent effects on transcript versus protein levels (e.g., p16Ink4a, p57Kip2) (Supplementary Figure 5C). These data imply that in colon epithelium protein levels of the Cdk inhibitory factors studied was largely post-transcriptional. Our results showing no change in p16INK4a protein levels in epithelium of two month old G22Cre KrasLSL-G12D mice are consistent with our analysis of 12 human HPPs, where we found p16INK4a expression in 1–5 cells per crypt of only 5–10% of crypts of 10 of 12 HPPs studied (Supplementary Figure 5D and Supplementary Table 1). The other 2 HPPs had no detectable p16INK4a staining. The data indicate p16INK4a protein is not substantially induced in human HPPs, though elevated p16INK4a protein expression has been reported in human serrated adenomas and carcinomas.18 Our findings of no significant increase in p16INK4a protein expression in epithelium of G22Cre KrasLSL-G12D mice contrasts with the reported significant increase in p16INK4a protein levels when a Villin-Cre transgene was used to activate Kras in small intestine and colon.18 Explanations for the divergent findings will be discussed below.
MEK (MAPK kinase) Inhibitor CI-1040 Treatment Blocks Hyperplastic Changes Induced by KrasG12D Activation
Ras oncogenes have been shown to engage multiple downstream pathways and effectors including the Raf-MAPK, phosphatidylinositol-3-kinase (PI3K), and Ral protein pathways.19, 20 Mutant KRAS/Kras proteins are poor activators of PI3K signaling,21 and some human CRCs with KRAS mutations also carry activating mutations in the PI3K signaling pathway.22 In CRCs, BRAF and KRAS mutations are mutually exclusive,22 arguing KRAS oncogenic signaling depends on downstream RAF-MAPK signaling. In a prior study where an intestinal fatty acid binding protein liver (Fabpl) regulatory element-driven Cre transgene targeted KrasG12D in small intestine and colon tissues, the mice showed colon epithelial hyperplasia that responded to MEK inhibitor CI-1040 treatment. 11 We found a four-day treatment with CI-1040 dramatically inhibited colon epithelial hyperplasia in G22Cre KrasLSL-G12D mice (Supplementary 6A and B) and restored p27Kip1 levels to those seen in control mice (Supplementary 6C).
KrasG12D Activation-Induced Effects on Cell Fate in Intestine and Colon Epithelium are Dependent on MAPK Signaling
Prior studies where a Fapbl-Cre transgene11 or a Villin-Cre transgene12 was used to activate the KrasLSL-G12D allele revealed increased goblet cells11, 12 and, in the case of the Villin-Cre transgene, reduced Paneth cells.12 We confirmed G22Cre-mediated KrasG12D activation increased goblet cell numbers per crypt in small intestine and colon (Figure 2, A–C), and led to a complete loss of lysozyme-expressing Paneth cells in small intestine (Figure 2A and B), and did not alter enteroendocrine cell numbers (data not shown). We obtained very similar effects on Paneth cells using the Lgr5-EGFP-IRES-CreERT2 transgene to activate KrasLSL-G12D (Supplementary Figure 7A). A 4-day treatment of G22Cre KrasLSL-G12D mice with the MEK inhibitor CI-1040 restored Paneth cells and reduced goblet cells (Figure 2, C–F), indicating the key role of MAPK signaling in Kras-induced effects on cell fate specification and differentiation. In light of the persistence of Kras-mutant small intestinal crypts lacking Paneth cells for up to 8 months, our findings raise questions about the recently suggested role for Paneth cells to maintain a crypt niche for intestinal stem cells. 23
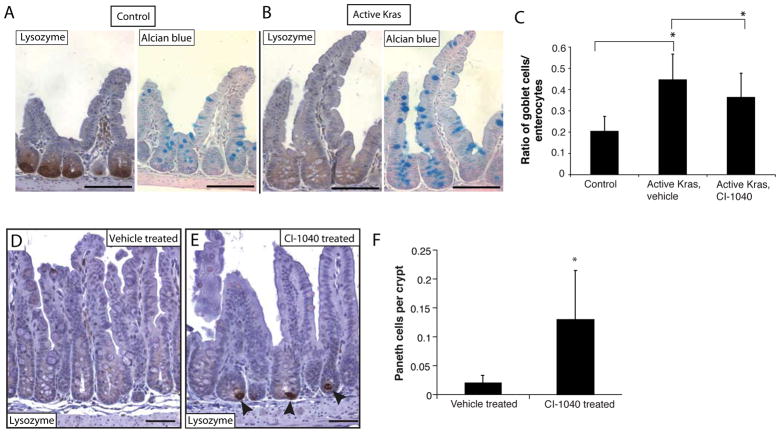
Figure 2.
Mutant Kras activation promotes increased goblet cells and Paneth cell loss, via RAS-MAPK-dependent signaling. Paneth cells, with lysozyme staining (left side, panels A and B) and goblet cells, with Alcian Blue staining (right side, panels A and B) in ileum. Scale bars, 100 μm. (C) Ratio of goblet cells vs. enterocytes in ileal crypts following CI-1040 treatment. (D and E) Recovery of Paneth cells in ileum of G22Cre KrasLSL-G12D mice following CI-1040 treatment. Scale bars, 50 μm. Arrowheads indicate lysozyme-positive Paneth cells. (F) Quantification of Paneth cells from G22Cre KrasLSL-G12D mice treated with vehicle or CI-1040. *P < .001.
MAPK and Notch Pathway Activation Collaborate in Mediating Effects on Goblet and Paneth Cells in Kras/KRAS-Activated Epithelium
Prior studies implicated Notch signaling in cell fate determination in intestinal epithelium.24, 25 We thus sought to address effects of Kras/KRAS activation on the Notch pathway target genes Hes1 and Hey1. Increased Hes1 and Hey1 transcripts were found in Kras-activated mouse ileal epithelium (Figure 3A). Increased Hes1 protein expression following Kras activation was confirmed by Western blot (Figure 3A) and by immunohistochemical (IHC) staining (Figure 3B). Human HPPs with KRAS or BRAF mutations showed strong HES1 staining throughout most hyperplastic crypts (Figure 3D and Supplementary table 1), whereas HES1 staining in normal colon epithelium was limited to the lower third of the crypt (Figure 3C), in the region populated mostly by transit amplifying cells.
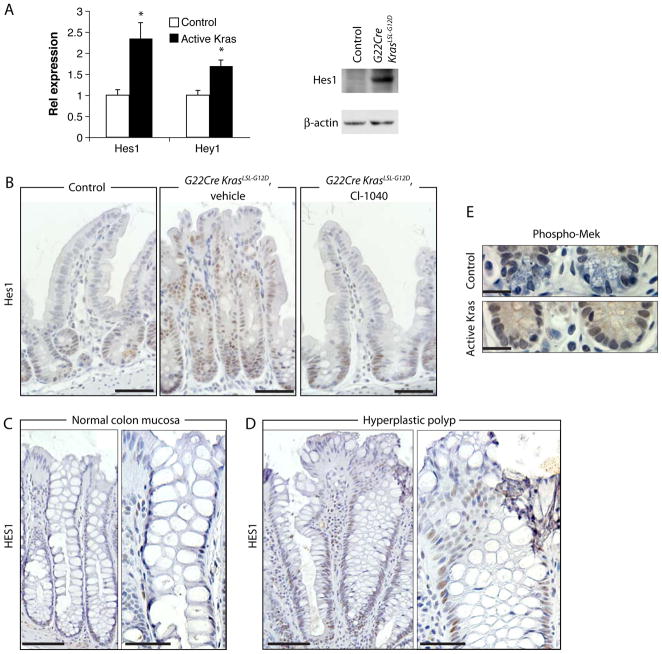
Figure 3.
MAPK and Notch pathway activation collaborate in mediating effects on goblet and Paneth cells in Kras-activated epithelium. RNA and proteins were isolated from distal ileum of four G22Cre KrasLSL-G12D mice and four control mice. (A) Hes1 and Hey1 expression was measured by qRT-PCR and normalized to U6 expression (left). *P < .01. Hes 1 Western blot analysis shown at right. (B) IHC staining for Hes1 in ileum of a wild type mouse (left), vehicle-treated (middle), or CI-1040-treated (right) G22Cre KrasLSL-G12D mice. Scale bars, 50 μm. (C and D) Hes 1 IHC analysis in normal human colon mucosa (C) or HPPs (D). Representative images of low (C and D, left) or higher magnification (C and D, right) are shown. Scale bars, 100 μm for panels C and D, left, and 50 μm for panels C and D, right. (E) IHC detection of phospho-MEK at the ileum crypt base. Scale bar, 20 μm.
In Kras-activated mouse intestinal epithelium, phospho-Mek-expressing cells replaced the phospho-Mek-negative Paneth cells normally present at the crypt base (Figure 3E). While Hes1 is a NOTCH pathway target gene, Hes1 has been reported to be activated via MAPK signaling in neuroblastoma cells.26 Given prior reports that Hes1−/− mice show premature Paneth cell development,27 we hypothesized MAPK-dependent Hes1 activation may underlie Paneth cell loss. Using rat IEC-6 intestinal cells stably expressing a G12V mutant KRAS cDNA, we found the MEK inhibitors PD98059 or U0126 blocked KRAS-induced activation of Hes1 and Notch1 (Figure 4A and B). Similarly, the MEK inhibitor CI-1040 blocked Hes1 induction in Kras-activated mouse epithelium in vivo (Figure 3B). We detected a strong increase in levels of the activated (cleaved) cytosolic domain of Notch1 in IEC-6/KRAS G12V cells (Figure 4C). To assess if KRAS-induced activation of Hes1 depends on Notch signaling, we blocked NOTCH signaling in IEC-6/KRAS G12V cells with the γ-secretase inhibitor, DAPT. DAPT potently blocked Notch activation (Figure 4C), but only partially inhibited the KRAS effect on Hes1 (Figure 4D). Our data indicate Kras/KRAS activation can induce Hey1/Hes1 expression via MAPK signaling, and MAPK-dependent induction of Hes1 is only partially dependent on Notch. Hes1 activation may contribute to the Paneth cell loss observed, but a role for Hes1 activation in goblet cell loss is inconsistent with prior data showing that NOTCH loss-of-function leads to increased goblet cell numbers.24, 25
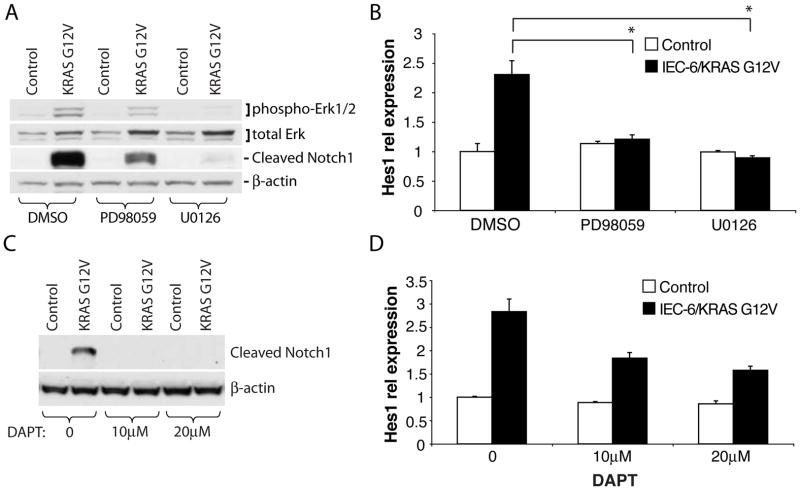
Figure 4.
IEC-6 cells stably expressing a human KRAS G12V mutant or control vector were treated for 24 hr with MEK inhibitors (50 μM PD98059, 20 μM U0126) or DMSO (A and B), or with DMSO or γ-secretase inhibitor DAPT at concentrations of 10μM, and 20μM (C and D). Western blot analysis of phospho-Erk, total Erk, cleaved Notch1 and β-actin in (A), or cleaved Notch1 and β-actin in (C). Hes1 expression was measured by qRT-PCR and normalized to β actin expression (B and D). In (A), *P < .01.
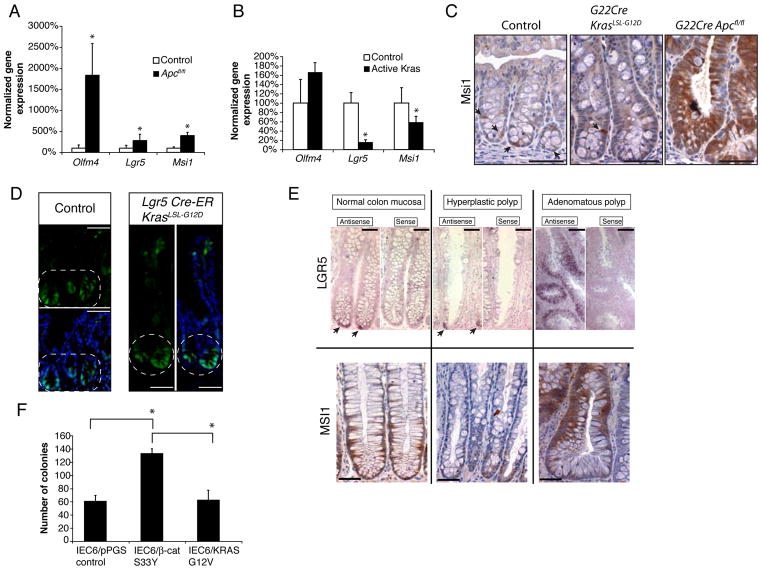
Kras/KRAS Activation Does Not Expand Expression of Crypt Stem Cell Markers in Colon Epithelium and Does Not Enhance Colony Formation In Vitro
Prior studies of van de Wetering et al suggested APC inactivation and the resultant β-catenin dysregulation in intestinal epithelium leads to a TCF-dependent transcription program resembling that in normal intestinal stem cells.7 Presumptive markers for intestinal and colon stem cells include the Wnt pathway-regulated target gene Lgr5,17 as well as the Musashi-1 (Msi1) and olfactomedin-4 (Olfm4) genes,28–30 neither of which is known to be regulated by Wnt signaling. We found dramatically induced expression of all three presumptive crypt stem cell markers in mouse colon epithelium when both Apc alleles were inactivated (G22Cre APCfl/fl, Figure 5A). In contrast, we observed a decrease in the relative abundance of Lgr5 and Msi1 gene expression in Kras-activated colon epithelium and no change in Olfm4 expression (Figure 5B). The relative decrease in Lgr5 and Msi1 expression following Kras activation reflects the dilution of the cell population in a crypt that express Lgr5 and Msi1, as Kras crypts were hyperplastic with substantially greater numbers of epithelial cells per crypt than normal crypts. Only a few cells near the crypt base in normal mouse colon epithelium and in Kras-activated colon epithelium expressed Msi1 (Figure 5C and Supplementary Figure 7B). In contrast, Msi1 expression was broadly activated in Apc-mutant crypts. Using the Lgr5-CreERT2 targeted allele, where integrated enhanced green fluorescence protein (EGFP) sequences mark cells with endogenous Lgr5-expression,17 we found Kras-activated epithelium showed only the few EGFP-positive cells at crypt base in colon and terminal ileum that were similarly visualized in normal epithelium (Figure 5D and Supplementary Figure 7A). Expression of LGR5 and MSI1 in normal human colon epithelium and HPPs was detected in a minority of cells at the crypt base (Figure 5E and Supplementary Table 1). In contrast, LGR5 and MSI1 were broadly expressed in glands of human colon adenomas (Figure 5E).
Figure 5.
Kras/KRAS activation does not increase presumptive colon stem cells. (A and B) RNA was extracted from proximal colon epithelium of four G22Cre Apcfl/fl, seven G22Cre KrasLSL-G12D, and seven control mice. Expression of Olfm4, Lgr5 and Msi1, relative to U6, was measured by qRT-PCR. (C) IHC staining for Msi1 in proximal colon of wild type, G22Cre KrasLSL-G12D, and G22Cre Apcfl/fl mice. Arrows indicate Msi1-expressing cells near the crypt base. Scale bars, 50 μm. (D) GFP imaging in proximal colon tissues counter-stained with Hoechst 33342. Lgr5-EGFP-IRES-CreERT2 KrasLSL-G12D mice and Lgr5-EGFP-IRES-CreERT2 control mice were analyzed after tamoxifen treatment to activate Cre. The circled region denotes Lgr5 expression restricted to the crypt base. Scale bars, 50 μm. (E) LGR5 and MSI1 expression in human HPPs, adenomas, and normal colon mucosa. (upper panels) In situ hybridization was performed with digoxigenin-labeled LGR5 antisense or control LGR5 sense riboprobes. Arrows indicate LGR5-expressing cells at the crypt base. Scale bars, 50 μm for normal mucosa and HPPs, and 100 μm for adenomas. (lower panels) IHC analysis of MSI1 expression. Scale bars, 50 μm. (F) Colony formation assay with IEC-6 cells expressing S33Y (serine 33 substitution with tyrosine) mutant β-catenin, KRAS G12V, or control vector. *P < .01.
To address the functional significance of these stem cell marker expression differences, we studied gene expression and colony forming ability following Kras activation or β-catenin dysregulation in IEC-6 cells. In IEC-6/KRAS G12V cells, reduced Msi1 expression was seen, though no significant inhibition of Lgr5 expression was found (Supplementary Figure 8A). The colony forming ability of IEC-6/KRAS G12V cells was similar to that of control IEC-6 cells, whereas expression in IEC-6 cells of a cancer-derived S33Y missense mutant form of β-catenin to model consequences of APC inactivation led to a 2-fold increase in the number and size of colonies (Figure 5F and Supplementary Figure 8B). Our studies and data do not conclusively define whether β-catenin dysregulation enhanced colony formation because of a general over-expression of Wnt target genes or because of a Wnt-induced intestinal cell phenotype. Nonetheless, our data indicate that while Apc/APC inactivation is associated with enhanced crypt stem cell gene expression and phenotypes, Kras/KRAS activation is not linked to an expansion of the crypt progenitor pool in intestinal and colon epithelium.
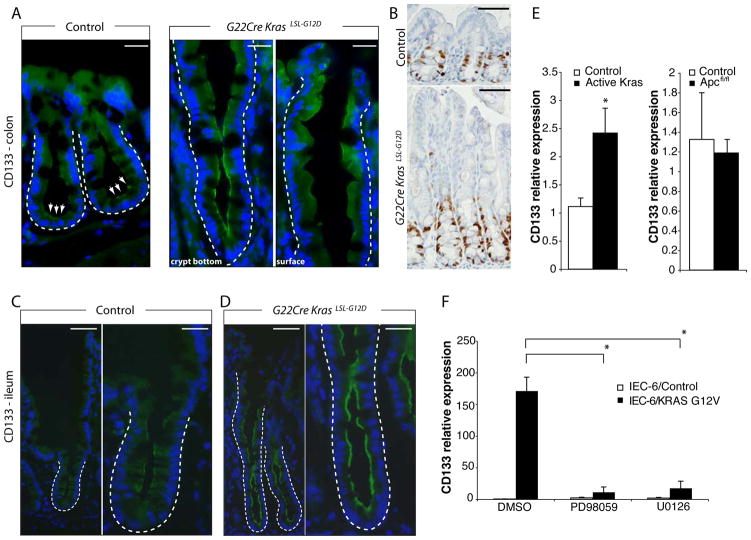
Kras Activation Enhances CD133 Expression, Marking a Potential Transit Amplifying Population, Via MAPK-Dependent Signaling
The CD133 (prominin-1) protein has been reported as a possible cancer stem cell marker, including in CRC.31 CD133 expression is not restricted to stem cells but is also prominently expressed in the transit amplifying pool in normal intestinal epithelium.32 In colon and terminal ileum epithelium from G22Cre KrasLSL-G12D mice, we found the fraction of cells expressing CD133 and also the staining intensity of the positive cells substantially increased (Figure 6A, C, and D). In line with this data, CD133 transcript levels were increased 2.5-fold (Figure 6E). Interestingly, in proximal colon of G22Cre KrasLSL-G12D mice, the proliferating cell population marked by Ki67 staining (Figure 6B) was only a subset of the CD133-positive population, while in small intestine, cells expressing detectable CD133 levels roughly corresponded to the transit amplifying cells and stem cells (Figure 6C and D, and data not shown). Thus, CD133 expression may mark different cell populations in small intestine and colon. In IEC-6 cells, KRAS G12V expression dramatically increased CD133 expression and the MEK inhibitors PD98059 and U0126 potently suppressed the induction (Figure 6 E and F).
Figure 6.
Kras activation induces CD133 expression. (A) Proximal colon tissues from wild type (left) or G22Cre KrasLSL-G12D (right) mice were stained for CD133 (green), and counter-stained with Hoechst 33342 (Blue). In panel A, right side, the crypt base (left) and a region close to the luminal surface (right) from a single crypt are shown. Each crypt is outlined with dashed lines. White arrows indicate the CD133-positive cells at the crypt base region in normal epithelium. Scale bars, 20 μm. (B) IHC staining for Ki67 in proximal colon of same mice as (A). Scale bars, 50 μm. CD133 (green) immunofluorescence in mouse distal ileum of wild type control (C) or G22Cre KrasLSL-G12D mice (D). Representative low (left) or high magnification (right) images are shown. The outlined areas indicate CD133-positive cells, which may include progenitor and transit amplifying cells. Scale bars, 50 μm for left panels, and 20 μm for right panels. (E) qRT-PCR analysis for CD133 expression in mouse proximal colon epithelium. (F) IEC-6/KRAS G12V or IEC-6/Control cells were treated with 50 μM PD98059, 20 μM U0126 or DMSO, and CD133 gene expression was measured by qRT-PCR and normalized to β-actin expression. For E and F, *P < .01.
No significant difference in CD133 transcript levels was seen between Apc-mutant and control colon epithelium (Figure 6E), in spite of the fact that Apc-mutant epithelium shows a major expansion of presumptive stem cells. We also found β-catenin activation in IEC-6 cells modestly inhibited CD133 expression (Supplementary Figure 8C). Our observations are therefore consistent with prior arguments that CD133 expression may not specifically mark colon crypt progenitor cells,32 but might instead mark a transit amplifying cell population. Overall, we conclude Kras-induced hyperplasia is caused by a MAPK-dependent increase of the transit amplifying cells marked by CD133 and Ki67, without concomitant increase of crypt stem cells.
Discussion
Roughly 40% of human CRCs and larger adenomas carry KRAS activating mutations.5, 33 Mutant KRAS alleles are functionally significant in advanced CRC cells.8 In light of these data on KRAS mutations in CRC pathogenesis, it has not previously been clear why human HPPs with somatic KRAS (or BRAF) mutations have nominal risk of progressing to CRC.2 One well-documented difference between colon adenomas, which are precursor lesions for CRC, and HPPs is that most adenomas have APC inactivation and resultant β-catenin dysregulation, whereas HPPs lack defects in APC or β-catenin.2
We hypothesized Kras/KRAS mutations might not increase stem cell numbers in the glands of hyperplastic colon epithelial lesions. Using the presumptive stem cell markers Lgr5, Msi1, and Olfm4, we demonstrated that, in contrast to Apc/APC inactivation, Kras/KRAS activation does not increase the presumptive colon stem cell pool in hyperplastic epithelium of the mouse or in human HPPs. Hence, while HPP epithelium is more proliferative than normal epithelium, because stem cell numbers are not increased in HPP crypts, HPP epithelium may be at no greater risk than normal epithelium to acquire rate-limiting mutations for CRC development. This stands in contrast to adenomatous epithelium, which appears to show a substantial increase in stem cells in each crypt. In the work here we used multiple markers presumed to define intestinal stem cells. While Lgr5 is both a presumptive marker of intestinal stem cells and a direct target of β-catenin/TCF action,17, 34 there are no data to suggest that Msi1 or Olfm4 are directly regulated by β-catenin/TCF. As such, increased Msi1 and Olfm4 expression in Apc/APC-mutant epithelium presumably reflects an increase in stem cells and not simply activation of β-catenin/TCF target genes. Furthermore, besides the effects on stem cell markers in mouse and human tissues, we found that in IEC-6 cells Kras/KRAS activation does not activate crypt stem cell marker expression or stimulate colony formation in vitro.
Our findings on hyperplasia and altered differentiation arising from expression of mutant KrasG12D allele are similar to data from Haigis and Trobridge,11, 12 where Villin-Cre or Fabpl-Cre transgenes were used to activate KrasG12D, respectively. However, our findings contrast with data from transgenic overexpression of mutant Kras in intestinal cells or activation of mutant Kras with the AhCre transgene, where essentially no proliferation or differentiation effects were reported.9, 10 Differences in phenotypes observed in the various studies with Cre recombinase transgenes likely reflect differences in the particular cells expressing the mutant Kras protein, and transgenic over-expression studies may also be influenced by expression level-dependent effects. Finally, we note our studies using the CDX2-G22Cre and Lgr5-Cre transgenes and those of Trobridge et al. using a Villin-Cre transgene to activate KrasG12D differ from those in the recent paper from Bennecke et al. where it was reported that KrasG12D activation with a Villin-Cre transgene led to hyperplasia and a serrated morphology but no observed differences in cell proliferation or death.18 The differences between the Trobridge et al. and Bennecke et al. papers, both using the Villin-Cre transgene, are difficult to reconcile, though subtle differences in mouse genetic backgrounds may have played a role. With regard to our results versus those of Bennecke et al., the CDX2-G22Cre and Lgr5-Cre transgenes we used are likely expressed in different subsets of epithelial cells than the Villin-Cre transgene, particularly progenitor cells at the crypt base. While the data of Bennecke et al. suggest a tumor suppressive role for p16INK4a-dependent cellular senescence in their particular model, the lack of clearly elevated p16INK4a expression in a significant fraction of the epithelial cells of the 12 human HPPs analyzed in our study argues against a major role for p16INK4a induction in most human HPPs. This raises the possibility that the studies of Bennecke and colleagues might better model the situation for dysplastic serrated adenomas, which are known to have malignant potential. p16INK4a-dependent cellular senescence could play a tumor suppressive role in progression of these particular lesions.18
In addition to hyperplastic changes, we also observed altered differentiation that was dependent on MAPK signaling, namely, increased goblet cells and Paneth cell loss. To our knowledge, our studies are the first to show increased HES1 expression in human HPPs. While Hes1 is a NOTCH pathway target gene, it is also activated via MAPK signaling.26 Kras-induced activation of HES1 in our in vitro system was completely dependent on MAPK signaling and only partially dependent on NOTCH1 signaling. While we saw NOTCH1 activation (as measured by its cleaved intracellular form) upon expression of mutant KRAS, in our model, HES1 expression is only partially driven by NOTCH signaling (possibly downstream of MAPK signaling). Hes1-deficient newborn (P0) mice show precocious differentiation of Paneth cells;27 thus, increased Hes1 expression could have contributed to the absence of Paneth cells in our studies. In turn, it seems unclear if increased NOTCH-independent expression of HES1 could increase goblet cell numbers, as goblet cell increases have previously been linked to NOTCH signaling loss-of-function.24, 25
The observation that Kras mutation can trigger malignant progression in some mouse tissues, such as lung,15 but not in colon, highlights tissue-specific differences in effects for particular oncogene lesions in the mouse and more than likely in man as well. Our findings do not implicate a tumor suppressive role for p16INK4a in the pathogenesis of human HPPs,18 but our data do offer an intriguing possible explanation for why KRAS mutations in human colon are not sufficient to generate a clonally related cell population with enhanced potential to progress to malignancy. Hence, future studies of the differing effects of Kras/KRAS in promoting tumor development in one tissue versus another should consider organ-specific mechanisms of stem cell expansion and not solely senescence-related effects of Kras/KRAS. Finally, insights into the presumed cooperative effects of APC, KRAS, and other mutations in human colorectal tumorigenesis will require further studies, as the effects of KRAS mutations in concert with other gene defects likely reflect complex tissue and genotype-dependent interactions.35
Material and Methods
Mice
KrasLSL-G12D/+ mice have been described previously.15 We crossed KrasLSL-G12D/+ mice to CDX2P9.5-G22Cre (G22Cre) transgenic mice to activate the mutant Kras allele in the intestinal tract.14 To monitor Cre expression, G22Cre mice or G22Cre KrasLSL-G12D/+ mice were intercrossed with mice carrying the Gt(ROSA)26Sor tm1(EYFP)Cos/J reporter allele (The Jackson Laboratory, Bar Harbor, ME). To target Apc alleles, G22Cre mice or CDX2P 9.5-NLS Cre mice were intercrossed with mice homozygous for Apc targeted alleles (ApcloxP/loxP, 580S) as described.14, 36 To study effects of Kras activation on Lgr5 expression, we crossed KrasLSL-G12D/+ and Lgr5-EGFP-IRES-creERT2 (B6.129P2-Lgr5tm1(cre/ERT2)Cle/J) mice.17 The resultant mice were injected twice intraperitoneally with tamoxifen (200 mg/kg weight; Sigma-Aldrich, St. Louis, MO) dissolved in corn oil. For MEK inhibition, 25-day-old G22Cre KrasLSL-G12D/+ mice were injected intraperitoneally with CI-1040 (50mg/kg weight) three times daily for 4 days.
Cell Lines and Plasmids
The rat intestinal epithelial line IEC-6 was obtained from American Type Culture Collection (ATCC). Polyclonal IEC-6 lines stably expressing mutant KRAS (a glycine to valine mutation at codon 12; G12V) or control vector were obtained following infection with pPGS-CMV-CITE-neo vector-based retroviruses.37 For MEK inhibitor studies, IEC-6/KRAS G12V cells and control cells were exposed to 50μM PD98059 (Cell Signaling Technology, Danvers, MA), or 20μM U0126 (Cell Signaling Technology) for 24 hr. To block Notch function, cells were treated with γ-secretase inhibitor, DAPT (Sigma-Aldrich) at concentrations of 10μM, and 20μM for 24 hr.
Immunohistochemistry and Immunofluorescence
Sections of paraffin-embedded human or mouse tissues were subjected to IHC analysis as described.37 Hes1 Immunostaining on human HPP samples was scored on a four level scale for intensity (−, absent; +/−, weak; +, moderate; ++, strong) in the cell nucleus. The Student t-test was used to determine statistical significance between groups with strong or moderate versus weak/no Hes1 staining. The de-identified human colon specimens were provided by Dr. Joel Greenson from the Department of Pathology at the University of Michigan Health System (Ann Arbor, MI). Standard immunofluorescence staining was performed on 6-μm frozen sections. See Supplementary Method for details.
In Situ Hybridization
A 969bp PCR product spanning Lgr5 nucleotides 24 to 992 was subcloned into the TA-cloning vector pGEM-T easy (Promega Corp., Madison, WI). Digoxigenin-labeled riboprobes were prepared using the DIG RNA labeling kit (Roche Applied Sciences). In situ hybridization assays on 4 human hyperplastic polyps, 3 human colon adenomas and 3 normal colon samples were performed as described.37
Western Blot Analysis
Western blot analyses on lysates from mouse colon mucosa or terminal ileum or IEC-6 cells were performed as described.37 See Supplementary Materials for information on antibodies.
Quantitative Reverse Transcription (RT)-PCR (qRT-PCR)
cDNA was synthesized using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). qRT-PCR was performed with an ABI Prism 7300 Sequence Analyzer using a SYBR green fluorescence protocol (Applied Biosystems). Primer sequences will be provided upon request.
Statistical Analysis
All data were evaluated by Student’s t test and asterisks denote significance with P < .05. Error bars denote S.D.
Supplementary Material
Acknowledgments
We thank Dr. Hideyuki Okano for generously providing the Msi1 antibody and Dr. Joel Greenson for providing human colon tissue. We thank Dr. Shuling Fan for help with immunofluorescence microscopy, Cameron Bothner for graphics assistance, and Brandon Fishman for comments on the paper.
This work was supported by NIH grant CA082223 and DOD award W81XWH-09-2-0014.
Abbreviations
- APC
adenomatous polyposis coli
- Cdk
cyclin dependent kinase
- EGFP
enhanced green fluorescent protein
- ERK
extracellular signal-regulated kinase
- EYFP
enhanced yellow fluorescent protein
- Fabpl
fatty acid binding protein liver
- H&E
hematoxylin and eosin
- HPP
hyperplastic polyp
- IHC
immunohistochemical
- ISH
in situ hybridization
- Lgr5
leucine-rich-repeat-containing G-protein coupled receptor 5
- MAPK
mitogen activated protein kinase
- MEK
MAPK kinase
- Msi1
Musashi-1
- Olfm4
Olfactomedin-4
- PI3K
phosphatidylinositol-3-kinase
- TCF
T-cell factor
- TSG
tumor suppressor gene
Footnotes
Contribution of authors
Study design and drafting of the manuscript: Ying Feng, Guido T. Bommer and Eric R. Fearon Acquisition, analysis and interpretation of data: all the authors
No conflicts of interest exist.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hamilton SR. Prevention and Early Detection of Colorectal Cancer. WB Saunders Ltd; 1996. Pathology and Biology of Colorectal Neoplasia. [Google Scholar]
- 2.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 3.East JE, Saunders BP, Jass JR. Sporadic and syndromic hyperplastic polyps and serrated adenomas of the colon: classification, molecular genetics, natural history, and clinical management. Gastroenterol Clin North Am. 2008;37:25–46. v. doi: 10.1016/j.gtc.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 6.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.van de Wetering M, Sancho E, Verweij C, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 8.Shirasawa S, Furuse M, Yokoyama N, et al. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 9.Kim SH, Roth KA, Moser AR, et al. Transgenic mouse models that explore the multistep hypothesis of intestinal neoplasia. J Cell Biol. 1993;123:877–893. doi: 10.1083/jcb.123.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sansom OJ, Meniel V, Wilkins JA, et al. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc Natl Acad Sci U S A. 2006;103:14122–14127. doi: 10.1073/pnas.0604130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haigis KM, Kendall KR, Wang Y, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trobridge P, Knoblaugh S, Washington MK, et al. TGF-beta receptor inactivation and mutant Kras induce intestinal neoplasms in mice via a beta-catenin-independent pathway. Gastroenterology. 2009;136:1680–1688. e7. doi: 10.1053/j.gastro.2009.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen KP, el-Marjou F, Pinto D, et al. Targeted expression of oncogenic K-ras in intestinal epithelium causes spontaneous tumorigenesis in mice. Gastroenterology. 2002;123:492–504. doi: 10.1053/gast.2002.34786. [DOI] [PubMed] [Google Scholar]
- 14.Akyol A, Hinoi T, Feng Y, et al. Generating somatic mosaicism with a Cre recombinase-microsatellite sequence transgene. Nat Methods. 2008;5:231–233. doi: 10.1038/NMETH.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivas S, Watanabe T, Lin CS, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 18.Bennecke M, Kriegl L, Bajbouj M, et al. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell. 2010;18:135–146. doi: 10.1016/j.ccr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 20.Mitin N, Rossman KL, Der CJ. Signaling interplay in Ras superfamily function. Curr Biol. 2005;15:R563–574. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Zhu T, Guan KL. Transformation potential of Ras isoforms correlates with activation of phosphatidylinositol 3-kinase but not ERK. J Biol Chem. 2004;279:37398–37406. doi: 10.1074/jbc.M405730200. [DOI] [PubMed] [Google Scholar]
- 22.Baldus SE, Schaefer KL, Engers R, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–799. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 23.Sato T, van Es JH, Snippert HJ, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fre S, Huyghe M, Mourikis P, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 25.van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 26.Stockhausen MT, Sjolund J, Axelson H. Regulation of the Notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp Cell Res. 2005;310:218–228. doi: 10.1016/j.yexcr.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki K, Fukui H, Kayahara T, et al. Hes1-deficient mice show precocious differentiation of Paneth cells in the small intestine. Biochem Biophys Res Commun. 2005;328:348–352. doi: 10.1016/j.bbrc.2004.12.174. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura S, Wakabayashi N, Toyoda K, et al. Expression of Musashi-1 in human normal colon crypt cells: a possible stem cell marker of human colon epithelium. Dig Dis Sci. 2003;48:1523–1529. doi: 10.1023/a:1024763723240. [DOI] [PubMed] [Google Scholar]
- 29.Potten CS, Booth C, Tudor GL, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 30.van der Flier LG, Haegebarth A, Stange DE, et al. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 31.Zhu L, Gibson P, Currle DS, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snippert HJ, van Es JH, van den Born M, et al. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136:2187–2194. e1. doi: 10.1053/j.gastro.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 34.Van der Flier LG, Sabates-Bellver J, Oving I, et al. The Intestinal Wnt/TCF Signature. Gastroenterology. 2007;132:628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 35.Janssen KP, Alberici P, Fsihi H, et al. APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology. 2006;131:1096–1109. doi: 10.1053/j.gastro.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Hinoi T, Akyol A, Theisen BK, et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007;67:9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 37.Feng Y, Bommer GT, Zhai Y, et al. Drosophila split ends homologue SHARP functions as a positive regulator of Wnt/beta-catenin/T-cell factor signaling in neoplastic transformation. Cancer Res. 2007;67:482–491. doi: 10.1158/0008-5472.CAN-06-2314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.