Abstract
Mechanisms regulating plant host differentiation of the nitrogen-fixing root nodules remain mostly unknown. Sinorhizobium meliloti induces this process in Medicago sativa in which the Mszpt2-1 gene is expressed in vascular bundles of roots and nodules. This gene codes for a Krüppel-like zinc finger protein, a class of transcription factors involved in many animal developmental processes. Expression of Mszpt2-1 in yeast cells conferred osmotic tolerance. Antisense plants grew normally but developed nonfunctional nodules, in which differentiation of the nitrogen-fixing zone and bacterial invasion were arrested. Hence, a vascular bundle-associated Krüppel-like gene is required for the formation of the central nitrogen-fixing zone of the root nodule.
Keywords: TFIIIA, nitrogen fixation, alfalfa
Bacteria of the Rhizobiaceae family can induce the development of a symbiotic nodule on the roots of leguminous host plants. In this organ rhizobia are able to fix atmospheric nitrogen. Symbiotic nodule development is controlled by specific signal exchanges between the two partners (see Schultze and Kondorosi 1998, and references therein). This organogenesis starts with the entry of rhizobia into the root through infection threads and the division of inner cortical cells. Bacteria are then released from the infection threads into the cytoplasm of the primordium cells and differentiate into nitrogen-fixing bacteroids. In alfalfa, nodules of indeterminate type are formed where distinct zones can be observed, following a differentiation gradient: a persistent apical meristem (zone I), an invasion zone (II) where infection by Sinorhizobium meliloti occurs, an interzone II–III that accumulates amyloplasts, a central or symbiotic zone (III) where nitrogen fixation takes place in the bacteroids, and a senescent zone (IV; Vasse et al. 1990). These central tissues are surrounded by peripheral cell layers, containing vascular bundles connecting the nodule to the root.
Plant mutants affected in their symbiotic behavior were isolated in several leguminous species (Schultze and Kondorosi 1998), but the first regulatory gene has only been cloned very recently (Schauser et al. 1999). Numerous genes coding for transcripts induced at different stages of nodule development, the so-called nodulin genes, have been characterized (Bladergroen and Spaink 1998). However, their role in nodulation has been investigated using functional approaches only in few cases (Cheon et al. 1993; Csanadi et al. 1996; Charon et al. 1999). Hence, little is known about regulatory genes controlling nodule organogenesis.
Krüppel-like (or TFIIIA) zinc finger proteins appear to be members of the largest family of eukaryotic transcription factors (300–500 are estimated to be encoded in the human genome; Bray et al. 1991), which play central roles in various tissue-specific differentiation as well as general housekeeping processes. In plants, several TFIIIA zinc finger proteins have been described and certain members of the petunia EPF-type family showed DNA-binding activity (Takatsuji 1998). Only two plant TFIIIA genes, SUPERMAN in Arabidopsis and INDETERMINATE in maize, have been functionally characterized. A mutation in SUPERMAN caused homeotic changes in floral organs (Sakai et al. 1995), whereas INDETERMINATE mutants showed delayed flowering, suggesting its involvement in the vegetative to floral transition of the meristem (Colasanti et al. 1998). Plant Krüppel-like genes are expressed predominantly in flowers, although transcripts of certain members were detected in vegetative tissues such as roots (Takatsuji 1998).
Here we demonstrate that a Krüppel-like zinc finger gene is strongly induced in the vascular tissues surrounding the symbiotic root nodule. Antisense Mszpt2-1 plants were affected in the formation of the central nitrogen-fixing region of this organ, suggesting that this gene is involved in a non-cell-autonomous mechanism controlling root nodule differentiation.
Results
Previously we have identified several transcripts induced during nodulation of alfalfa (Medicago sativa; Frugier et al. 1998). One of these, Mscp17 (renamed Mszpt2-1), codes for a putative Krüppel-like Cys-2/His-2 zinc finger protein (Fig. 1A). Sequence analysis of the complete Mszpt2-1 cDNA revealed best similarities to Zpt2-3 from petunia (61% identity, 70% homology; Takatsuji et al. 1994), to Stz and Atzpt2-10 (renamed from Zat10) from Arabidopsis (53% identity, 65% homology; Lippuner et al. 1996; Meissner and Michael 1997), and to Scof-1 from soybean (67% identity, 75% homology; Genbank accession no. U68763), indicating that it belongs to the EPF-type family. These genes code for Krüppel transcription factors divergent from SUPERMAN and INDETERMINATE, and contain two canonical Cys2/His2 zinc finger motives (CX2CX3FX5LX2HX3H consensus; Fig. 1A, residues 86–106 and 142–162), carrying the invariant QALGGH motif (residues 97–102 and 152–157) specific to plant EPF-type zinc finger proteins (Takatsuji 1998). Mutation in this domain of the homologous EPF-type gene Zpt2-2 abolished the DNA-binding activity of the encoded protein (Kubo et al. 1998), whereas in SUPERMAN, it led to altered flower development (Sakai et al. 1995). Other features of the Mszpt2-1 protein are (1) the presence of a hydrophobic region in the carboxy-terminal part (the DLNL box, residues 192–202) conserved in most TFIIIA plant zinc fingers and likely involved in protein–protein interactions, and (2) a putative nuclear localization signal in the amino-terminal part (NLS box, consensus: KR/KXR/K, Chelsky et al. 1989; residues 34–40). Southern blot analysis under stringent conditions with the Mszpt2-1 probe revealed one strongly and another faintly hybridizing band in the diploid Medicago truncatula, whereas two main bands associated to two fainter ones were detected in the tetraploid M. sativa (data not shown). Thus, Mtzpt2-1 seems to be present as a single copy gene in the diploid species, even though a related gene might exist.
Figure 1.
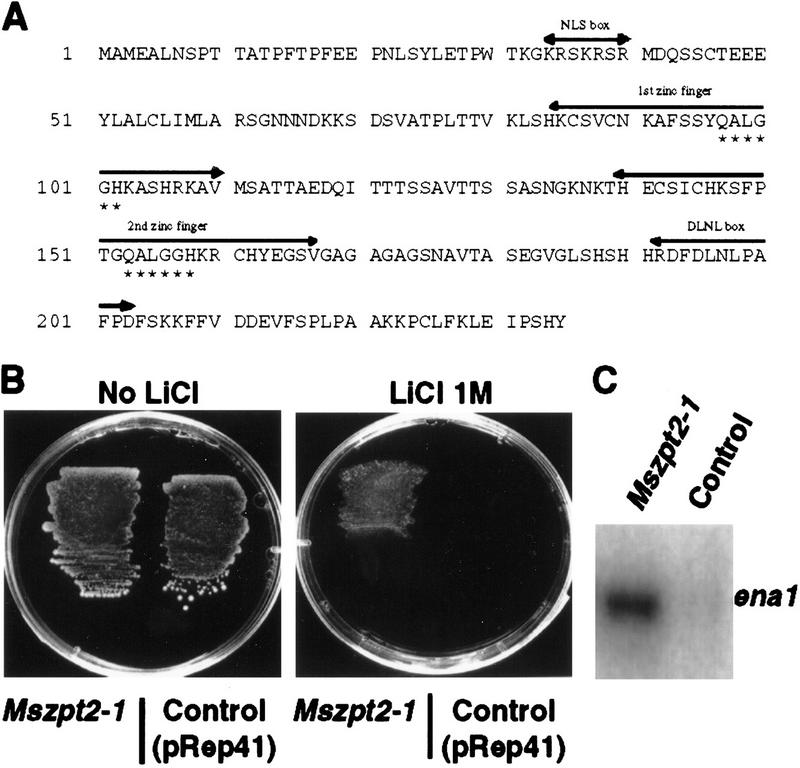
(A) Deduced amino acid sequence from the Mszpt2-1 cDNA (EMBL accession no. Y18788). The NLS box corresponds to a putative nuclear localization signal (basic residues; consensus, KR/KXR/K). The two Krüppel-like or TFIIIA zinc finger motifs are shown (consensus, CX2CX3FX5LX2HX3H); underlined amino acids indicate the highly conserved residues of EPF-type zinc fingers; asterisks show the plant-specific QALGGH motif. The DNLN box is a hydrophobic region corresponding to a putative protein–protein interaction motif. (B) Yeast cells carrying Mszpt2-1 in pRep41 or only the empty vector (control) were grown in a medium with or without 1 m LiCl. (C) RT–PCR analysis of ena1 expression in yeast cells corresponding to the strains shown in B grown in the absence of LiCl.
The closest Arabidopsis Mszpt2-1 homolog, Stz, has been found to induce osmotic tolerance in yeast cells (Lippuner et al. 1996), a function that we tested by expressing Mszpt2-1 (using the yeast-expressing vector pREP41; Maundrell et al. 1990) under the control of a promoter repressible by thiamine (data not shown). On selective minimum medium containing either 1.4 m NaCl (not shown) or 1 m LiCl (Fig. 1B), the strain expressing Mszpt2-1 was able to grow efficiently, in contrast to the same strain grown under noninducing conditions, or the strain transformed with pREP41. This result indicates that ectopic expression of Mszpt2-1 is able to increase osmotic stress tolerance in yeast. Moreover, transcriptional activation of the osmotic tolerant yeast gene ena1, coding for the major sodium/lithium efflux system (Rudolph et al. 1989), was detected in the Mszpt2-1-expressing strain (Fig. 1C).
Mszpt2-1 expression was analyzed in different alfalfa organs (Fig. 2A). Basal expression levels were detected in vegetative tissues, for example, roots, stems, and leaves. Higher levels of the Mszpt2-1 transcript were present in flowers, as well as in spontaneous nodules and nodules induced by S. meliloti but not in roots infected with the nonnodulating strain ZB138 (Nod−). The expression pattern of Mszpt2-1 in wild-type nodules, as well as in those arrested at different stages of development, was investigated by using the wild type (Sm41) and different mutants of S. meliloti. The surface polysaccharide mutant strain AK1492 (Eps−Kps−) is unable to invade root cortical cells and forms nodules devoid of bacteria, whereas the Sm8368 strain (Bac−) cannot differentiate into nitrogen-fixing bacteroids but is able to infect and invade plant cells (Kondorosi et al. 1984; Glazebrook et al. 1993). Upon infection with the invading strains Sm41 or Bac−, accumulation of the Mszpt2-1 transcripts was strongly enhanced in the root region adjacent to the nodule (Fig. 2B) In contrast, the noninvading Eps−Kps− strain showed similar Mszpt2-1 transcript levels in both roots and nodule tissues. These results indicate that even though Mszpt2-1 induction is associated with nodulation, the strong activation observed in the surrounding root tissues required bacterial invasion of the plant cells. The spatial expression pattern of Mszpt2-1 was determined using in situ hybridization (Fig. 3). A signal associated with the vascular bundles from both roots and nodules was observed. Notably, the closest petunia homolog, Zpt2-3, is expressed in vascular bundles of floral tubes and sepals (Takatsuji et al. 1994).
Figure 2.
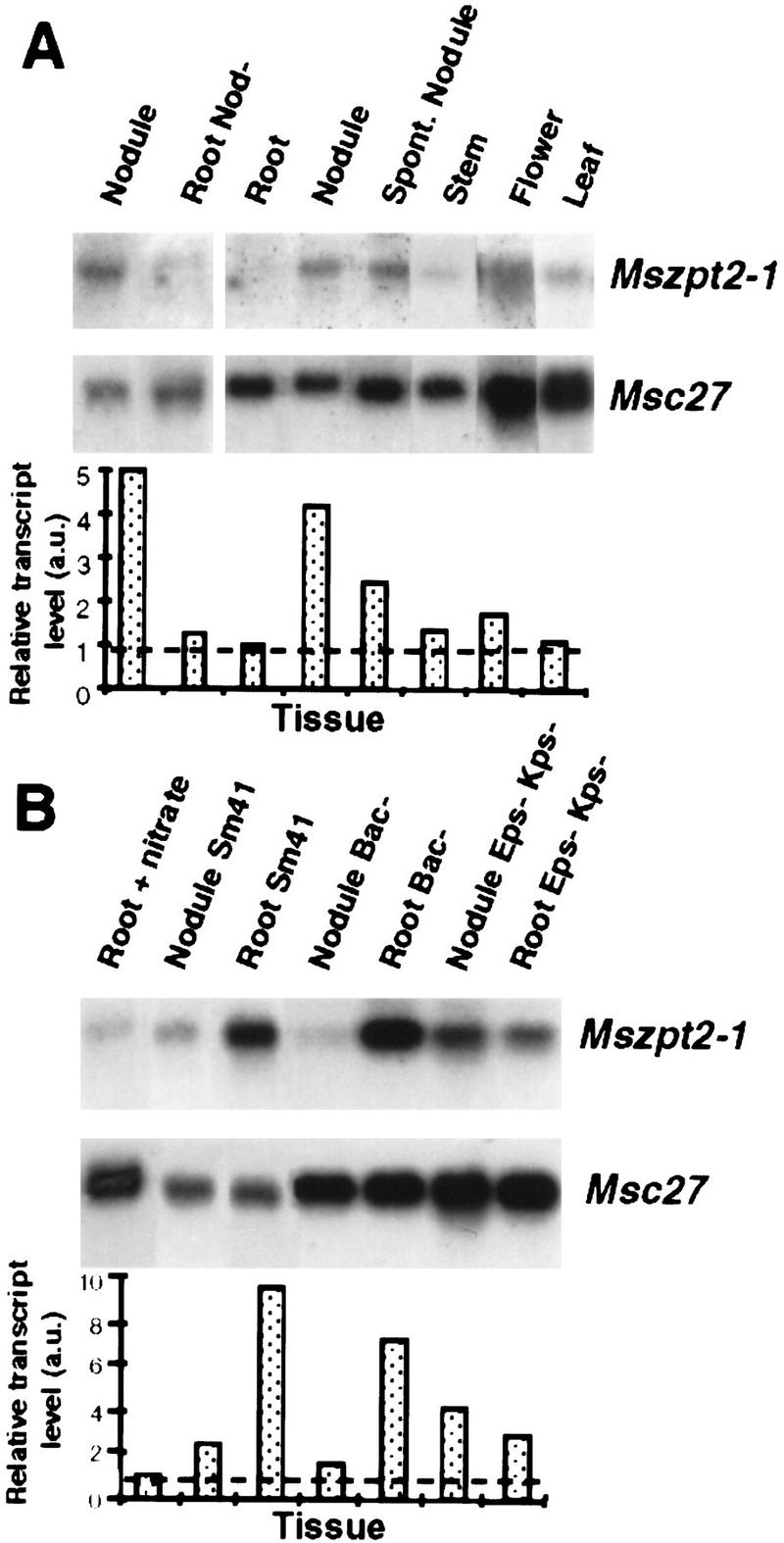
Northern blot analysis of Mszpt2-1 expression in different organs of alfalfa plants. Total RNAs (10 μg) were extracted from nodules and roots infected with a non-nodulating strain (Root Nod−) (A, left) and roots, mature S. meliloti-induced symbiotic nodules, spontaneous nodules (NAR phenotype: Nodulation in Absence of Rhizobium, Truchet et al. 1989), stems, and flowers (A, right), as well as roots grown on a nitrate-containing medium, and roots and nodules 3 weeks after infection with a wild-type Sm41, a Bac− (Sm8368), or an Eps−Kps− (AK1492) mutant strain (all infections were done in low nitrogen medium). (B) In all cases, Msc27 was used as control of RNA loading (Pay et al. 1992), and histograms (below) show the relative quantification of Mszpt2-1 transcript levels in arbitrary units (a.u.). The broken line indicates the transcript level corresponding to the uninoculated root RNA sample (1 a.u.).
Figure 3.
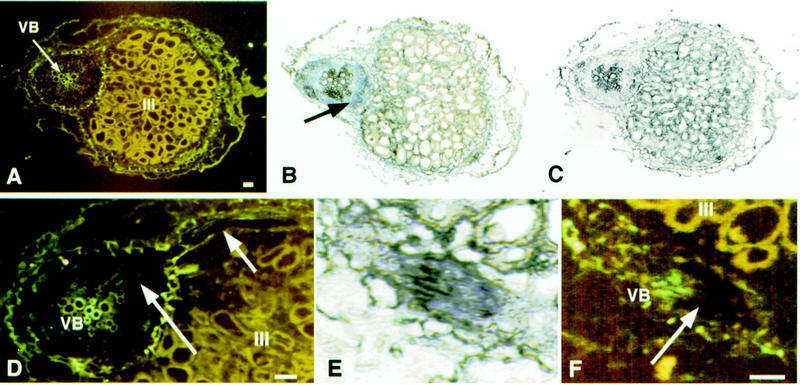
Localization of Mszpt2-1 expression in alfalfa roots and nodules infected with Sm41 by in situ hybridization. Sections (8 μm) were hybridized with digoxigenin-labeled Mszpt2-1 riboprobe. (A–C) longitudinal sections of mature symbiotic nodules (20 d.p.i.), containing transversal sections of contiguous roots. (A) Epifluorescence illumination, showing the nitrogen-fixing zone (III, Vasse et al. 1990) and the root vascular bundles (VB); (B) Consecutive section of A hybridized with the Mszpt2-1 antisense probe, viewed in bright-field illumination (arrow indicates signal localization); (C) the same section as A hybridized with the Mszpt2-1 sense control probe, viewed in bright-field illumination. (D–F) Higher magnification of sections hybridized with the Mszpt2-1 antisense probe, showing details of root and nodule vascular bundles. Under epifluorescence illumination (D,F), signals (arrows) appear as quenching of autofluorescence due to precipitation of the alkaline phosphatase product. (E) Bright-field illumination of section F. Bar, 50 μm.
To analyze potential functions of this Krüppel-like gene in plant organogenesis, we constructed M. truncatula lines expressing Mszpt2-1 in sense orientation or an antisense DNA fragment of the homologous Mtzpt2-1 gene (for M. truncatula, see Materials and Methods), in both cases under the control of the strong constitutive 2x35S CaMV promoters. After in vitro regeneration, the primary transformants of either sense or antisense plants were not disturbed in their growth characteristics when compared with plants transformed with the control vector alone. However, 15 primary transformants (T0) of 16 sense plants showed an abnormal flower phenotype. These flowers had an unusually long style compared to the anther size that became dehydrated rapidly (data not shown), leading to inefficient pollinization. Only a few pods were produced (18 on average, vs. ∼150 in the wild type) containing few seeds per pod (1 on average, vs. 4–7 in wild type). The antisense plants did not exhibit any abnormal flower morphology. These results suggest that the floral phenotype observed on the sense plants is likely due to the ectopic overexpression of Mszpt2-1 in flowers.
Because Mszpt2-1 expression was enhanced during nodule development and no alterations in growth or floral phenotypes were detected on the antisense plants, nodulation assays were performed on their progeny. Descendants (T1) from eight independent T0 lines (carrying one to three T-DNA insertion loci, as confirmed by Southern analysis; data not shown) were nodulated with Sm41. In three separate experiments, nodules from several T1 plants (25%–29%) obtained from three independent T0 lines were unable to fix nitrogen (Fig. 4A), in contrast to the control plants. Fifteen days after infection, both plant types had similar growth characteristics (Fig. 4A, left), and no significant differences either in leaf number or root length could be observed between antisense and control plants (data not shown). Nevertheless, only small white Fix− nodules could be detected on roots of these antisense plants, whereas typical Fix+ nodules (that emerged about 12 d.p.i.) were seen on all plants carrying the control vector construct (Fig. 4B). The Fix− phenotype was not due to any delay in nodulation, as even 1 month after infection, these plants showed severe starvation symptoms (Fig. 4A, right) and reduced weight of their aerial parts (0.18 ± 0.04 gram/plant in contrast to 1.1 ± 0.3 gram/plant for the controls), indicating strong limitations in their nitrogen nutrition. However, after transfer to a medium containing combined nitrogen, the plants were completely recovered and continued their normal development until flowering (data not shown). This indicated that the plant phenotype was due to nitrogen starvation induced by a defect in symbiotic nitrogen fixation.
Figure 4.
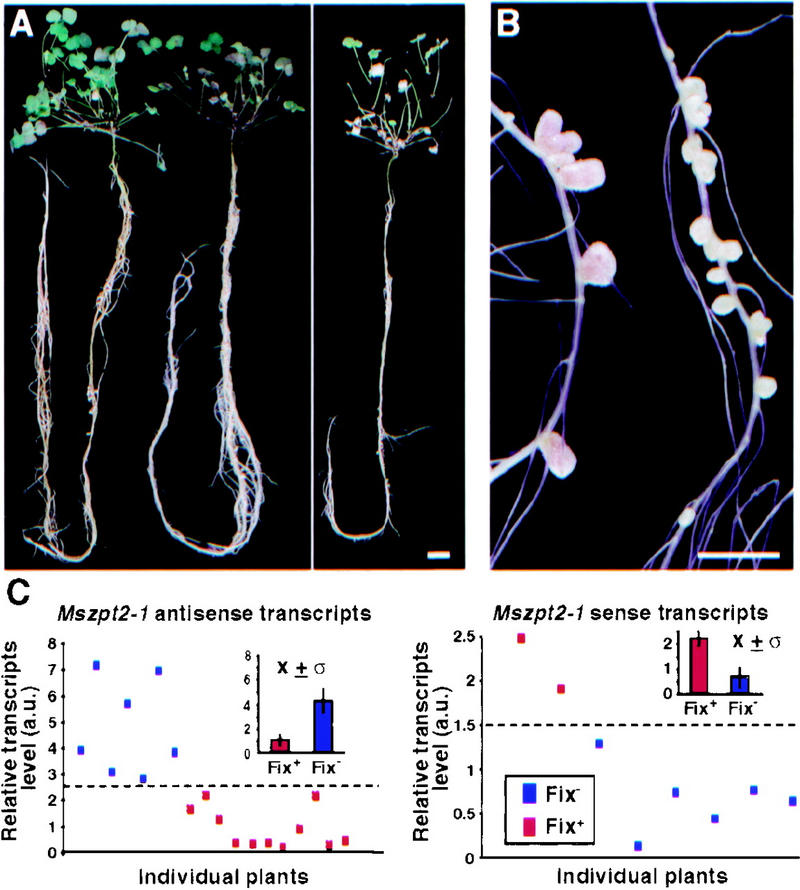
Nodulation phenotype of M. truncatula expressing Mszpt2-1 in antisense orientation. Plants were inoculated with the wild-type strain Sm41. (A) Whole view of a control vector-transformed plant (left) and a typical Mszpt2-1 antisense plant (right), 15 d.p.i. (left). One month after infection, a Fix− antisense plant displayed characteristic nitrogen-deprived appearance (right). Plants were grown in aeroponical conditions, optimal for root and nodule development. (B) Higher magnification of roots from plants 30 d.p.i. showing pink nitrogen-fixing nodules (left) on control plants, compared to white Fix− nodules on a Mszpt2-1 antisense plant (right). Bar, 0.5 cm. (C) Correlation of Mtzpt2-1 expression and Fix+ or Fix− nodule phenotype in individual plants from the progeny of several T0 lines. Total RNAs (10 μg) from roots and nodules 30 d.p.i. were probed with an Mszpt2-1 RNA probe, and Msc27 was used as RNA-loading control. Points and inset histograms depict, respectively, individual values and averages of the relative quantification of Mtzpt2-1 antisense (left) or sense (right) transcript levels in the plants analyzed by Northern blot. Fix+ (red) or Fix− (blue) values are indicated. Progeny of the three different antisense Fix− plant lines were tested. The broken line marks a threshold separating the two plant populations (Fix− and Fix+).
The antisense fragment used spans the zinc finger regions of Mtzpt2-1. To analyze the specificity of our approach for Mtzpt2-1, (1) we searched for the expression of homologous gene(s) in nodules or root RNA samples by RT–PCR, Northern analysis, or library screening even under low stringency conditions (data not shown), and (2) Northern hybridizations with strand-specific probes were performed to measure the amounts of antisense and sense Mtzpt2-1 transcripts in infected roots and nodules of several individuals originating from different plant lines. In these experiments, expression of homologous genes could not be detected in nodules. Moreover, a direct correlation was found between the Fix− phenotype, the reduction of the sense transcript level, and the increase in antisense expression (Fig. 4C). A threshold level of Mtzpt2-1 expression could be observed beyond which the plants were Fix−. The differences in the antisense and sense transcript levels between the Fix+ and Fix− populations were shown to be statistically significant (P < 0.01, using Student's t-test). Furthermore, the cosegregation of the Fix− phenotype with the T-DNA was studied. In the T1 generation (54 plants from the 3 independent lines tested), all Fix− plants (14) were also herbicide resistant. The phenotype was stable in the T2 generation, and no intermediate phenotypes were detected in our experiments even 1 month after infection (in the same plant, only nitrogen-fixing or -nonfixing nodules were found). Overall, these results suggest strongly that inactivation of Mtzpt2-1 was responsible for the arrest of root nodule development in these plants.
Further analysis of the nodulation phenotype was carried out using S. meliloti strains containing a constitutively expressed (δ-ala) or a bacteroid-specific (nifH) lacZ fusion (Leong et al. 1985; Szeto et al. 1987). Bacterial infection as well as the early stages of nodule development took place normally in these plants, whereas no lacZ expression was detected in the central zone of young and mature nodules for both strains (data not shown), suggesting that S. meliloti was not able to reach these regions or to express the nitrogenase gene nifH. This analysis confirmed the lack of nitrogen fixation at any nodule developmental stage. By histological analysis, no typical zone III containing strongly autofluorescent cells invaded by bacteroids was detected, even though only few infected cells were present in the apical region of these nodules (Fig. 5A,B). Using lugol staining, the central region of the Fix− nodules was shown to contain a high number of amyloplasts, in contrast to Fix+ nodules, where amyloplasts were observed only in the interzone II–III (Fig. 5C,D). Detailed analysis using optical (Fig. 5E,F) and electron (Fig. 5G,H) microscopy showed that in the Fix− nodules, the bacteria present in apical infected cells did not develop beyond the early stages of bacteroid differentiation and did not display the typical radial pattern organized around a central vacuole, observed for the nitrogen-fixing symbiosomes in the central region. In addition, no differences in the morphology of vascular bundles between Fix+ and Fix− nodules were detected. In both cases, xylem vessels, sieve elements, and transfer cells showing characteristic wall ingrowths (Joshi et al. 1993) were observed under electron microscopy (data not shown). These results indicate that expression of the Mtzpt2-1 gene in antisense orientation affected nodule development by reducing bacterial invasion of the plant cells and preventing differentiation of the nitrogen-fixing zone III cells.
Figure 5.

Microscopy analysis of Fix− nodules on Mszpt2-1 antisense plants. (A–D) Longitudinal sections (8 μm) of Fix+ and Fix− nodules observed by optical microscopy (paraffin embedding). (A) Fix+ nodule from a wild-type plant, showing typical zonation of nitrogen-fixing nodules (I, II, III; Vasse et al. 1990); (B) Fix− nodule from an Mtzpt2-1 antisense plant, showing abnormal central tissue differentiation and an apical infected zone (IZ), both viewed under epifluorescence illumination. (C,D) Higher magnifications of equivalent samples stained with lugol (amyloplast coloration), viewed in bright-field illumination: (C) A Fix+ nodule section, showing amyloplast accumulation (A) restricted to the interzone II–III; (D) a Fix− nodule, showing amyloplast accumulation (A) in a large central region. (E,F) Semithin sections of nodules stained with toluidine blue showing cells from E, the central region (III) of a Fix+ nodule, and F, the apical region of a Fix− nodule (IZ), indicated in A and B. Asterisks indicate one infected cell. Bar, 50 μm. (G,H) Ultrathin sections analyzed by electron microscopy showing details of one infected cell originating from the regions shown in E and F, respectively. (A) Amyloplast; (B) bacteroid. Bar, 1 μm.
Discussion
The analysis of antisense plants indicates that Mszpt2-1 is specifically involved in a vegetative organogenesis, the formation of the nitrogen-fixing nodule, and suggests that this transcription factor family may act in several plant-specific developmental programs (Takatsuji 1998). Overexpression of Mszpt2-1 likely affected, either by ectopic interaction or eventually by a cosuppression phenomenon, the action of homologous genes involved in flower differentiation. Most members of EPF-type TFIIIA zinc finger proteins are expressed predominantly in flower tissues and tightly regulated in specific floral organs (Takatsuji et al. 1994). Our work also suggests that certain aspects of flower and nodule differentiation might share similar mechanisms. Nodulin genes might have derived from homologs expressed in other organs that were recruited to participate in nodule organogenesis (Bladergroen and Spaink 1998).
To maximize the specificity of our antisense approach we used a fragment of the Mtzpt2-1 gene in M. truncatula, although we cannot completely exclude that inactivation of a related gene (undetectable by the different approaches undertaken; see Results) is responsible for the observed phenotype. Nevertheless, the fact that Mtzpt2-1 transcript levels were specifically reduced in the antisense plants, together with the induction of this gene during this process, strongly suggests that Mtzpt2-1 is the target of our approach. Moreover, the high frequency of detection of the Fix− phenotype in independent antisense lines (three of eight) and the linkage of the phenotype with the transgene in the T1 and T2 generation confirmed that this antisense construction is responsible for the Fix− phenotype. As a means of comparison, in a large scale transformation experiment carried out in our laboratory a maximum frequency of 1 Fix− per 1000 plants was found. Numerous plant mutants affected in symbiotic nitrogen fixation have been isolated, indicating that different genes must be specifically involved in controlling this process. In M. truncatula, at least nine Nod+Fix− mutants have been isolated using γ-ray mutagenesis, but the reported phenotypes do not correspond to the defect induced by antisensing Mtzpt2-1 (Sagan et al. 1995). The reduction of Mtzpt2-1 expression might evoke the Fix− phenotype also because of a slowly accumulating effect in these nodules, and a loss-of-function allele might produce a more severe phenotype. However, even in one plant showing very low Mtzpt2-1 expression, we could not detect any alteration of the phenotype.
Histological analysis of Fix− nodule development on the Mszpt2-1 antisense plants suggests that initial root infection proceeds normally, whereas bacterial invasion of plant cells and nodule central zone differentiation are arrested. Accumulation of amyloplasts in the empty central region of these nodules indicates that the carbon source is not metabolized by the bacterial partner. These nodules resemble those obtained with surface polysaccharide mutant rhizobia where severe reduction of infection blocks nodule differentiation at early stages (Dickstein et al. 1988). Low levels of invasion occurring in the antisense plants were restricted to only a few cells in the nodule apex, further supporting that Mtzpt2-1 is required for proper development of zone III. Because the expression of nifH (coding for the structural gene of the enzyme nitrogenase) was not detected at any developmental stage or in the mature nodules, we could discard any premature senescence process of the nodules formed on these plants.
The association of bacterial invasion events with the late induction of this gene in root and nodule vascular bundles, together with the phenotype of the antisense plants in the central region of the nodule, suggests a role of this transcription factor in a cell to cell communication process between these tissues required for the development of the nitrogen-fixing zone. This could be related to the translocation of specific metabolites in and out of the vascular phloem involved in sensing the carbon/nitrogen metabolic status of the plant (Streeter 1993). Phloem transfer cells are likely involved in the short distance transport of solutes (Joshi et al. 1993), and Mszpt2-1 may be expressed in these cells. The exchange of metabolites through the vascular tissue may require osmotic adaptations of these cells. We showed that Mszpt2-1 conferred salt tolerance in yeast, like its closest Arabidopsis homolog. Signaling mediated by carbon/nitrogen metabolites affecting osmotic conditions of the connecting vascular bundles might be crucial for the regulation of symbiotic nitrogen fixation (Streeter 1993). Interestingly, a function in a diffusible signaling process has been proposed for INDETERMINATE, another member of the plant Krüppel-like gene family, as it was expressed in leaves but influenced flower meristem formation (Colasanti et al. 1998). Although several explanations for the reported effects are possible, we propose that Mtzpt2-1 may be also involved in a long-distance cell–cell communication process between root vascular bundles and the central region of the nodule.
Our results show that Mtzpt2-1 is involved in symbiotic nodule organogenesis, permitting its invasion by rhizobia. Thus, its function in a vegetative developmental process indicates that Krüppel-like zinc finger proteins have various roles in plant organogenesis.
Materials and methods
Bacterial strains
S. meliloti mutant strains used were (1) the wild-type strain Sm41; (2) the AK1492 mutant strain (Eps−Kps−Fix−), a spontaneous Eps− (exoB) derivative of Sm41, in which a Tn5 insertion in the fix-23 region resulted in a Kps− and Fix− phenotype (Kondorosi et al. 1984); (3) the ZB138 Nod− mutant strain, a derivative of Sm41 where all of the nod genes of the pSym megaplasmid are deleted (Kondorosi et al. 1984); and (4) the Sm8368 mutant strain (Bac−Fix−), a derivative of Sm1021 obtained by TnphoA mutagenesis in the bacA gene (Glazebrook et al. 1993). A Sm41 strain carrying the pXLGD4 plasmid with a δ-ala::lacZ fusion (Leong et al. 1985) or an Sm1021 strain harboring a nifH::lacZ fusion (Szeto et al. 1987) have been used to follow bacterial infection and differentiation in transgenic plants. All strains were grown at 30°C on TA agar medium (Orosz et al. 1973).
Plants and nodulation assays
Alfalfa (M. sativa ssp. sativa cv. Sitel) or M. truncatula cv. 108-R4 seeds were sterilized, germinated, and grown for 1 week as described (Charon et al. 1999). Plants were then transferred to an aeroponic culture system (Coba de la Peña et al. 1997). After 1 additional week, plants were infected with a stationary-phase culture of a S. meliloti strain. Spontaneous nodules were obtained from M. sativa ssp. sativa genotype A2 lines as described (Coba de la Peña et al. 1997).
Nucleic acid techniques
Genomic DNA (15 μg), prepared from leaf material of M. sativa ssp. varia genotype A2 and M. truncatula 108-1, according to Sambrook et al. (1989), was used for Southern analysis. Total RNAs (10 μg) from different tissues, extracted as described in Sambrook et al. (1989), were used in Northern analysis. Hybridizations were carried in Church buffer (Church and Gilbert 1984) with a Mszpt2-1 probe (the complete cDNA) overnight at 65°C, and washed under high stringent (65°C; 2× SSC; 0.1% SDS for 30 min; 1× SSC; 0.1% SDS for 30 min; 0.2× SSC; 0.1% SDS for 15 min) or low stringent (65°C; 2× SSC; 0.1% SDS for 30 min) conditions. Single-stranded RNA probes were synthesized with a DIG RNA labeling kit and revealed using chemiluminiscence according to the manufacturer's instructions (Boehringer Mannheim Biochemica, Meylan, France). Sequencing was carried out with an automatic sequencer 373A (Applied Biosystems, Foster City, CA) using the dideoxy chain termination method (Pharmacia kit, Uppsala, Sweden). Sequence analysis was done using the GCG software package (Genetic Computer Group, University of Wisconsin, Madison, WI) and BLAST algorithm (Gish and States 1993).
Mszpt2-1 expression in yeast
Expression of Mszpt2-1 in yeast was done by cloning the complete Mszpt2-1 cDNA in the NdeI–SalI sites of the yeast expression vector pREP41 under the control of the thiamine-repressed promoter nmt1 (Maundrell et al. 1990). Transformation of Saccharomyces cerevisiae strain YRG-2 (Stratagene) was done as described in Ausubel et al. (1989). Transformed yeast cells were selected for leucine auxotrophy (leu2 marker) on agar plates and tested for osmotic tolerance by incubating for 6 days with or without 2 μm thiamine in the presence or absence of 1.4 m NaCl or 1 m LiCl.
RT–PCR experiments (Charon et al. 1999) were done with total RNAs (extracted using RNeasy kit; Quiagen, Hilden, Germany) from yeast containing the control vector (pRep41) or expressing Mszpt2-1. cDNAs (6 ng) were PCR-amplified 30 cycles with specific oligonucleotides against the ena1 gene (Rudolph et al. 1989). Membranes were hybridized to a ena1 probe.
Sense and antisense expression in transgenic plants
For construction of transgenic plants a binary plasmid containing a strong constitutive chimeric promoter, comprising two 35S CaMV transcriptional enhancer elements and a translational enhancer from AMV, and the 3′ nopaline synthetase region was used (Datla et al. 1991). Sense constructs were prepared using the complete Mszpt2-1 cDNA (EMBL accession no. Y18788), whereas antisense constructs contained a 598-bp region (nucleotides 15–612) of the M. truncatula Mtzpt2-1 gene cloned by RT–PCR. The sequence amplified was at least 88% identical to Mszpt2-1 (data not shown), and no other Mszpt2-1-derived sequences could be detected in M. truncatula nodule RNA samples. M. truncatula 108-R4 plants were transformed with sense, antisense, or control vector according to Trinh et al. (1997).
In situ hybridization and microscopic analysis
In situ hybridization, starch staining, and histological techniques using paraffin-embedded material were performed as described in Coba de la Peña et al. (1997). In situ lacZ assays on nodule samples were done as described in Boivin et al. (1990). For electron microscopy, nodules were fixed for 1 hr under vacuum in 3% glutaraldehyde, 1% paraformaldehyde, 0.1 m Na cacodylate (pH 6.8), rinsed several times in cacodylate buffer, postfixed with 1% tannic acid for 1 hr, rinsed again and treated with 1% osmium for 1 hr. After washing, samples were incubated overnight at 4°C with 0.5% uranyl acetate, dehydrated using successive ethanol treatments (10%, 20%, 30%, 50%, 70%, 90%, each step 20 min), followed by 100% ethanol (30 min and 1 hr), and embedded in Spurr resin. After polymerization, either semithin (0.5 μm; stained with toluidine blue) or ultrathin (80 nm) sections were cut using a Reichert Jung ultramicrotome UltracutE and analyzed with an electron microscope Philips CM10.
Acknowledgments
F.F. was supported by a fellowship of the Ministère Français de l'Enseignement Supérieur et de la Recherche, and by a grant from the Académie d'Agriculture de France. We thank Nathalie Mansion for photographic work, Anne-Lise Franquemagne for help in transgenic plant characterization, Jacques Batut for providing the S. meliloti strain carrying the nifH::lacZ fusion, and Pascal Ratet for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL adam.kondorosi@isv.cnrs-gif.fr; FAX 33-01 69 82 3695.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Vol. 2. New York, NY: Greene Publishing Associates; 1989. Basic techniques of yeast genetics: preparation of yeast media; p. 13.1.2. [Google Scholar]
- Bladergroen MR, Spaink HP. Genes and signal molecules involved in the Rhizobia-Leguminoseae symbiosis. Curr Opin Plant Biol. 1998;1:353–359. doi: 10.1016/1369-5266(88)80059-1. [DOI] [PubMed] [Google Scholar]
- Boivin C, Camut S, Malpica CA, Truchet G, Rosenberg C. Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell. 1990;2:1157–1170. doi: 10.1105/tpc.2.12.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray P, Lichter P, Thiesen H-J, Ward DC, Dawid IB. Characterization and mapping of human genes encoding zinc finger proteins. Proc Natl Acad Sci. 1991;88:9563–9567. doi: 10.1073/pnas.88.21.9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon C, Sousa C, Crespi M, Kondorosi A. Alteration of enod40 expression modifies Medicago root nodule development induced by Sinorhizobium meliloti. Plant Cell. 1999;11:1953–1965. doi: 10.1105/tpc.11.10.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D, Ralph R, Jonak G. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol Cell Biol. 1989;9:2487–2492. doi: 10.1128/mcb.9.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon C-, III, Lee N-G, Siddique A-BM, Bal AK, Verma DPS. Roles of plant homologs of rab1p and rab7p in the biogenesis of the peribacteroid membrane, a subcellular compartment formed de novo during root nodule symbiosis. EMBO J. 1993;12:4125–4135. doi: 10.1002/j.1460-2075.1993.tb06096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coba de la Peña T, Frugier F, McKhann HI, Bauer P, Brown S, Kondorosi A, Crespi M. A carbonic anhydrase gene is induced in the nodule primordium and its cell-specific expression is controlled by the presence of Rhizobium during development. Plant J. 1997;11:407–420. doi: 10.1046/j.1365-313x.1997.11030407.x. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V. The INDETERMINATE gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell. 1998;93:593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- Csanadi G, Szecsi J, Kalo P, Kiss P, Endre G, Kondorosi A, Kondorosi E, Kiss GB. Enod12, an early nodulin gene, is not required for nodule formation and efficient nitrogen fixation in alfalfa. Plant Cell. 1996;6:201–213. doi: 10.1105/tpc.6.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datla RS, Hammerlindl JK, Pelcher LE, Crosby WL, Selvaraj G. A bifunctional fusion between beta-glucuronidase and neomycin phosphotransferase: A broad-spectrum marker enzyme for plants. Gene. 1991;101:239–246. doi: 10.1016/0378-1119(91)90417-a. [DOI] [PubMed] [Google Scholar]
- Dickstein R, Bisseling T, Reinhold VN, Ausubel FM. Expression of nodule-specific genes in alfalfa root nodules blocked at an early stage of development. Genes & Dev. 1988;2:677–687. doi: 10.1101/gad.2.6.677. [DOI] [PubMed] [Google Scholar]
- Frugier F, Kondorosi A, Crespi M. Identification of novel putative regulatory genes induced during alfalfa nodule development with a cold-plaque screening procedure. Mol Plant-Microbe Interact. 1998;11:358–366. doi: 10.1094/MPMI.1998.11.5.358. [DOI] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ichige A, Walker GC. A Rhizobium meliloti homolog of the Escherichia coli peptide antibiotic transport protein sbma is essential for bacteroid development. Genes & Dev. 1993;7:1485–1497. doi: 10.1101/gad.7.8.1485. [DOI] [PubMed] [Google Scholar]
- Joshi P, Caetano-Anollés G, Graham E, Gresshoff P. Ultrastructure of transfer cells in spontaneous nodules of alfalfa (Medicago sativa) Protoplasma. 1993;172:64–76. [Google Scholar]
- Kondorosi E, Banfalvi Z, Kondorosi A. Physical and genetic analysis of a symbiotic region of Rhizobium meliloti: Identification of nodulation genes. Mol & Gen Genet. 1984;193:445–452. [Google Scholar]
- Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Nishino T, Takatsuji H. Cys2/His2 zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 1998;26:608–615. doi: 10.1093/nar/26.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong SA, Williams PH, Ditta GS. Analysis of the 5′ regulatory region of the gene for δ-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 1985;13:5965–5976. doi: 10.1093/nar/13.16.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V, Cyert MS, Gasser CS. Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J Biol Chem. 1996;271:12859–12866. doi: 10.1074/jbc.271.22.12859. [DOI] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. J Biol Chem. 1990;265:10857–10854. [PubMed] [Google Scholar]
- Meissner R, Michael AJ. Isolation and characterization of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs. Plant Mol Biol. 1997;33:615–624. doi: 10.1023/a:1005746803089. [DOI] [PubMed] [Google Scholar]
- Orosz L, Svab Z, Kondorosi A, Sik T. Genetic studies on Rhizobiophage 16-3 T; genes and function on the chromosome. Mol & Gen Genet. 1973;125:341–350. [PubMed] [Google Scholar]
- Pay A, Heberle-Bors E, Hirt H. An alfalfa cDNA encodes a protein with homology to translationally controlled human tumor protein. Plant Mol Biol. 1992;19:501–503. doi: 10.1007/BF00023399. [DOI] [PubMed] [Google Scholar]
- Rudolph H, Antebi A, Fink G, Buckley C, Dorman T, LeVitre J, Davidow L, Mao J, Moir D. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a calcium ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Sagan M, Morandi D, Tarenghi E, Duc G. Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after γ-ray mutagenesis. Plant Sci. 1995;111:63–71. [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;378:199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- Schultze M, Kondorosi A. Regulation of symbiotic root nodule development. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- Streeter JG. Translocation—A key factor limiting the efficiency of nitrogen fixation in legume nodules. Physiol Plant. 1993;87:616–623. [Google Scholar]
- Szeto WW, Tracy Nixon B, Ronson CW, Ausubel FM. Identification and characterization of the Rhizobium meliloti ntrC gene: R. meliloti has separate regulatory pathways for activation of nitrogen fixation genes in free-living and symbiotic cells. J Bacteriol. 1987;169:1423–1432. doi: 10.1128/jb.169.4.1423-1432.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji T. Zinc-finger transcription factors in plants. Cell Mol Life Sci. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H, Nakamura N, Katsumoto Y. A new family of zinc finger proteins in Petunia: Structure, DNA sequence recognition, and floral organ-specific expression. Plant Cell. 1994;6:947–958. doi: 10.1105/tpc.6.7.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh TH, Ratet P, Kondorosi E, Durand P, Kamaté K, Bauer P, Kondorosi A. Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp. falcata lines improved in somatic embryogenesis. Plant Cell Rep. 1997;17:345–355. doi: 10.1007/s002990050405. [DOI] [PubMed] [Google Scholar]
- Truchet G, Barker DG, Camut S, Billy FD, Vasse J, Huguet T. Alfalfa nodulation in the absence of Rhizobium. Mol & Gen Genet. 1989;219:65–68. [Google Scholar]
- Vasse J, de Billy F, Camut S, Truchet G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol. 1990;172:4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


