Abstract
Global leptospirosis disease burden estimates are hampered by the lack of scientifically sound data from countries with probable high endemicity and limited diagnostic capacities. We describe the seroepidemiologic and clinical characteristics of the leptospirosis outbreak in 2008 in Sri Lanka. Definitive/presumptive case definitions proposed by the World Health Organization Leptospirosis Epidemiology Reference Group were used for case confirmation. Of the 404 possible cases, 155 were confirmed to have leptospirosis. Highest titers of patient seum samples reacted with serovars Pyrogenes (28.7%), Hardjo (18.8%), Javanica (11.5%), and Hebdomadis (11.5%). Sequencing of the 16S ribosomal DNA gene identified six infections: five with Leptospira interrogans and one with L. weilli. In this patient population, acute renal failure was the main complication (14.8%), followed by myocarditis (7.1%) and heart failure (3.9%). The case-fatality rate was 1.3%. This report strengthens the urgent need for increasing laboratory diagnostic capabilities to determine the causes of epidemic and endemic infectious diseases in Sri Lanka, a finding relevant to other tropical regions.
Introduction
Leptospirosis is a globally widespread, neglected, and emerging zoonotic disease,1 posing important public health threats in the developing and developed world alike. Millions of persons are estimated to be affected annually, and increasing number of outbreaks have been reported recently from several countries.2 The disease is endemic in humid, tropical, and subtropical areas of the world where most of the developing countries are located.3 Emerging leptospirosis mostly affects vulnerable communities living in resource poor settings. Often the disease is either not suspected under or misdiagnosed in marginalized populations because of the need for laboratory resources to confirm leptospirosis; typically, such resources are neither accessible nor affordable. From the clinical perspective, better diagnostics are needed to prevent severe complications and death. From the public health perspective, the lack of reliable and efficient diagnostics tests makes assessing the burden of disease, whether regionally or globally, impossible.
With the recent emerging global threat of leptospirosis worldwide, the first meeting of Leptospirosis Epidemiology Reference Group (LERG) was held in December 2009 with guidance of the World Health Organization (WHO). LERG conducted a systematic review of literature on existing evidence to obtain accurate estimates of global leptospirosis incidence and prevalence. Of 12,033 reports reviewed, only 64 studies fulfilled the inclusion criteria and had low or moderate risk of bias. One major concern of the LERG group was the geographic bias of the selected data that might cause regional gaps in global estimates. Some of the countries such as Sri Lanka in which leptospirosis has been declared as highly endemic were not included in the systematic review because of lack of scientifically sound data.4
Despite lack of scientific publications, routinely reported data published by the epidemiology unit of Sri Lanka suggests that Sri Lanka has experienced one of the largest global leptospirosis outbreak reported in recent years.5 In 2008, the total number of clinically suspected cases reported to the surveillance system was 7,406 and 204 deaths.6 The reported incidence rate based on notification data was 35.7 per 100,000 population. In 2009, 4,980 cases and 145 deaths were reported7 and the outbreak persisted in 2010 with 4,553 cases and 121 deaths.8 The probable case incidence remains more than 22.5 per 100,000 population, making it the second highest reported incidence of leptospirosis worldwide; the highest has been reported from Seychelles with 43.2 per 100,000 population.9 However, these cases are reported on the basis of clinical suspicion and fewer than 10% were laboratory confirmed because of lack of diagnostic capacity. These data available in the national surveillance program did not conform with definitive or presumptive case definitions proposed by LERG and were not published in scientific literature, making the utility of such data questionable for disease burden estimates.
Sri Lanka is a tropical island with a population of approximately 20 million persons. The economic activities, environment, hygienic conditions, recent ecological changes, and climate in this country combine to provide an ideal environment for leptospirosis transmission. A large part of the economy in Sri Lanka is based on agriculture, especially rice farming. Farming activities depend on two monsoons where the average rainfall is approximately 200 cm per year. Water buffalo and cattle are commonly used in rice-farming activities. A small holder dairy industry is a major component of the rural economy. Rodents and small mammals that could harbor Leptospira are abundant in and around households and rice fields. Rapid unplanned urbanization during the past two decades has brought wild and domesticated animals into close contact. Rapid urbanization has also led to the proliferation of rodent populations in human population centers.
Despite having the second highest reported incidence worldwide on a per capita basis and a significant number of deaths, little has been published about leptospirosis in Sri Lanka. Laboratory confirmation of the disease is not routinely made because of lack of diagnostic capabilities. The Medical Research Institute of Sri Lanka in Colombo provides diagnostic capacity for the entire country and uses an abbreviated form of the microscopic agglutination test (MAT) with only genus-specific L. biflexa serovar Patoc strain for diagnosis, but does not use a broad panel of serovars. Only one published report is available on disease confirmation during the 2008 outbreak, which confirmed 26 cases in 107 suspected cases using a single-sample MAT. In the same report, molecular diagnosis and species identification was reported for three cases and the deduced leptospirosis species were L. interrogans (2 samples) and L. kirschneri (1 sample).10 In the same sample of suspected leptospirosis patients, the authors confirmed eight cases of hantavirus infection, which clinically mimics leptospirosis.11
We report a study conducted in three leptospirosis-endemic districts of Sri Lanka during the 2008 outbreak of leptospirosis using WHO LERG case definitions and a disease model. The objective of this study was to obtain scientifically sound data on leptospirosis in Sri Lanka within the ascertainment limits of a resource-poor setting and to place these limitations within the scope of emerging issues in assessing the global burden of this disease. The lessons drawn from this analysis are applicable to tropical regions where the burden of leptospirosis is likely to be substantial but remains unknown because of limited diagnostics capacity.
Methods
Study settings, study samples, and patient selection.
The present study was conducted in the districts of Kegalle, Kandy, and Matale in Sri Lanka. All three districts have been identified as leptospirosis-endemic by the Epidemiology Unit of Sri Lanka. One hospital in each of the three districts was selected as a study site, namely, District General Hospital-Kegalle, Teaching Hospital - Kandy, and District General Hospital - Matale.
The study population consisted of consecutively identified male and female patients with acute febrile illness admitted to medical wards in the three hospitals during August 20, 2008–January 6, 2009. All fever patients admitted to selected wards were screened. Patients conforming to the study inclusion criteria (Table 1) were invited to participate in the study. Inclusion criteria for possible cases were designed to increase the sensitivity of screening procedure. Thus, the stringent case definition proposed by the WHO and adopted by the Epidemiology Unit of Sri Lanka for surveillance activities was not used at this initial screening stage. Less stringent criteria to include undifferentiated fever, which reduced specificity, were used to detect even mild-to-moderate cases. A dedicated interviewer administering a clinical checklist carried out initial screening and obtained standardized clinical data. Socio-demographic profiles, exposure histories, and environmental data were obtained by using a pre-tested, interviewer-administered, questionnaire.
Table 1.
Inclusion criteria for selecting fever patients during leptospirosis outbreak in Kegalle, Kandy and Matale, Sri Lanka, 2008
| Inclusion criteria |
|---|
| Criterion 1 |
| 1. Patients admitted to medical wards in the selected hospitals |
| 2. Presenting complaint is acute febrile illness (fever less than 15 days and temperature > 37.8°C) |
| 3. With any one of the following major symptoms |
| Headache |
| Myalgia |
| Prostration |
| 4. Associated with any of the following signs (at least one) |
| Conjunctival suffusion/conjunctival hemorrhage |
| Meningeal irritation |
| Anuria or oliguria/proteinuria/hematuria |
| Jaundice |
| Hemorrhages |
| Purpuric rash |
| Cardiac arrhythmia or failure |
| Criterion 2 |
| Any other patient suspected and treated as a leptospirosis patient by a treating physician |
Sample collection and transport.
On admission, 5 mL of whole blood was obtained in a plain tube to obtain serum for enzyme-linked immunosorbent assay (ELISA) and MAT; an additional 5 mL was collected into a tube containing EDTA for polymerase chain reaction (PCR) on whole blood. Another 5-mL blood sample was obtained at least 7–10 days later as the convalescent-phase sample for serologic testing, which was conducted at least 10 days after the onset of symptoms. All samples were stored at 4–8°C12 and transported in cooled boxes to the study laboratory within 24 hours. In the laboratory, serum samples were separated by centrifugation and IgM ELISA testing was carried out by using a commercially available kit (Panbio, Sinnamon Park, Queensland, Australia)13 for all cases. Remaining samples were stored at –60°C until send in dry ice to Thailand and Australia for further testing.
Case definitions for leptospirosis.
Two case definitions were used in diagnosing leptospirosis on the basis of the WHO LERG report. A definitive case was classified by any one of the following: 1) seroconversion (negative first sample and a titer ≥ 1:100 in the second sample), or a 4-fold increase in MAT titer between acute-phase and convalescent-phase samples; 2) Leptospira DNA detected by PCR. A presumptive case definition was defined as a patient with symptoms consistent with leptospirosis and presence of IgM against Leptospira shown by ELISA. Presumptive case definition was necessary because MAT was carried out only for patients with paired serum samples, and PCR was applied only to a sample of patients because of logistic issues. For remaining patients with only acute-phase serum samples, the ELISA-based presumptive case definition was used to confirm the diagnosis. For the presumptive diagnosis, a patient with symptoms consistent with leptospirosis was defined according to WHO recommended surveillance case definition of leptospirosis; “A suspected case involves a person presenting with acute febrile illness; headache, myalgia and prostration; associated with any of the following symptoms: conjunctival suffusion; meningeal irritation; anuria, oliguria, or proteinuria; jaundice; hemorrhage (from intestines; lung bleeding often notorious); cardiac arrhythmia or failure; skin rash; a history of exposure to infected animals or an environment contaminated with infected animal urine.”14
Sample processing and analysis.
The MAT was performed by the WHO/Food and Agricultural Organization/World Organization for Animal Health Collaborating Center for Reference and Research on Leptospirosis, Western Pacific Region (Archerfield, Queensland, Australia) on 167 paired serum samples. The reference panel represented 5 species and 16 serogroups of Leptospira spp. and included four isolates from Sri Lanka (Table 2).
Table 2.
Panel of Leptospira serovars used for MAT analysis and the rationale of including these serovars in the MAT panel to diagnose leptospirosis cases during the leptospirosis outbreak in Kegalle, Kandy and Matale, Sri Lanka, 2008*
| Serogroup | Serovar | Strain | Species | Reasons |
|---|---|---|---|---|
| Icterohaemorrhagiae | Copenhageni | M20 | L. interrogans | Classic Weil's disease organism, common in Rattus norvegicus |
| Hebdomadis | Hebdomadis | Hebdomadis | L. interrogans | Common in cattle throughout Asia |
| Autumnalis | Autumnalis | Akiyami A | L. interrogans | Common in rodents in rice-growing areas |
| Autumnalis | Alice | Alice | L. santarosai | Isolated from human source in Sri Lanka in 1968 |
| Autumnalis | Weerasinghe | Weerasinghe | L. interrogans | Isolated from human source in Sri Lanka in 1965 |
| Pyrogenes | Pyrogenes | Salinem | L. interrogans | Common in various Rattus spp. throughout Asia |
| Bataviae | Bataviae | Swart | L. interrogans | Common in various Rattus spp. throughout Asia |
| Grippotyphosa | Ratnapura | Wumalasena | L. kirschneri | Isolated from human source in Sri Lanka in 1966 and has since been isolated from cattle/buffalo |
| Canicola | Canicola | Hond Utrecht IV | L. interrogans | Often associated with disease in dogs worldwide |
| Australis | Australis | Ballico | L. interrogans | Common in rodents near rice-growing areas in Asia and can have severe pulmonary symptoms |
| Pomona | Pomona | Pomona | L. interrogans | Found in domestic pigs |
| Javanica | Ceylonica | Piyasena | L. borgpetersenii | Isolated from human source in Sri Lanka in 1964 |
| Sejroe | Geyaweera | Geyaweera | L. borgpetersenii | Isolated from human source in Sri Lanka in 1965 |
| Sejroe | Hardjo | Hardjoprajitno | L. interrogans | Found in domestic cattle worldwide |
| Tarassovi | Tarassovi | Perepelitsin | L. borgpetersenii | Found in domestic pigs |
| Ballum | Ballum | Mus127 | L. borgpeterseni | Common in mice and some rat species |
| Loisiana | Lanka | R740 | L. interrogans | Isolated from a human source in Sri Lanka in 1967 |
| Celledoni | Celledoni | Celledoni | L. weilii | Representing species L. weilii |
| Sarmin | Sarmin | Sarmin | L. weilii | Representing species L. weilii |
| Semaranga | Patoc | Patoc 1 | L. biflexa | Used in MRI MAT panel |
MAT = microscopic agglutination test; MRI = magnetic resonance imaging.
Molecular diagnosis was carried out at the Oxford-Mahidol Welcome Trust Research Unit (Bangkok, Thailand). DNA was extracted from the 400 μL of EDTA blood sample by using the QIAamp DNA Blood Mini Kit (QIAGEN GmbH, Hilden, Germany) and eluted in 200 μL of Tris-EDTA buffer. DNA samples were stored at –20°C prior to use. A single-tube nested PCR was used to amplify a region of the 16S ribosomal DNA gene specific for pathogenic and intermediate Leptospira spp. The PCR primers were rrs-outer-F (5-CTCAGAACTAACGCTGGCGGCGCG-3′), rrs-outer-R (5′-GGTTCGTTACTGAGGGTTAAAACCCCC-3′), rrs-inner-F (5′-CTGGCGGCGCGTCTTA-3′), and rrs-inner-R (5′-GTTTTCACACCTGACTTACA-3′).
The resulting amplicon was predicted to be approximately 547 basepairs. A 25-μL PCR reaction contained 4.5 mM MgCl2, 200 μM dNTP, 1.25 units of Tag DNA polymerase (Roche, Indianapolis, IN), 0.150 pmol of each outer primer, 1.25 pmol of rrs-inner-F, 5 pmol of rrs-inner-R, 1 M betaine (Sigma-Aldrich, St. Louis, MO) and either 1 μL of DNA extracted from laboratory cultures or 5 μL of DNA extracted from EDTA blood samples taken from febrile patients. The PCR was performed in duplicate for each sample by using a PTC-200 Peltier Thermal Cycler (MJ Research, Boston, MA) and the following conditions: one cycle at 95°C for 2 minutes; 40 cycles at 95°C for 10 seconds, 67°C for 15 seconds, and 72°C for 30 seconds; 40 cycles at 95°C for 10 seconds, 55°C for 15 seconds, and 72°C for 30 seconds; and one cycle at 72°C for 7 minutes. Positive and negative controls were included in each test. The positive control was genomic DNA extracted from L. interrogans serovar Lai strain Lai spiked with human DNA. Human DNA was extracted from a 5-mL blood sample obtained from one person and extracted as described above for clinical samples. The negative control was reaction mixture minus DNA template. Amplicons were visualized by using 1.5% gel electrophoresis and staining with ethidium bromide. A positive PCR result was defined as the visualization of a band of the predicted size for one or both samples. Purified PCR products were sequenced by Macrogen Inc. (Seoul, South Korea) by using the rrs-inner-F and rrs-inner-R primers. Sequences were aligned using SeqManII software (DNASTAR Inc., Madison, WI), and reduced to a 443-basepair region of the ribosomal DNA gene (positions 89–531 of the ribosomal DNA gene of L. interrogans serovar Lai strain 56601 (GenBank accession no. NC_004342).
Sequelae.
All case-patients were followed-up until the end of hospitalization to obtain data on complications and sequelae.
Data analysis.
Data were managed and analyzed by using Epi-Info version 6.04 (Centers for Disease Control and Prevention, Atlanta, GA) and Statistical Package for Social Sciences version 13.0 (SPSS Inc., Chicago, IL), respectively. The proportion of definitive and presumptive cases among suspected cases and the 95% confidence intervals (CIs) for proportions were calculated. Descriptive analysis of the socio-demographic profile of patients and common clinical symptoms and signs was conducted. Results are presented as percentages with 95% CIs.
Ethical approval.
The protocol was reviewed and approved by the Post Graduate Institute of Medicine and the Ethical Review Board of the Faculty of Medicine, University of Peradeniya, Sri Lanka.
Results
Of 746 fever patients screened, 401 patients were enrolled in the study (Figure 1). Of these patients, 370 fulfilled the inclusion criteria and were clinical treated as cases of leptospirosis by the primary physicians. The remaining 31 patients were recruited on the basis of inclusion criteria alone and were treated by the primary physicians for a range of other conditions. Only 167 (41.6%) patients returned for follow-up and provided a second serum sample.
Figure 1.
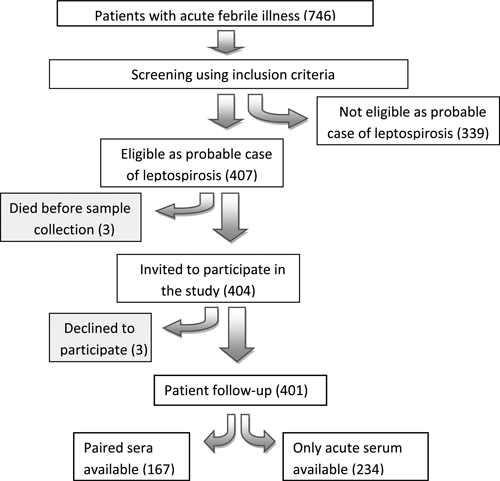
Selection of probable cases of leptospirosis, during the 2008 outbreak of leptospirosis in Kegalle, Kandy and Matale, Sri Lanka.
Disease confirmation.
Of the 401 probable cases, 155 (38.7%) were identified as having leptospirosis (Table 3) either by definitive (n = 92) or presumptive (n = 63) criteria according to LERG criteria (outlined above). Of the 167 patients for whom paired serum samples were available, 154 patients were managed clinically as having leptospirosis by treating physicians, of whom only 81 (52.6%) showed the required 4-fold increase/seroconversion in titer. Of the 13 patients treated as having other conditions, but recruited based on inclusion criteria, six (46.2%) had a positive MAT result.
Table 3.
Summary of case confirmation for 401 probable cases of leptospirosis during the leptospirosis outbreak in Kegalle, Kandy and Matale, Sri Lanka, 2008*
| Category | Diagnosis criteria | No. | % |
|---|---|---|---|
| Definitive cases (n = 92, 23.0%) | MAT, PCR positive | 3 | 0.8 |
| MAT positive | 84 | 21 | |
| PCR positive | 5 | 1.2 | |
| Presumptive cases | ELISA, surveillance case definition positive | 63 | 15.7 |
| Unconfirmed | 246 | 61.3 | |
| Total | 401 | 100.0 |
MAT = microscopic agglutination test; PCR = polymerase chain reacrion; ELISA = enzyme-linked immunosorbent assay. Total number tested with MAT = 167; PCR = 100; ELISA = 368.
Serologic data.
Of the battery of 22 serovars used in the MAT panel, 17 were reactive for one or more serum samples (Table 4). The most frequent antibody response (based on the highest titer for a given patient) was against L. interrogans serogroup Pyrogenes serovar Pyrogenes, which accounted for 28.7% of positive serum samples. Three isolates in the test panel (representing L. interrogans [n = 2] and L. borgpetersenii [n = 1]) each had the highest titer for 10 patients.
Table 4.
Frequency of MAT antibody titers to the test panel of Leptospira spp. among 87 MAT-positive cases during the leptospirosis outbreak, Kegalle, Kandy, and Matale, Sri Lanka, 2008*
| Species | Serogroup | Serovar | No. | % | Range of antibody titers in MAT† |
|---|---|---|---|---|---|
| L. interrogans | Pyrogenes | Pyrogenes | 25 | 28.7 | 200–6,400 |
| L. borgpetersenii | Javanica | Ceylonica | 10 | 11.5 | 100–3,200 |
| L. interrogans | Sejroe | Hardjo | 10 | 11.5 | 100–6,400 |
| L. interrogans | Hebdomadis | Hebdomadis | 10 | 11.5 | 100–6,400 |
| L. borgpetersenii | Sejroe | Geyaweera | 6 | 6.9 | 400–3,200 |
| L. interrogans | Loisiana | Lanka | 5 | 5.8 | 100–3,200 |
| L. interrogans | Autumnalis | Weerasinghe | 4 | 4.6 | 400–6,400 |
| L. santarosai | Autumnalis | Alice | 2 | 2.3 | 400–3,200 |
| L. interrogans | Australis | Australis | 2 | 2.3 | 200–400 |
| L. interrogans | Autumnalis | Autumnalis | 2 | 2.3 | 100–800 |
| L. interrogans | Icterohaemorrhagiae | Copenhageni | 2 | 2.3 | 1,600–3,200 |
| L. interrogans | Pomona | Pomona | 2 | 2.3 | 100–400 |
| L. kirschneri | Grippotyphosa | Ratnapura | 2 | 2.3 | 200–1,600 |
| L. interrogans | Canicola | Canicola | 2 | 2.3 | 100–200 |
| L. interrogans | Bataviae | Bataviae | 1 | 1.2 | 800 |
| L. weilii | Celledoni | Celledoni | 1 | 1.2 | 200 |
| L. weilii | Sarmin | Sarmin | 1 | 1.2 | 400 |
MAT = microscopic agglutination test.
Results represent highest titer for each patient.
Species identification.
Of the 8 PCR-positive samples, 16S rDNA sequencing was carried out on 6 samples. The deduced Leptospira species were L. interrogans (5) and L. weilli (1) (Figure 2).
Figure 2.
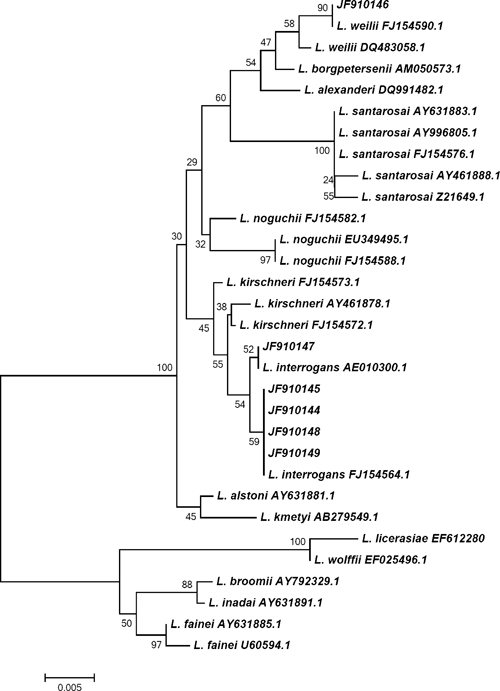
Phylogenetic tree based on the Leptospira 16S ribosomal DNA gene. Sequences obtained in this study are indicated in sample numbers 334, 68, 109, 229, 75, and 82. Sequences were aligned in MEGA4 using CLUSTALW, and phylogenetic distances were calculated in MEGA4 using maximum-likelihood. Numbers at nodes are bootstrap support after 500 replicates.
Clinical profile.
Median duration of fever before admission among confirmed cases was 5 days (interquartile range = 4–7), and the median duration of hospital stay was 4 days (interquartile range = range 3–6). The classical clinical picture of leptospirosis was not observed in most of the cases (Table 5).
Table 5.
Prevalence of clinical symptoms and signs among 155 confirmed case-patients with leptospirosis during the leptospirosis outbreak, Kegalle, Kandy, and Matale, Sri Lanka, 2008
| Clinical symptoms/signs | Definitive cases (n = 92) | Presumptive cases (n = 63) | ||
|---|---|---|---|---|
| No. | % | No. | % | |
| Headache | 76 | 82.6 | 55 | 87.3 |
| Myalgia | 71 | 77.2 | 47 | 74.6 |
| Arthralgia | 67 | 72.8 | 45 | 71.4 |
| Prostration | 55 | 59.8 | 52 | 82.5 |
| Nausea/vomiting | 57 | 62.0 | 45 | 71.4 |
| Anorexia | 46 | 50.5 | 35 | 55.6 |
| Conjunctival suffusion | 42 | 45.7 | 33 | 52.4 |
| Chills | 35 | 38.0 | 26 | 41.3 |
| Jaundice | 32 | 34.8 | 25 | 39.7 |
| Oliguria | 33 | 35.9 | 25 | 39.7 |
| Abdominal pain | 25 | 27.2 | 22 | 34.9 |
| Muscle tenderness | 28 | 30.4 | 18 | 28.6 |
| Diarrhea | 20 | 21.7 | 10 | 15.9 |
| Hepatomegaly | 16 | 17.4 | 8 | 12.7 |
| Shortness of breathing | 11 | 12.0 | 4 | 6.3 |
| Neck stiffness | 6 | 6.5 | 7 | 11.1 |
| Mental confusion | 5 | 5.4 | 2 | 3.2 |
Validity of presumptive case definition.
Diagnostic validity of proposed presumptive case definition (clinical leptospirosis and ELISA) was evaluated among 167 patients with paired serum samples, assuming paired sample MAT as the gold standard. Of these patients, definitive ELISA results (omitting the equivocal results) was available for 147 patients. The diagnostic sensitivity of the presumptive case definition was 66.2% and the specificity was 89.0% (Table 6).
Table 6.
Validity of the presumptive case definition of leptospirosis among 147 possible cases for whom paired serum samples were obtained during the leptospirosis outbreak in Kegalle, Kandy and Matale, Sri Lanka, 2008*
| Parameter | Estimate | 95% CI |
|---|---|---|
| Sensitivity | 66.2% | 54.9–76.0 |
| Specificity | 89.0% | 79.8–94.3 |
| Positive predictive value | 86.0% | 74.7–92.7 |
| Negative predictive value | 72.2% | 62.2–80.4 |
| Diagnostic accuracy | 77.5% | 70.1–83.5 |
| Likelihood ratio of a positive test result | 6.04 | 4.63–7.88 |
| Likelihood ratio of a negative test result | 0.38 | 0.35–0.41 |
| Diagnostic odds | 15.93 | 6.62–38.3 |
Eighteen patients with equivocal results in the enzyme-linked immunosorbent assay (ELISA) and two patients in whom ELISA results were available were not included in analysis. CI = confidence interval.
Sequelae.
Acute renal failure was classified based on the report of the Second International Consensus Conference of the Acute Dialysis Quality Initiative (ADQI) Group.15 Serum creatinine levels were greater than 1.5 μmol/L in 23 (14.8%) of 155 patients, and 11 of these patients had acute bilateral renal paranchymal disease detected by ultrasonography. For 76 (49%) patients, serum creatinine levels were not available. On the basis of the final diagnosis provided by the treating physicians, 11 (7.1%) patients had myocarditis and 6 (3.9%) patients had heart failure. Of the patients with myocarditis, 7 patients had T wave inversions, 3 had ST wave abnormalities, and 3 had tachyarrhythmia. We did not observe hyperbilirubinemia or pulmonary hemorrhage in this patient population. Three deaths were recorded among the 155 patients, giving a case-fatality rate of 1.9%. The three patients who died were hospitalized on the seventh day of illness and died within 24 hours of admission because of fulminant myocarditis. The mean ± SD duration of fever on admission among patients who had complications (n = 33) was 7.7 ± 3.8 days compared with 5.25 ± 2.2 days among other patients (t = 4.714, degrees of freedom = 153, P < 0.001).
Discussion
Leptospirosis is a globally important disease with a broad range of clinical manifestations that mimic a wide variety of acute infectious diseases. Because of the variable clinical manifestations, laboratory methods are essential for disease confirmation. The purpose of this analysis was to obtain scientifically sound information on leptospirosis in Sri Lanka and to discuss the implications of diagnosing leptospirosis in resource-poor settings in assessing the global burden of this disease. A central issue in leptospirosis is the difficulty in readily establishing the diagnosis. In the present study, of patients treated as if they did have leptospirosis, 52.6% (95% CI = 43.8–59.3%) were laboratory confirmed to have leptospirosis. Paired serum samples required for optimal serologic diagnosis (MAT) were available for only 167 of 404 patients. Serologic diagnosis ultimately only retrospectively confirms diagnosis and does not contribute actively to patient care. Because of this limitation of MAT, patients may not be efficiently referred for leptospirosis diagnosis outside a research setting. These data yet again argue that the lack of point of care diagnostics for leptospirosis not only impairs clinical care but also significantly limits regional and global public health assessments of the burden of this disease. This lesson is supported by the findings in leptospirosis outbreaks in Sri Lanka, a region where the disease is highly endemic and diagnosis remains a challenge, even with close cooperation with governmental health ministry laboratory resources.
Specifically with regard to the leptospirosis outbreak in Sri Lanka in 2008, the differential diagnosis of acute febrile illness is problematic. Even if one considers the 167 patients with paired serum samples, the confirmed diagnosis of only approximately 50% of suspected leptospirosis cases during this epidemic shows the limitations of clinical diagnosis of the syndrome of acute undifferentiated fever. One probable explanation is that there may have been a concurrent outbreak of another infectious disease with similar clinical characteristics, which may have been masked by the leptospirosis outbreak. A number of tropical diseases such as dengue,16–18 hantavirus infection,19,20 rickettsial infection18,21 and malaria22,23 may mimic the clinical presentation of leptospirosis. In Sri Lanka, dengue is highly endemic but not efficiently diagnosed. Rickettsial diseases are also common in small pockets in all three districts but difficult to diagnosis. Hantavirus infection is usually not considered in the differential diagnosis of leptospirosis patients in Sri Lanka, the incidence is not known, and diagnostic testing is not routinely available. Nevertheless, in 1995–1996, Sunil Chandra and others showed that 34.5% of patients with leptospirosis-like illness had hantavirus infections.24 During the 2008 outbreak, Gamage and others also described eight cases of hantavirus infection among 103 suspected cases of leptospirosis in Peradeniya.11 These data suggest that hantavirus infection among patients with leptospirosis-like illness in Sri Lanka is a possibility.24 The major drawback of the lack of definitive diagnosis is that a large number of patients developed a leptospirosis-like illness, 10–20% of whom had complications and occasional deaths. If confirmatory diagnostic testing was available and the correct diagnosis was made, some of the deaths caused by other conditions and complications may have been prevented.
Serologic data showed that multiple serovars likely circulate in Sri Lanka. However, proper interpretation of these data is difficult because of probable cross-reactivity among different serogroups. Smythe and others have shown that correct detection of serovars by using a panel of standard serovars in MAT was as low as 33% when compared with titers against the homologous isolate obtained from a patient.25 Levett reported similar findings but a slightly higher sensitivity (46.4% detection rate for 151 isolates) and that high MAT titer did not necessarily predict the serovar isolated from clinical samples.26 Thus, the results of the present study should not be used as definitive evidence of the serovars circulating in Sri Lanka. Nevertheless, it seems evident that a range of serovars are causing human leptospirosis in Sri Lanka. Prevalence of serovars Hardjo, Hebdomadis, and Sejroe in the present study might be an indication of the important role of cattle/buffaloes in human leptospirosis in Sri Lanka.
This study expands our knowledge of circulating Leptospira in Sri Lanka. The first report of species identification was published by Koizumi and others, which reported two cases of infection with L. interrogans and one case of infection with L. krischneri.10 The present study is consistent with a diversity of Leptospira infection in Sri Lanka. However, the molecular epidemiology of Leptospira species in Sri Lanka needs further investigation.
The proportion of renal complications among the confirmed cases of leptospirosis in this sample was 14.8% (95% CI = 9.8–21.1%). In some studies, rates of renal complications are as high as 44–67%27 whereas most studies report a range of renal complications ranging from 10% to 40%.17,28,29 In previously reported studies in Sri Lanka, renal complication rates ranged from 25%30 to 70%.31 The most probable explanation for the low complication rates is that the present study involved active case finding, which resulted in detecting mild-to-moderate cases, which could have missed in routine clinical practice. It is also possible that the set of infecting Leptospira in the region of study had different virulence capacities, which led to varying complications. Interestingly, pulmonary hemorrhage and other pulmonary complications were not observed among these confirmed cases. Previous studies in the National Hospital of Sri Lanka (Colombo) reported acute lung injury among 31% of cases,30 and at Peradeniya Hospital (Central Province) at least 6–8 deaths because of pulmonary hemorrhage during this study period were attributed to leptospirosis.32 This finding could be caused by varying virulence of infecting serovars in different studies. Different clinical manifestations caused by specific serovars are well documented in the literature. Leptospira interrogans serovars Icterohaemorrhagiae, Copenhageni, Javanica, and Bataviae are reported as causing severe icteric, typical Weil's disease, and most of the other serovars are associated with mild disease.33
The frequency of leptospirosis with undifferentiated fever and the absence of the classical clinical picture of leptospirosis in this study complicates the accurate disease burden estimate in countries where proper diagnostic facilities are not available. Use of presumptive diagnosis was having a diagnostic sensitivity of only 60% in this study, if the proposed case definition for surveillance activities is used. Use of this proposed presumptive case definition carries a risk of systematically underestimating the disease burden in countries with no access to diagnostic facilities and also missing considerable number of patients who will end up with severe complications. Clinicians need more efficient, precise, and specific point of care diagnostic tools, which would enable prompt individual treatment. Contrary to popular belief, accurate and more sensitive diagnostic tests are equally important to epidemiologists and public health professionals for rapid outbreak response and better disease surveillance. We proposed widening of clinical criteria for surveillance purposes to improve the sensitivity of presumptive case definition and urge scientific community and funding agencies to develop rapid and accurate diagnostic tools.
The present study was carried out in only three districts in Sri Lanka and was based in only three large hospitals. A large network of other primary and secondary care hospitals exists within these three districts. Patients with mild-to-moderate leptospirosis could be visiting these other hospitals. Furthermore, because of the protean manifestations of leptospirosis, a large number of patients could be having only mild symptoms for which they visit general practitioners or outpatient departments and are not treated as in-ward patients. Thus, this study is not representative of all patients with leptospirosis in these three districts. The clinical manifestations discussed are only for persons with moderate-to-severe cases who required hospitalization. The percentage of patients having specified clinical features is an overestimation because of this selection bias. Diagnosis of patients with a single serum sample was achieved mainly by ELISA, for which the sensitivity is not optimal. The probability of missing several cases with leptospirosis because of this diagnostic procedure could not be excluded. Selecting hospital controls makes selection bias likely. Furthermore, hospitalized patients are a population that are sicker than ambulatory patients, and thus represents a biased sampling. Therefore, any generalization of the case-control study results should be made cautiously.
ACKNOWLEDGMENTS
We thank all physicians, medical officers, and nursing staff in three hospitals and the epidemiology unit for their support during this study. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Footnotes
Financial support: This study was supported by World Health Organization research grants through the epidemiology unit of Sri Lanka and Welcome Trust through Oxford-Mahidol Medicine Research Unit, Bangkok, Thailand; the U.S. Public Health Service (grants K24AI068903 and D43TW007120); and the University of California San Diego School of Medicine. The ELISA kits for were provided by Inverness Medicals (Sinnamon Park, Queensland, Australia).
Authors' addresses: Suneth B. Agampodi, Department of Community Medicine, Faculty of Medicine and Allied Sciences, Rajarata University of Sri Lanka, Saliyapura, 50008, Sri Lanka and Department of Medicine, School of Medicine, University of California, George Palade Laboratories, La Jolla, CA, E-mails: sunethagampodi@yahoo.com or suneth1@mobileemail.vodafone.ik. Sharon J. Peacock, Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Ratchathewi, Bangkok, Thailand, Department of Microbiology and Immunology, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, and Department of Medicine, University of Cambridge, Addenbrooke's Hospital, Cambridge CB2 0QQ, United Kingdom. Vasanthi Thevanesam, Thamarasi Senaratne, and Athula Kumara, Department of Microbiology, Faculty of Medicine, University of Peradeniya, Peradeniya, 20400, Sri Lanka. Danaseela B. Nugegoda and Sahan Perera, Department of Community Medicine, Faculty of Medicine and Allied Sciences, Rajarata University of Sri Lanka, Saliyapura, 50008, Sri Lanka. Lee Smythe, Mary Ann Burns, and Michael Dohnt, World Health Organization/Food and Agricultural Organization/World Organization for Animal Health, Collaborating Centre for Reference and Research on Leptospirosis, Archerfield, Queensland 4108, Australia. Janjira Thaipadungpanit and Siriphan Boonsilp, Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Ratchathewi, Bangkok 10400, Thailand. Scott B. Craig, World Health Organization/Food and Agricultural Organization/World Organization for Animal Health Collaborating Centre for Reference and Research on Leptospirosis, Archerfield, Queensland, Australia and Faculty of Science, Health and Education, University of the Sunshine Coast, Sippy Downs, Queensland, Australia. Paba Palihawadana, Epidemiology Unit, Colombo, Sri Lanka. Joseph M. Vinetz, Department of Medicine, School of Medicine, University of California, George Palade Laboratories, La Jolla, CA.
References
- 1.1999. Leptospirosis worldwide Wkly Epidemiol Rec. 1999;74:237–242. [PubMed] [Google Scholar]
- 2.Cachay ER, Vinetz JM. A global research agenda for leptospirosis. J Postgrad Med. 2005;51:174–178. [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks GF, Butel JF, Morse SA. Leptospira and Leptospirosis. Medical Microbiology. Singapore: McGraw Hill Education (Asia); 2004. pp. 338–340. [Google Scholar]
- 4.World Health Organization . Report of the First Meeting of Leptospirosis Burden Epidemiology Reference Group. Geneva: World Health Organization; 2010. pp. 1–34. [Google Scholar]
- 5.Agampodi S, Peacock SJ, Thevanesam V. The potential emergence of leptospirosis in Sri Lanka. Lancet Infect Dis. 2009;9:524–526. doi: 10.1016/S1473-3099(09)70211-7. [DOI] [PubMed] [Google Scholar]
- 6.Unit E Selected notifiable diseases reported by Medical Officers of Health. Wkly Epidemiol Rec. 2009;36:4. [Google Scholar]
- 7.Unit E Selected notifiable diseases reported by Medical Officers of Health. Wkly Epidemiol Rec. 2010;37:4. [Google Scholar]
- 8.Unit E Selected notifiable diseases reported by Medical Officers of Health. Wkly Epidemiol Rec. 2011;38:4. [Google Scholar]
- 9.Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008;12:351–357. doi: 10.1016/j.ijid.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Koizumi N, Gamage CD, Muto M, Kularatne SA, Budagoda BD, Rajapakse RP, Tamashiro H, Watanabe H. Serological and genetic analysis of leptospirosis in patients with acute febrile illness in Kandy, Sri Lanka. Jpn J Infect Dis. 2009;62:474–475. [PubMed] [Google Scholar]
- 11.Gamage CD, Yasuda SP, Nishio S, Kularatne SA, Weerakoon K, Rajapakse J, Nwafor-Okoli C, Lee RB, Obayashi Y, Yoshimatsu K, Arikawa J, Tamashiro H. Serological evidence of Thailand virus-related hantavirus infection among suspected leptospirosis patients in Kandy, Sri Lanka. Jpn J Infect Dis. 2011;64:72–75. [PubMed] [Google Scholar]
- 12.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, Apiwatanaporn A, Slack AT, Suputtamongkol Y, White NJ, Feil EJ, Day NP, Peacock SJ. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1:e56. doi: 10.1371/journal.pntd.0000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levett PN, Branch SL. Evaluation of two enzyme-linked immunosorbent assay methods for detection of immunoglobulin M antibodies in acute leptospirosis. Am J Trop Med Hyg. 2002;66:745–748. doi: 10.4269/ajtmh.2002.66.745. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization . Leptospirosis. Communicable Disease Epidemiological Profile. Geneva: World Health Organization; 2010. pp. 102–108. [Google Scholar]
- 15.Bellomo R, Ronco C, Kellum JA, Mehta RL, Palevsky P. Acute renal failure—definition, outcome measures, animal models, fluid therapy and information technology needs: the Second International Consensus Conference of the Acute Dialysis Quality Initiative (ADQI) Group. Crit Care. 2004;8:R204–R212. doi: 10.1186/cc2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders EJ, Rigau-Perez JG, Smits HL, Deseda CC, Vorndam VA, Aye T, Spiegel RA, Weyant RS, Bragg SL. Increase of leptospirosis in dengue-negative patients after a hurricane in Puerto Rico in 1996. Am J Trop Med Hyg. 1999;61:399–404. doi: 10.4269/ajtmh.1999.61.399. [DOI] [PubMed] [Google Scholar]
- 17.Cruz LS, Vargas R, Lopes AA. Leptosoirosis: a worldwide resurent zoonoses and important cause of acute renal failure and death in developing nations. Eth Dis. 2009;19((Suppl 1)):S137–S141. [PubMed] [Google Scholar]
- 18.Deepak NA, Patel ND. Differential diagnosis of acute liver failure in India. Ann Hepatol. 2006;5:150–156. [PubMed] [Google Scholar]
- 19.Santos VM, Rocha de Sa DA, Turra TZ, Ferreira Borges NM, Nascimento UM, Damasceno EA. Hantavirus pulmonary syndrome in Brasilia periphery: a diagnostic challenge. J Infect Dev Ctries. 2009;3:639–643. doi: 10.3855/jidc.558. [DOI] [PubMed] [Google Scholar]
- 20.Akritidis N, Boboyianni C, Pappas G. Reappearance of viral hemorrhagic fever with renal syndrome in northwestern Greece. Int J Infect Dis. 2010;14:e13–e15. doi: 10.1016/j.ijid.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 21.Nachega JB, Bottieau E, Zech F, Van Gompel A. Travel-acquired scrub typhus: emphasis on the differential diagnosis, treatment, and prevention strategies. J Travel Med. 2007;14:352–355. doi: 10.1111/j.1708-8305.2007.00151.x. [DOI] [PubMed] [Google Scholar]
- 22.Leelarasamee A, Chupaprawan C, Chenchittikul M, Udompanthurat S. Etiologies of acute undifferentiated febrile illness in Thailand. J Med Assoc Thai. 2004;87:464–472. [PubMed] [Google Scholar]
- 23.Murav'Ev NV. Problem of differential diagnosis of malaria from leptospirosis [in German] Zh Mikrobiol Epidemiol Immunobiol. 1956;27:97–101. [PubMed] [Google Scholar]
- 24.Sunil-Chandra NP, Premaratne R, Somasiri DA, de Silva HJ. Evidence of leptospira and hanta virus co-infections amongst pateints hospitalized for leptospirosis-like illness. Bulletin of the Sri Lankan Colleage of Microbiology. 2003;1:32–33. [Google Scholar]
- 25.Smythe LD, Wuthiekanun V, Chierakul W, Suputtamongkol Y, Tiengrim S, Dohnt MF, Symonds ML, Slack AT, Apiwattanaporn A, Chueasuwanchai S, Day NP, Peacock SJ. The microscopic agglutination test (MAT) is an unreliable predictor of infecting Leptospira serovar in Thailand. Am J Trop Med Hyg. 2009;81:695–697. doi: 10.4269/ajtmh.2009.09-0252. [DOI] [PubMed] [Google Scholar]
- 26.Levett PN. Usefulness of serologic analysis as a predictor of the infecting serovar in patients with severe leptospirosis. Clin Infect Dis. 2003;36:447–452. doi: 10.1086/346208. [DOI] [PubMed] [Google Scholar]
- 27.Hurst FP, Neff RT, Katz AR, Buchholz AE, Sasaki DM, Berg BW, Abbott KC. Acute kidney injury requiring hemodialysis in patients with anicteric leptospirosis. Clin Nephrol. 2009;72:186–192. doi: 10.5414/cnp72186. [DOI] [PubMed] [Google Scholar]
- 28.Merien F, Perolat P. Public health importance of human leptospirosis in the South Pacific: a five-year study in New Caledonia. Am J Trop Med Hyg. 1996;55:174–178. doi: 10.4269/ajtmh.1996.55.174. [DOI] [PubMed] [Google Scholar]
- 29.Yersin C, Bovet P, Merien F, Wong T, Panowsky J, Perolat P. Human leptospirosis in the Seychelles (Indian Ocean): a population-based study. Am J Trop Med Hyg. 1998;59:933–940. doi: 10.4269/ajtmh.1998.59.933. [DOI] [PubMed] [Google Scholar]
- 30.Gunawardhana SA, Sellahewa KH. Clinical features of leptospirosis: a prospective descriptive study at the National Hospital of Sri Lanka (NHSL) in 2007. Ceylon Med J. 2008;53:155–156. doi: 10.4038/cmj.v53i4.293. [DOI] [PubMed] [Google Scholar]
- 31.Rajasuriya K, Munasinghe DR, Vitarne UT, Ratnaike VT, Peiris OA. Leptospirosis in Ceylon: a Clinical Study. Ceylon Med J. 1964;93:136–153. [PubMed] [Google Scholar]
- 32.Kularatne SA. Zoonotic and Emerging Zoonotic Diseases and their Prevention in the SAARC Region. Peradeniya, Sri Lanka: University of Peradeniya; 2008. (Human leptospirosis: case studies). Faculty of Veterinary Medicine and Animal Science. [Google Scholar]
- 33.Faine S. Leptospira and Leptospirosis. Boca Raton, FL: CRC Press; 1994. [Google Scholar]


