Abstract
MAP kinases phosphorylate specific groups of substrate proteins. Here we show that the amino acid sequence FXFP is an evolutionarily conserved docking site that mediates ERK MAP kinase binding to substrates in multiple protein families. FXFP and the D box, a different docking site, form a modular recognition system, as they can function independently or in combination. FXFP is specific for ERK, whereas the D box mediates binding to ERK and JNK MAP kinase, suggesting that the partially overlapping substrate specificities of ERK and JNK result from recognition of shared and unique docking sites. These findings enabled us to predict new ERK substrates and design peptide inhibitors of ERK that functioned in vitro and in vivo.
Keywords: MAP kinase, ERK, JNK, KSR, ETS transcription factor
Mitogen-activated protein (MAP) kinases are components of signaling cascades that regulate normal development and pathological processes such as oncogenesis. MAP kinases were identified during biochemical searches for serine/threonine-specific protein kinases stimulated by growth factors in vertebrate cells (for review, see Sturgill and Wu 1991). MAP kinases were also identified in screens for mutations that affect intercellular signaling in yeast, worms, and flies (for review, see Ferrell 1996). Together, these investigations revealed that MAP kinases function in many cell types, are regulated by a diverse group of extracellular stimuli, and mediate a wide variety of cellular responses. MAP kinases can be divided into subfamilies based on specific conserved residues, particularly a TXY motif in the activation loop (Ferrell 1996). The three best-characterized subfamilies in vertebrates are named extracellular-regulated kinase (ERK), c-Jun amino-terminal kinase (JNK, also called stress-activated protein kinase), and p38. There are probably several additional vertebrate MAP kinase subfamilies, since Saccharomyces cerevisiae contains six different MAP kinases (Madhani and Fink 1998). Here we use the name MAP kinase to refer to all members of the family, and the names ERK, JNK, and p38 to refer to members of those subfamilies.
MAP kinases function in modules composed of three protein kinases (for review, see Marshall 1994). MAP kinase kinase kinases, such as Raf-1, phosphorylate and thereby activate MAP kinase kinases, such as MEK (MAP kinase kinase or ERK kinase). MAP kinase kinases are serine/threonine and tyrosine-specific protein kinases that phosphorylate the TXY motif and thereby activate MAP kinases. In general, MAP kinases in different subfamilies are members of separate modules and are regulated by distinct extracellular stimuli (for review, see Whitmarsh and Davis 1996). For example, ERK is activated strongly by receptor tyrosine kinases (RTK) such as the epidermal growth factor receptor, whereas JNK is activated strongly by stress stimuli such as ultraviolet light. Several of the signaling pathways leading from extracellular stimuli to the activation of a MAP kinase module are well defined, whereas others have yet to be characterized in detail.
Whereas the upstream signaling events that regulate MAP kinases have been characterized extensively, considerably less is known about how MAP kinases regulate cell fates and contribute to the specificity of signaling pathways. Important questions that remain largely unanswered include: (1) How do MAP kinases recognize specific proteins as substrates? (2) What proteins are phosphorylated by a particular MAP kinase in different cell types and in different organisms? Answers to these questions will illuminate how the same MAP kinase mediates different cell fates in different developmental contexts and how MAP kinases from separate subfamilies mediate different cellular responses.
In the case of ERK, >50 different proteins have been reported to be substrates (for reviews, see Davis 1993; Karin 1995; Treisman 1996; Whitmarsh and Davis 1996; Madhani and Fink 1998). These include signaling proteins likely to function upstream of ERK such as Son-of-sevenless (Sos) guanine nucleotide exchange factor and MEK; signaling proteins likely to function downstream of ERK such the protein kinase pp90rsk; transcription factors such as c-Fos, GATA-2, c-Myc, and ETS proteins including Elk-1, LIN-1, and Aop/Yan; and proteins involved in a wide variety of other processes. These findings suggest that ERK plays a central role in signal propagation and feedback regulation. Furthermore, ERK is a transition point between signaling proteins and regulators of differentiation, suggesting it makes an important contribution to the specificity of RTK–Ras–ERK signaling pathways. Although a large number of ERK substrates have been identified, the understanding of ERK function remains fragmentary, as ERK probably phosphorylates different substrates in different cell types and the cellular context of most substrates has yet to be defined. In addition, many ERK substrates probably have not been identified. Substrates of JNK have been characterized less extensively, but it is notable that they include proteins that are also phosphorylated by ERK, such as Elk-1, as well as unique substrates (for review, see Minden and Karin 1997).
Little is known about how ERK recognizes such a diverse group of substrates. Although the structure of ERK was determined using X-ray crystallography (Zhang et al. 1994; Canagarajah et al. 1997), this approach has not revealed how ERK interacts with substrate proteins, because the structure of ERK bound to a substrate has yet to be determined. Studies of residues in substrate proteins that are phosphorylated by ERK and assays of peptide substrates identified a serine or threonine followed by a proline (S/TP) as the minimal consensus sequence for phosphorylation by ERK (for review, see Davis 1993; Songyang et al. 1996). In addition, a proline at position −2 is favorable, whereas a proline at position −1 is unfavorable (the phosphoacceptor S/T is position 0). However, this information is not sufficient to explain how ERK recognizes specific proteins as substrates, because many proteins that contain S/TP sequences are not phosphorylated by ERK. Studies of the interaction of JNK with its substrate c-Jun have identified a sequence positioned amino-terminal to the S/TP sites that is required for efficient phosphorylation (the δ domain) (Adler et al. 1992, 1994; Hibi et al. 1993; Kallunki et al. 1996). This led to the hypothesis that a docking site on the substrate protein that is separate from the phosphorylation sites mediates the interaction with JNK (Karin 1995). Other protein kinases, such as cyclin–cdk2, also appear to interact with a docking site on substrate proteins (Adams et al. 1996). Although >50 proteins have been reported to be ERK substrates, only recently has one such docking site for ERK been identified (Yang et al. 1998a,b). This docking site, a domain of Elk-1 called the D box, is similar in sequence to the δ domain of c-Jun, and these two domains are interchangeable functionally, suggesting that the δ domain/D box is a docking site for both ERK and JNK.
Here we describe the identification and characterization of a different docking site, the amino acid sequence FXFP, which mediates interactions with ERK but not JNK. These two docking sites define three classes of substrates: Proteins that contain only FXFP, only the δ domain/D box, or both. These findings suggest that a modular system of docking sites regulates interactions of the different MAP kinases with various substrates. In substrates that contain both docking sites, the sites function additively to create a high-affinity interaction with ERK. Thus, this system also modulates the affinity of substrates for ERK and may determine which residues are phosphorylated. We used this information to develop peptide inhibitors of ERK and identify new ERK substrates, including the kinase suppressor of ras (KSR) family of protein kinases.
Results
FQFP is necessary for high-affinity interactions between ERK and ETS proteins in the Elk subfamily
The Caenorhabidtis elegans LIN-1 protein contains an ETS DNA-binding domain and presumably regulates transcription (Beitel et al. 1995). LIN-1 appears to be regulated directly by ERK, as LIN-1 is efficiently phosphorylated by Erk2 in vitro and lin-1 is regulated negatively by RTK–Ras–ERK pathways in vivo (Jacobs et al. 1998; Tan et al. 1998). We identified and characterized six gain-of-function (gf) mutations that impair the ability of lin-1 to be regulated negatively by RTK–Ras–ERK pathways and disrupt vulval development (Jacobs et al. 1998). Each mutation alters or eliminates FQFP, a sequence located in the carboxy-terminal region of LIN-1, suggesting this motif is important for LIN-1 regulation (Fig. 1a). We analyzed the sequences of other ETS proteins and found FQFP in vertebrate Elk-1, SAP-1a, and Net/ERP/SAP-2, highly related proteins that comprise the Elk subfamily of ETS proteins (Treisman 1994). FQFP is positioned near the carboxyl terminus of a conserved region named the C box that contains multiple S/TP motifs that are phosphorylated by ERK (Fig. 1a; Marais et al. 1993; Price et al. 1995). In addition, we found FQFHP in a comparable position of Drosophila Aop/Yan (Fig. 1a). Aop/Yan also appears to be regulated directly by ERK (O’Neill et al. 1994). This combination of sequence and functional similarities led us to propose that LIN-1 and Aop/Yan are members of the Elk subfamily of ETS proteins (Jacobs et al. 1998). Based on these observations, we hypothesized that FQFP is an evolutionarily conserved docking site that mediates ERK binding to these ETS proteins. According to this model, the lin-1(gf) mutations diminish phosphorylation of LIN-1 by ERK because they alter or eliminate FQFP, resulting in constitutively active LIN-1.
Figure 1.
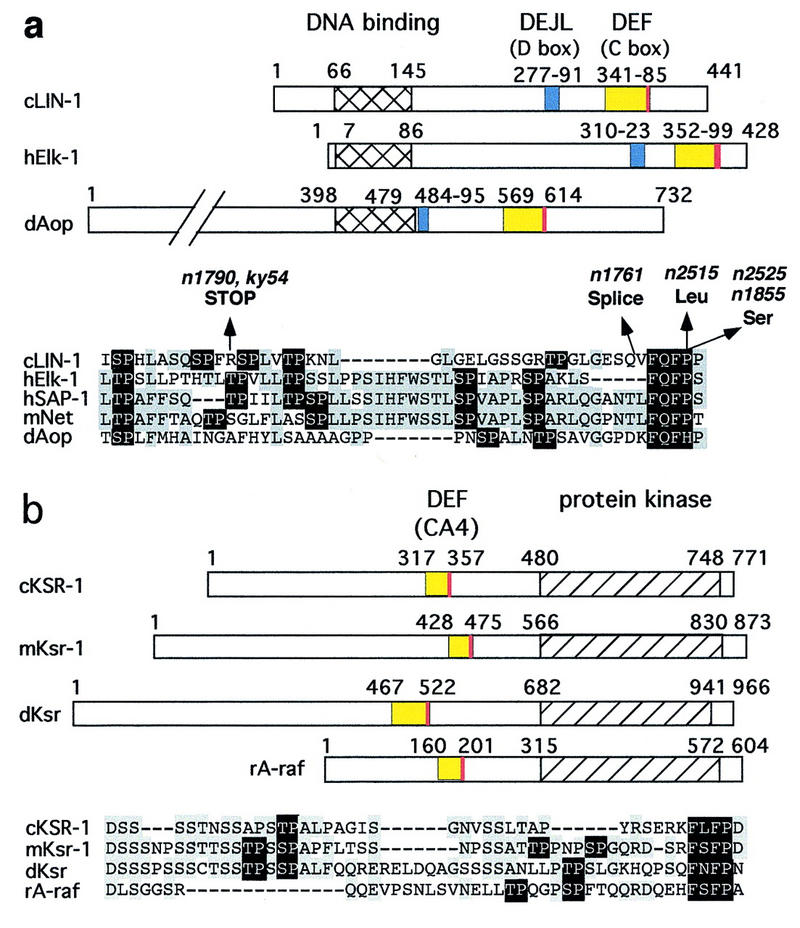
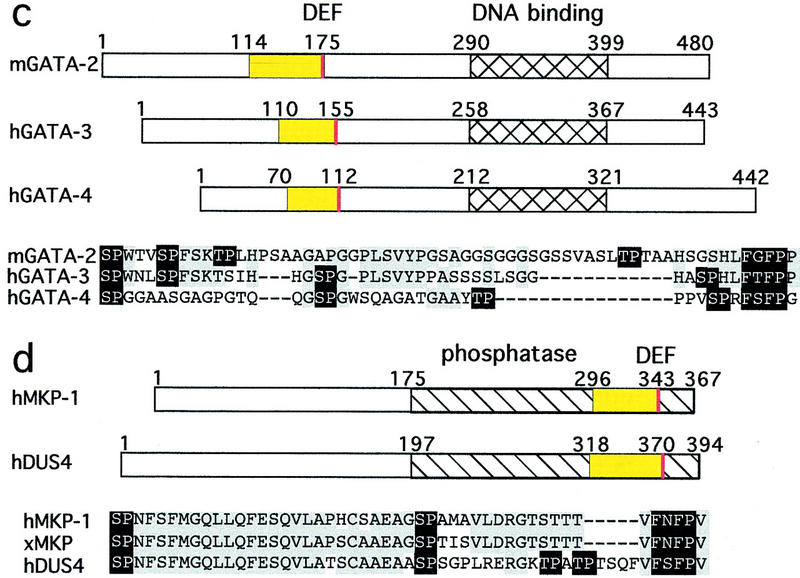
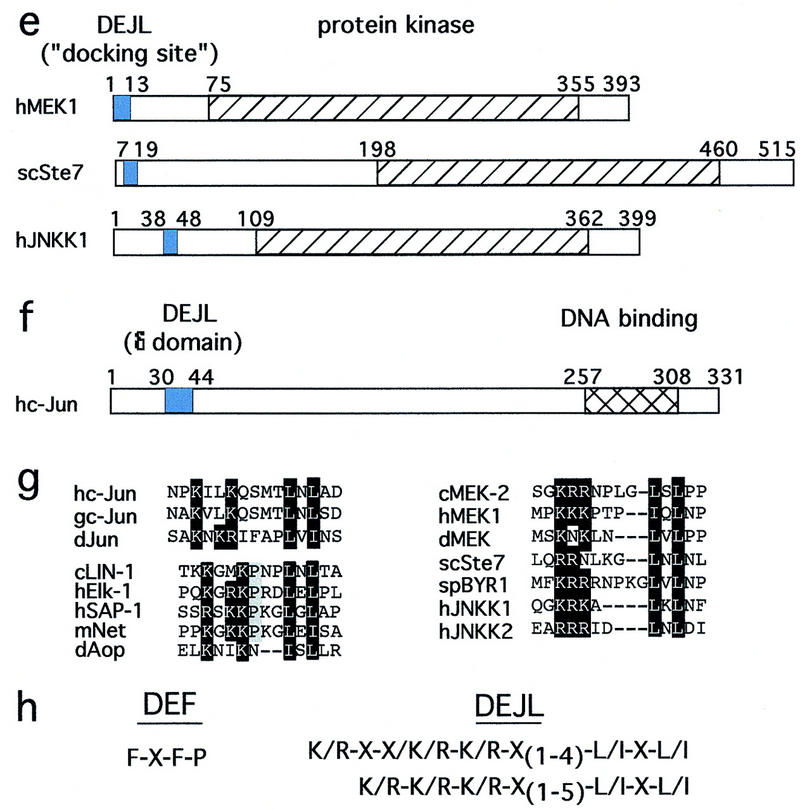
Multiple protein families contain docking sites for MAP kinases. Numbers indicate the first and last residue in each protein and domain. DEFs are red, and adjacent amino-terminal regions containing multiple S/TP sites are yellow. In alignments of these regions, residues conserved in two or more proteins are gray, and S/TP and FXFP sequences are black. DEJLs are blue. Alternate names for conserved regions are shown in parentheses. DNA-binding domains are hatched. Protein kinase domains are left diagonals. Phosphatase domains are right diagonals. (a,g) The Elk subfamily of ETS transcription factors: C. elegans LIN-1 (GenBank accession no. (g) 3158478), human Elk-1 (g119291), human SAP-1a (DEF, residues 353–402; DEJL, residues 316–329; g730711), murine Net (DEF, residues 328–380; DEJL, residues 290–303; g3041683), and D. melanogaster Aop/Yan (g418341). The positions and types of defect caused by the six lin-1(gf) mutations are shown above (Jacobs et al. 1998). n1790 and ky54 encode truncated proteins that terminate at residue 351. n1761 alters a splice site and probably results in ∼50 new amino acids following residue 379. n2515, n2525, and n1855 are missense mutations that change FQFP to FQFL or FQFS. (b) KSR protein kinases and A-raf: C. elegans KSR-1 (g1245976), murine Ksr-1 (g1171250), D. melanogaster Ksr (g1171240), and rat A-raf (g92443). (c) GATA transcription factors: murine GATA-2 (g2494682), human GATA-3 (g120962), and human GATA-4 (g1169845). (d) Dual-specificity protein phosphatases: human MAP kinase phosphatase-1 (MKP-1) (g1346900), Xenopus MKP (residues 298–345, g1050849), and human dual-specificity protein phosphatase-4 (DUS4, g2499745). (e,g) ERK-specific MAP kinase kinases include human MEK1 (g400274), C. elegans MEK-2 (residues 3–16; g2133469), D. melanogaster MEK (residues 1–12; g2499636), S. cerevisiae Ste7 (g134968), and Schizosaccharomyces pombe BYR1 (residues 1–15, g115194). JNK-specific MAP kinase kinases include human c-Jun amino-terminal kinase kinase 1 (JNKK1) (g1170596) and human JNKK2 (residues 23–34; g2558889). (f,g) c-Jun transcription factors: human c-Jun (g135298), chicken c-Jun (residues 26–40, g135295), and D. melanogaster Jun (residues 68–82; g135297). (g) Alignments of DEJLs, highly conserved positions are black. (h) The DEF consensus sequence based on 15 proteins (a–d). Two slightly different DEJL consensus sequences; the upper is based on eight DNA-binding proteins (g, left columns) and the lower is based on seven MAP kinase kinases (g, right column). Similar motifs in otherwise unrelated proteins might be descendants of a common ancestral motif that was dispersed during evolution by a mechanism such as exon shuffling, or they might be descendants of separate ancestral sequences and represent convergent evolution.
To test this hypothesis, we produced wild-type LIN-1 (residues 281–441), Elk-1 (residues 307–428), Aop/Yan (residues 480–732), or mutant proteins containing AAAP instead of FQFP in Escherichia coli. To minimize alterations of the overall protein structure, we did not mutate the proline. Proteins contained an amino-terminal glutathione S-transferase (GST) moiety to facilitate purification by affinity chromatography (Fig. 2). Each protein was assayed as a substrate for purified, recombinant, murine Erk2. Increasing the concentration of substrate protein resulted in saturation of phosphorylation. These data were used to determine Km, a measure of the binding affinity of a substrate and an enzyme (Fig. 2d).GST:LIN-1(281–441FQF) had a Km of 0.8 μm, which is fourfold lower than the 3.3 μm Km of myelin basic protein (MBP), a relatively good ERK substrate that is used frequently to assay ERK activity. By contrast, GST:LIN-1(281–441AAA) had a Km of 7 μm, which is ninefold higher than the Km for GST:LIN-1(281–441FQF). The relative acceptor ratio (Vmax/ Km) is an overall measure of the ability of a protein to function as a substrate. GST: LIN-1(281–441AAA) had a relative acceptor ratio that was 15-fold lower than the value for GST:LIN-1(281–441FQF). For Elk-1, changing FQF to AAA increased the Km threefold and decreased the relative acceptor ratio threefold (Fig. 2d). For Aop/Yan, changing FQF to AAA increased the Km fourfold and decreased the relative acceptor ratio sixfold (Fig. 2d). Thus, the FQF motif is necessary for high-affinity interactions between Erk2 and ETS proteins from worms, flies, and humans. However, the mutant proteins retained some ability to function as ERK substrates, suggesting other features of these proteins are also important for their interactions with ERK.
Figure 2.
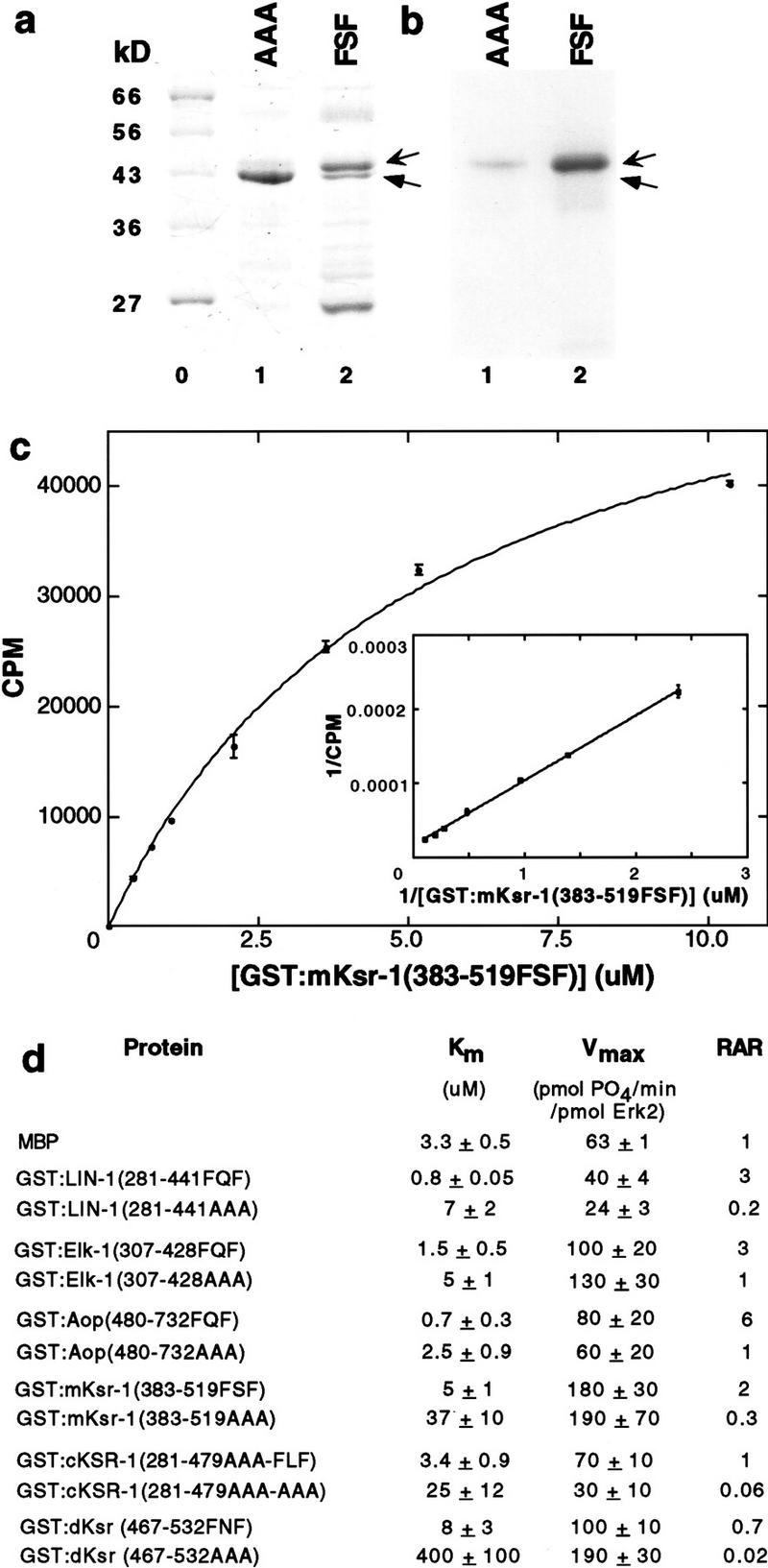
FXFP is necessary for ETS and KSR proteins to be high-affinity Erk2 substrates. Each protein listed in d was analyzed using the same experimental approaches; (a–c) show examples of the data using mKsr-1. (a,b) An approximately equal amount of GST:mKsr-1(383–519AAA) (lane 1, AAA) or GST: mKsr-1(383–519FSF) (lane 2, FSF) was incubated with Erk2 and [32P]ATP, fractionated by SDS-PAGE, stained with Coomassie blue (a) and exposed to film (b). Lane 0 contains protein markers; sizes are in kilodaltons (kD). Radioactive (phosphorylated) protein (upper arrows) migrated more slowly than nonradioactive (unphosphorylated) protein (lower arrows). Arrows indicate intact fusion proteins, because their size determined by gel mobility corresponds well with their predicted size of 40 kD, and proteins of this size were not present in similarly purified extracts of control cells (data not shown). Other bands are likely to be endogenous E. coli proteins or fragments of GST:mKsr-1 fusion proteins produced by proteolysis. (c) A kinetic analysis showing incorporated 32P measured by filter binding and scintillation counting (counts per minute, CPM) using increasing concentrations of GST:mKsr-1(383–519FSF). Values are the average of two samples; a bar indicates the range. The inset shows a Lineweaver-Burke plot of the data. (d) Km and Vmax were calculated from kinetic analyses like that shown in c; in each case a Lineweaver-Burke plot of the data closely approximated a straight line. Values are the mean ± s.d. of two to five separate experiments. To determine Vmax, we calculated total phosphate incorporated using the measured CPM and the specific activity of the [32P]ATP, and factored in the assay time and the amount of Erk2. Relative acceptor ratio (RAR) is Vmax/ Km; values were normalized by assigning a value of 1.0 to myelin basic protein (MBP). C. elegans KSR-1 contains FQFP beginning at position 309, eight residues upstream of CA4; to eliminate a potential redundant effect, we changed this FQF to AAA.
FQFP is sufficient to mediate high-affinity interactions with ERK
If FQFP is a docking site, then it might be sufficient to increase the affinity of other proteins and/or peptides for ERK. To test this prediction, we used an amino-terminal fragment of LIN-1 that lacks the C box (Fig. 3a). Although it contains nine S/TP sequences, GST:LIN-1(1–278+ +) was an extremely poor substrate with a Km of 40 μm (Fig. 3b–e). Introducing one copy of FQFP at an interior or carboxy-terminal position reduced the Km to 4.8 μm and 6 μm, respectively (Fig. 3b–e). Introducing two copies of FQFP reduced the Km 29-fold to 1.4 μm (Fig. 3b–e); this value is lower than the Km of MBP and similar to the Km of ETS proteins that are physiological ERK substrates (Fig. 2d). Thus, FQFP is sufficient to increase the affinity of a protein for Erk2 dramatically. These motif insertions also resulted in a greater Vmax, so that the relative acceptor ratio of GST:LIN-1(1–270FQF FQF) was 290-fold higher than that of the starting fusion protein (Fig. 3e).
Figure 3.
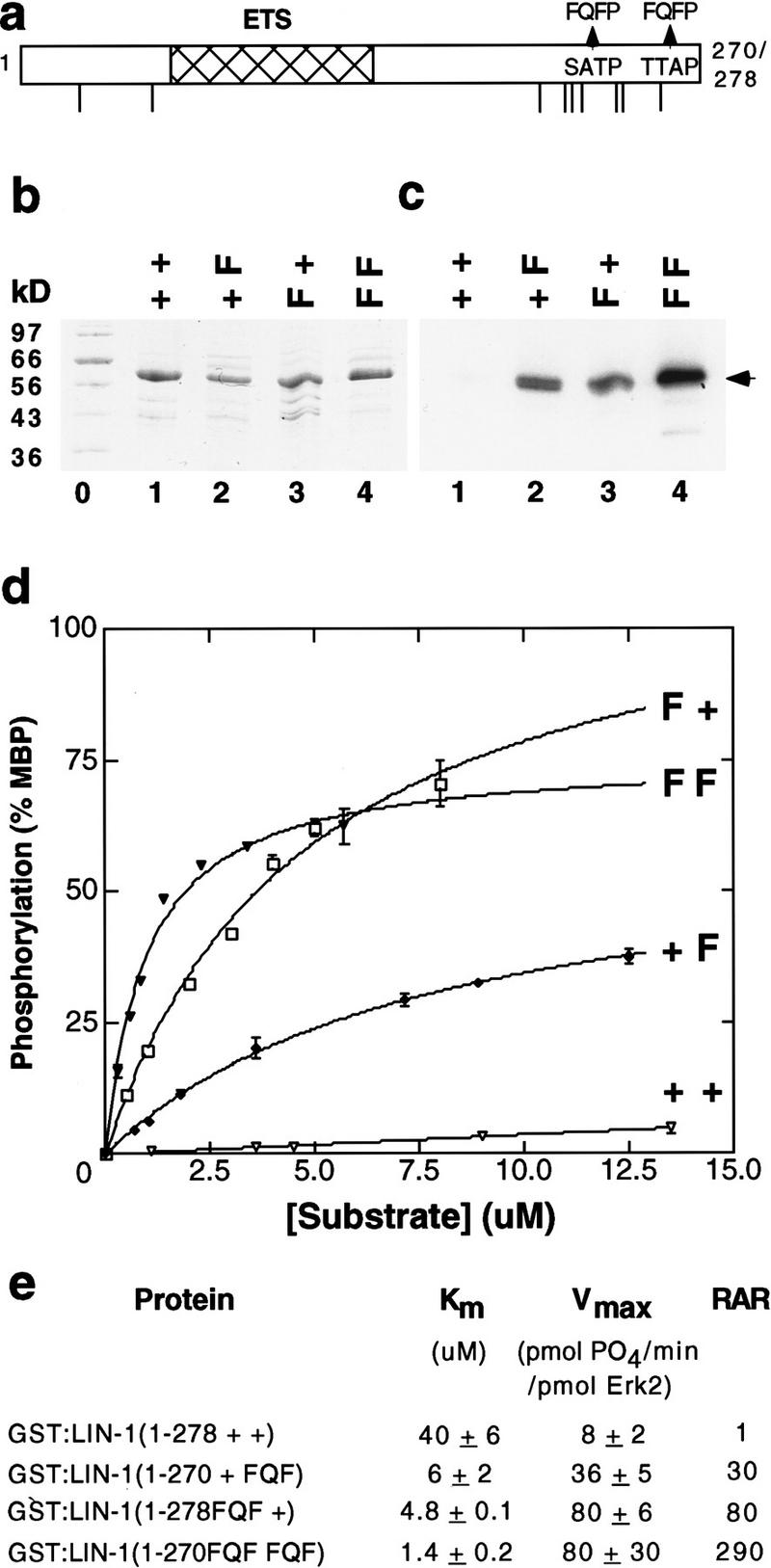
FQFP is sufficient to increase the affinity of a protein substrate for Erk2. (a) The amino-terminal region of C. elegans LIN-1 (residues 1–270 or 278) contains an ETS domain (hatched) and nine S/TP sequences (lines). Arrows indicate amino acids that were changed in the mutant proteins. To minimize changes to the tertiary structure, FQFP motifs included prolines present in LIN-1. (b,c) GST:LIN-1(1–278+ +) contained wild-type LIN-1 (lane 1, + +), GST:LIN-1(1–270+ FQF) contained FQFP beginning at position 267 (lane 2, + F), GST:LIN-1(1–278FQF +) contained FQFP beginning at position 239 (lane 3, F +), and GST:LIN-1(1–270FQF FQF) contained FQFP beginning at positions 239 and 267 (lane 4, F F). An approximately equal amount of each intact protein was incubated with Erk2 and [32P]ATP, fractionated by SDS-PAGE, stained with Coomassie blue (b), and exposed to film (c). Lane 0 contained protein size markers. An arrow indicates intact fusion proteins that displayed the predicted molecular weight of ∼56 kD. (d) A kinetic analysis showing incorporated 32P measured by filter binding using increasing concentrations of the proteins defined above. Values are the average of two samples; a bar indicates the range. Values were normalized by assigning a value of 100 to the amount of 32P incorporated by MBP assayed in parallel. (e) Km and Vmax were calculated from kinetic analyses; in each case, a Lineweaver-Burke plot of the data closely approximated a straight line. Values are the mean ± s.d. of two to four separate experiments. RARs were normalized by assigning a value of 1.0 to GST:LIN-1(1–278+ +).
To investigate whether FQFP was sufficient to increase the affinity of peptide substrates, we synthesized a series of pentadecapeptides based on the primary sequence of Elk-1 (residues 387–399). A peptide containing a PRSP phosphoacceptor sequence separated by four residues from FQFP had a Km of 130 μm (Fig. 4a,d; line 1). Changing FQFP to ATAP increased the Km 85-fold (Fig. 4a,d; line 6). Thus, FQFP is sufficient to increase the affinity of a peptide substrate for Erk2 dramatically.
Figure 4.
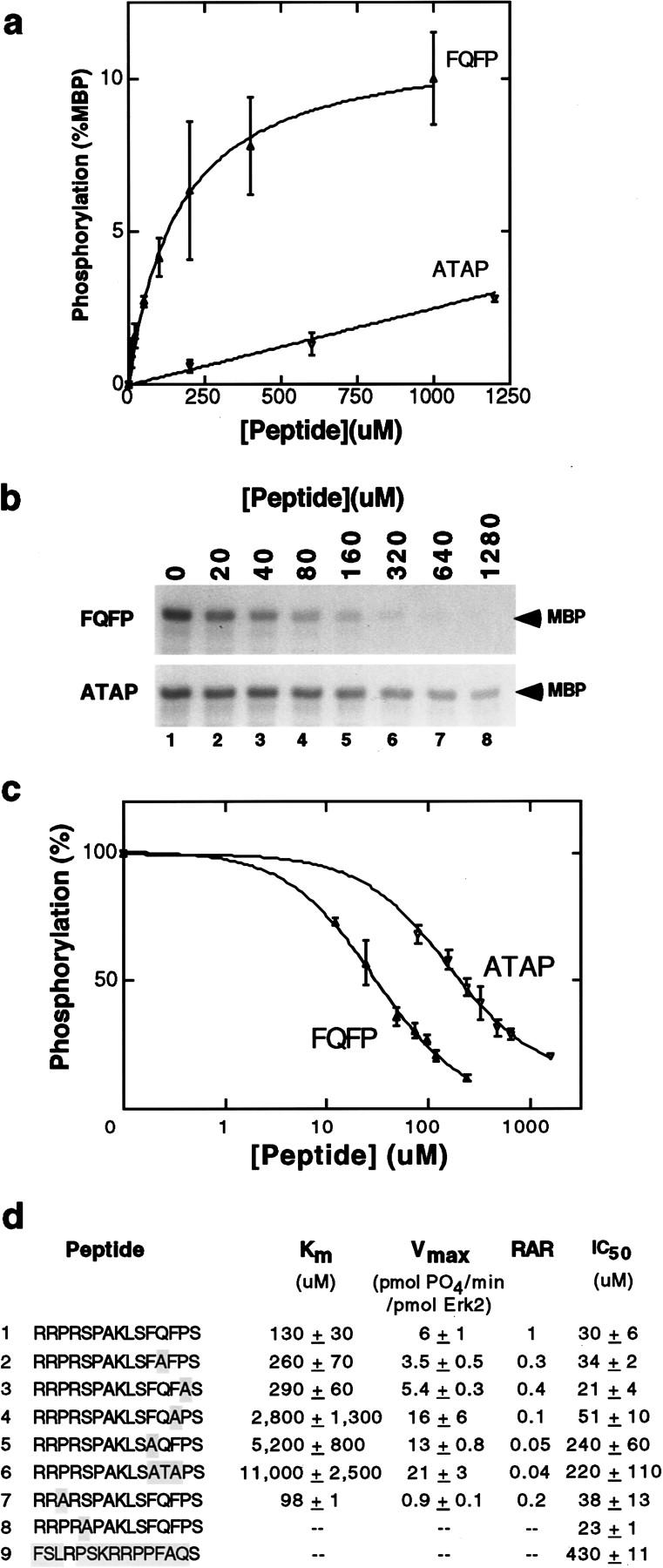
FQFP is sufficient to increase the ability of peptides to function as substrates and inhibitors of Erk2. (a) A kinetic analysis showing incorporated 32P measured by filter binding using increasing concentrations of RRPRSPAKLSFQFPS (closed triangles, FQFP) or RRPRSPAKLSATAPS (open triangles, ATAP). Values are the average of two samples; a bar indicates the range. Values were normalized by assigning a value of 100 to the amount of 32P incorporated by MBP assayed in parallel. The ATAP peptide displayed saturable phosphorylation at much higher concentrations than illustrated (data not shown). (b) Inhibition of Erk2 phosphorylation of MBP was measured by adding increasing concentrations of the FQFP peptide (top) or the ATAP peptide (bottom) to reactions containing [32P]ATP and 18 μm MBP. Reactions were fractionated by SDS-PAGE, and phosphorylated MBP was visualized by autoradiography (arrow). (c) Inhibition assays like those shown in b were quantified using a PhosphorImager (Molecular Dynamics). Signals for the FQFP peptide (closed triangles) or the ATAP peptide (open triangles) were normalized by assigning a value of 100% to assays containing no peptide and plotted on a logarithmic scale. Values are the averages of two samples; a bar indicates the range. (d) Highlighted residues in peptide sequences are differences compared to line 1. The peptide in line 9 contains the same residues as line 1, but the order was scrambled. Km and Vmax were calculated from kinetic analyses like those shown in a; in each case, a Lineweaver-Burke plot of the data closely approximated a straight line. Values are the mean ± s.d. of two independent experiments. RARs were normalized by assigning a value of 1.0 to the peptide in line 1. Peptides in lines 8 and 9 lack SP and were not phosphorylated significantly. Inhibition curves like those shown in c were used to determine IC50—the concentration of peptide that reduced Erk2 phosphorylation of MBP by 50%. Values are the mean ± s.d. of two independent experiments.
We used this assay to investigate the contributions of individual residues. Changing FQFP to FAFP or FQFA increased the Km about twofold (Fig. 4d; lines 2,3), indicating that the glutamine and proline residues play a minor role in binding affinity. However, this proline may have a structural function that is more important in proteins than peptides, as missense mutations that alter this proline in LIN-1 cause a vulval defective phenotype in worms and changing this proline to leucine increases the Km of full-length LIN-1 by fourfold (Jacobs et al. 1998). Changing FQFP to FQAP or AQFP increased the Km 21-fold and 40-fold, respectively (Fig. 4d; lines 4,5), indicating that both phenylalanines play a major role in binding affinity.
A proline at position −2 increases phosphorylation by ERK (Davis 1993). In contrast to alterations of FQFP, which affected primarily Km, changing PRSP to ARSP did not change the Km significantly and reduced the Vmax sixfold (Fig. 4d; line 7). Therefore, this proline is not an important determinant of binding affinity, but rather it accelerates Erk2 catalysis.
Peptides containing FQFP inhibit ERK in vitro and in vivo
To investigate whether our findings could be used to develop ERK inhibitors, we measured the ability of peptides to inhibit Erk2 phosphorylation of MBP. A peptide containing PRSP separated by four residues from FQFP inhibited phosphorylation of MBP, and the concentration necessary to inhibit phosphorylation by 50% (IC50) was 30 μm (Fig. 4b,c,d; line 1). A control peptide containing the same amino acids in a random order had an IC50 of 430 μm, a 14-fold increase, indicating that the residue order is essential for effective inhibition (Fig. 4d; line 9).
We used this assay to investigate the significance of individual residues. Changing FQFP to FAFP or FQFA did not affect inhibition significantly (Fig. 4d; lines 2,3). Changing FQFP to FQAP, AQFP, or ATAP increased the IC50 two-, eight-, and sevenfold, respectively (Fig. 4d; lines 4–6). These results indicate that FQFP mediates binding of peptides to Erk2 and peptide binding inhibits Erk2 from phosphorylating a protein substrate. These findings are consistent with the experiments using peptides as substrates for Erk2, as they also suggest that the two phenylalanine residues are the most important determinants of binding.
The SP acceptor site did not cause significant inhibition, because changing PRSP to PRAP or ARSP did not affect IC50 significantly (Fig. 4d; lines 7–8). These data demonstrate that FQFP-mediated binding to ERK does not require a phosphoacceptor site and support the model that FQFP is a docking site that interacts with an ERK binding pocket that is functionally distinguishable from the ERK active site.
To determine if pentadecapeptides containing FQFP can inhibit ERK in vivo, we assayed meiotic maturation of Xenopus oocytes. Treating immature oocytes with insulin activates the Ras–ERK pathway (Deshpande and Kung 1987; Korn et al. 1987; Nebreda et al. 1993), which results in morphological changes such as germinal vesicle breakdown (GVBD) and biochemical changes such as the activation of cdc2 kinase (Fig. 5; columns 1–2). We injected insulin-stimulated oocytes with a peptide containing FQFP (RRPRSPAKLSFQFPS) or a control peptide lacking FQFP (RRPRSPAKLSATAPS). The peptide containing FQFP reduced cdc2 kinase activity and the fraction of oocytes that displayed GVBD significantly, whereas the peptide lacking FQFP had no significant affect (Fig. 5; columns 3–4). To investigate further which step in this signaling pathway was inhibited by the peptide containing FQFP, we analyzed the degree of phosphorylation of ERK, an indirect measure of MEK activity. Extracts of oocytes injected with a peptide containing FQFP or a control peptide were analyzed by Western blotting using an antibody that recognizes phosphorylated ERK specifically. A similar amount of phosphorylated ERK was observed in both extracts, indicating that the peptide containing FQFP did not affect the activity of MEK or more upstream signaling events (data not shown). These results demonstrate that FQFP is sufficient to cause a peptide to function as an inhibitor of oocyte maturation. Furthermore, the peptide appeared to function at a step downstream of MEK and upstream of cdc2 kinase activation, consistent with the model that the peptide inhibited ERK activity.
Figure 5.
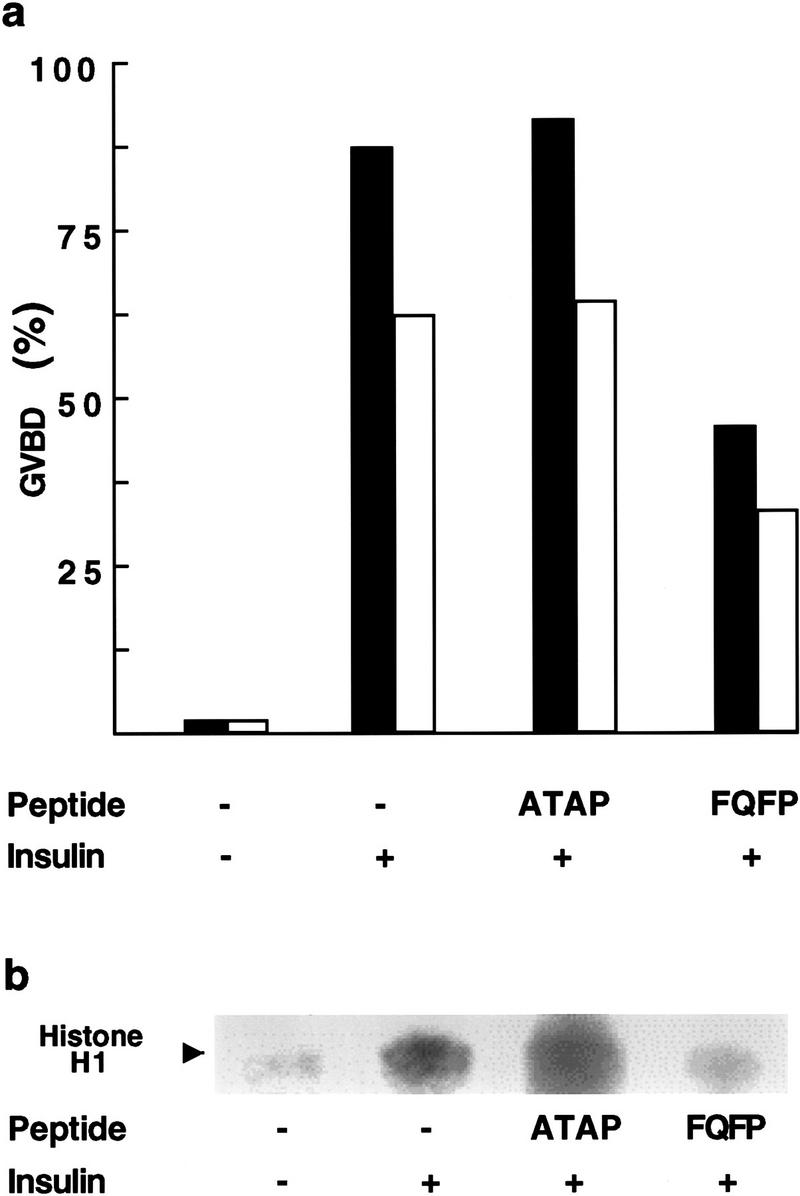
A peptide containing FQFP can inhibit Xenopus oocyte maturation. Immature Xenopus oocytes were microinjected with water alone (−) or water containing peptide RRPRSPAKLSFQFPS (FQFP) or peptide RRPRSPAKLSATAPS (ATAP). Some oocytes were treated with 8.25 μg/ml insulin (+). (a) Each oocyte was evaluated for the appearance of a white spot on the animal pole 24 hr after injection, an indication of germinal vesicle breakdown (GVBD). The black and white columns are data from two separate experiments; each column represents the average of ∼60 oocytes. The fraction of oocytes that were stimulated to undergo GVBD was somewhat higher in the first experiment (black columns), a reflection of variability in batches of oocytes. However, in both experiments the FQFP peptide reduced GVBD ∼50% whereas the ATAP peptide had no significant effect. (b) To measure cdc2 kinase activity, we prepared extracts 24 hr after injection, partially purified cdc2 kinase using p13suc beads, added the substrate protein histone H1 and [32P]ATP, and visualized radiolabeled histone H1 by SDS-PAGE and autoradiography (arrow). These results are representative of two separate experiments.
FXFP can be used to identify candidate ERK substrates, including KSR proteins that are high-affinity ERK substrates
To investigate whether FQFP motifs mediate interactions between ERK and substrates besides ETS proteins in the Elk subfamily and whether FQFP can be used to predict ERK substrates, we analyzed the sequences of proteins involved in vulval induction and found that C. elegans KSR-1 contains FLFP. This motif is conserved in murine Ksr-1 (FSFP) and Drosophila Ksr (FNFP) (Fig. 1b). These motifs are positioned at the carboxy-terminal end of conserved area 4 (CA4; Therrien et al. 1995). These CA4 regions are characterized by one or more S/TP sequences and thus resemble the C boxes of ETS proteins in the Elk subfamily (Fig. 1a,b).
To develop a systematic approach for identifying candidate ERK substrates, we defined a consensus sequence based on the C boxes of ETS proteins and the CA4 regions of KSR proteins (S/T-P-X(1–25)-S/T-P-X(1–25)-F-Q/N/L/S-F-P) and designed a computer algorithm to search available databases for proteins with this consensus sequence. This search identified a relatively small number of proteins, including A-raf, GATA transcription factors, and dual-specificity protein phosphatases (Fig. 1b–d).
To investigate the prediction that KSR proteins are ERK substrates, we generated in E. coli proteins containing an amino-terminal GST moiety and CA4-containing fragments of murine Ksr-1 (residues 383–519), C. elegans KSR-1 (residues 281–479), or Drosophila Ksr (residues 467–532) and assayed phosphorylation by Erk2 in vitro. All three proteins were phosphorylated efficiently by Erk2, indicating that the ability to function as an ERK substrate is an evolutionarily conserved property of KSR proteins (Fig. 2). These fragments of KSR contain S/TP sequences in CA4 only, demonstrating that phosphorylation must occur in CA4. To explore the role of the FXFP motif, we generated mutant forms of murine Ksr-1, C. elegans KSR-1, or Drosophila Ksr that had AAA instead of FXF; these proteins had Km values that were 7-, 7-, and 50-fold higher, respectively, than the values for wild-type proteins (Fig. 2d). Thus, the FXFP motif is necessary for high-affinity interactions between ERK and KSR proteins from three species.
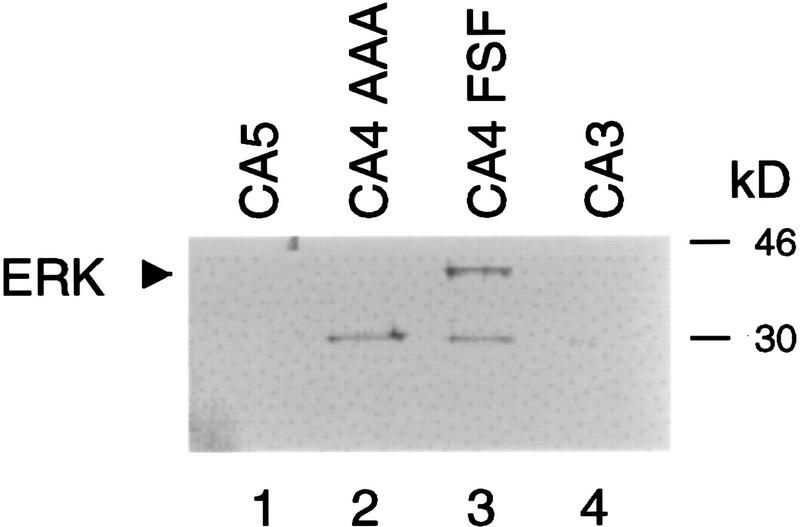
The model that FXFP is a docking site that mediates binding to ERK predicts that the enzyme–substrate complex might be stable enough to detect using a nonenzymatic assay. To investigate this possibility, we mixed GST:mKsr-1 fusion proteins immobilized on glutathione–Sepharose with a lysate of NIH 3T3 cells that contained endogenous ERK, washed extensively to remove nonspecifically bound proteins, and visualized retained ERK on a Western blot with an anti-ERK1 polyclonal antibody. ERK bound the CA4 region of mKsr-1 but not the amino-terminal CA3 region or the carboxy-terminal CA5 region, indicating that the interaction was specific (Fig. 6; lanes 1,3,4). This interaction required the FXFP motif, because no interaction was detected between ERK and a mutant protein containing AAAP (Fig. 6; lanes 2–3). These data are consistent with the model that ERK interacts with CA4 directly, but it is also possible that additional proteins mediated or facilitated this interaction. The in vitro phosphorylation experiments and these results together provide strong evidence for a direct and stable interaction between ERK and the CA4 region of KSR.
Figure 6.
The CA4 region of mKsr binds ERK. Approximately equal amounts of GST:mKsr-1(520–873) (lane 1, CA5), GST: mKsr-1(383–519AAA) (lane 2, CA4 AAA), GST:mKsr-1(383–519FSF) (lane 3, CA4 FSF), and GST:mKsr-1(312–392) (lane 4, CA3) were immobilized on glutathione–Sepharose, incubated with lysates of NIH 3T3 cells that contained ERK, and washed extensively. ERK retained by the glutathione–Sepharose was visualized on this Western blot using anti-ERK1 polyclonal antibody. The upper band (arrow) represents ERK, because it displayed the predicted mobility (42–44 kD) compared to protein size standards (indicated at right). The lower band represents a crossreacting protein. These results are representative of three separate experiments.
FQFP and the D box function independently and additively as docking sites for ERK
The ETS proteins Elk-1, SAP-1a, and Net contain a conserved region called the D box that is characterized by the amino acid sequence LXL carboxy-terminal to a cluster of basic residues (Lopez et al. 1994). We identified sequences similar to the D box in comparable positions of C. elegans LIN-1 and Drosophila Aop/Yan (Fig. 1a,g). These observations support our hypothesis that LIN-1 and Aop are members of the Elk subfamily and indicate that the ancestral gene encoded a protein with an ETS domain, a D box, and a C box. The D box of Elk-1 was reported recently to function as a docking site for ERK and JNK (Yang et al. 1998a,b). To investigate the relationship between the D box and the FQFP motif, we analyzed the ability of Erk2 to phosphorylate Elk-1 proteins with mutations that disrupt the D box, the FQFP motif, or both.
To disrupt the D box, we mutated the LEL sequence of Elk-1 to AEA; this change reduces the function of the D box as a docking site for ERK significantly (Yang et al. 1998b). Wild-type GST:Elk-1(307–428) had a Km of 1.5 μm (Fig. 7a,b,e). Changing FQFP to AAAP increased by 3-fold the Km of Elk-1 with a wild-type D box (Fig. 7e; lines 1,2) and by 12-fold the Km of Elk-1 with a mutant D box (Fig. 7e, lines 3,4). Thus, FQFP mediated an interaction with ERK in the presence or absence of a D box. Changing LEL to AEA increased by 14-fold the Km of Elk-1 with wild-type FQFP (Fig. 7e; lines 1,3) and by 50-fold the Km of Elk-1 with a mutant FQFP motif (Fig. 7e; lines 2,4). Thus, the D box mediated an interaction with ERK in the presence or absence of FQFP. The double mutant protein had a Km of 260 μm, showing that the two sites in combination reduced the Km 170-fold. Together, these results indicate that the two docking sites function independently and additively.
Figure 7.
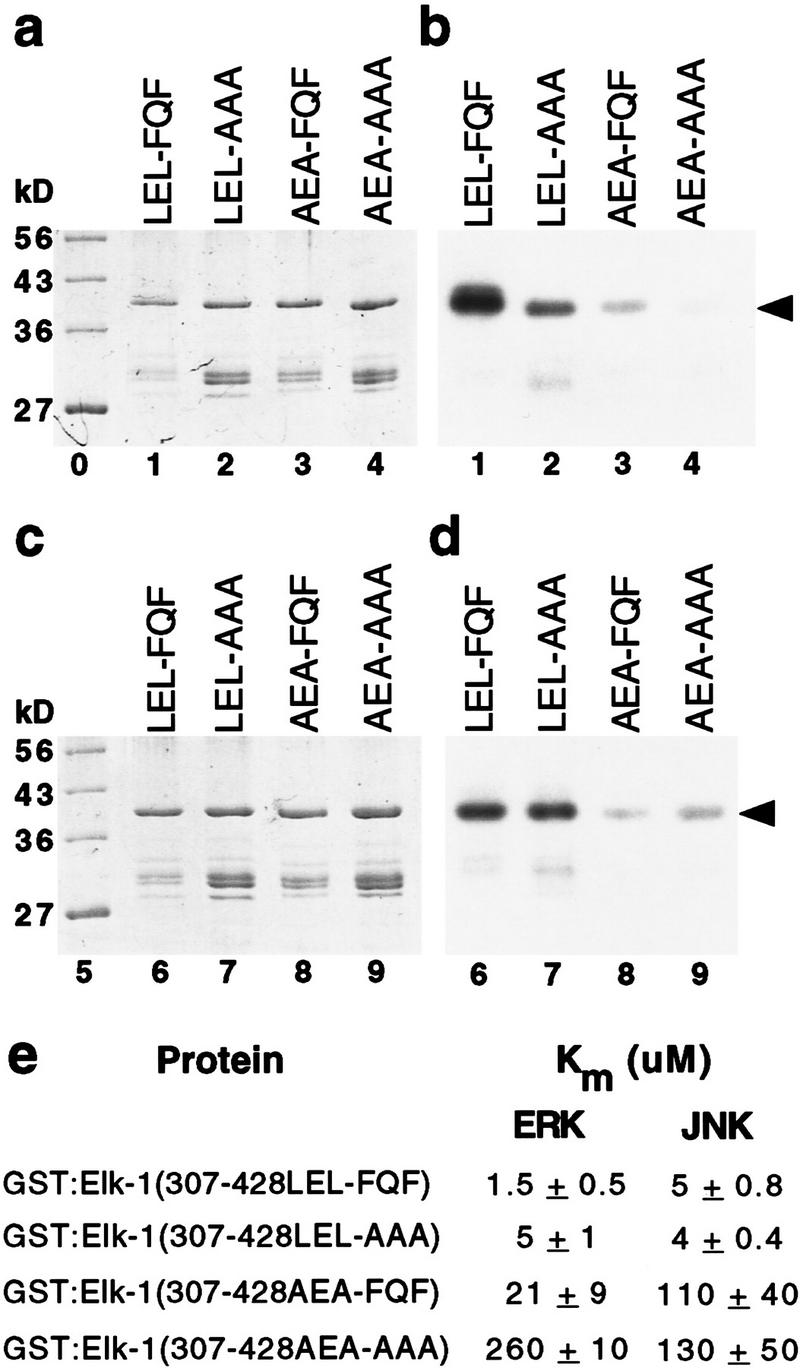
FQFP and the D box mediate additive interactions with ERK; FQFP does not mediate interactions with JNK. GST:Elk-1(307–428LEL-FQF) contained wild-type Elk-1 (lanes 1 and 6, LEL-FQF), GST:Elk-1(307–428LEL-AAA) contained a substitution of AAA for FQF (lanes 2 and 7, LEL-AAA), GST:Elk-1(307–428AEA-FQF) contained a substitution of AEA for LEL (lanes 3 and 8, AEA-FQF), and GST:Elk-1(307–428AEA-AAA) contained both substitutions (lanes 4 and 9, AEA-AAA). Proteins were incubated with Erk2 (a,b) or JNKβ (c,d) and [32P]ATP, and approximately equal amounts were fractionated by SDS-PAGE, stained with Coomassie blue (a,c), and exposed to film (b,d). Lanes 0 and 5 contain protein size markers. Arrows indicate intact fusion proteins that migrated with the predicted molecular weight of ∼39 kD. (e) Km was calculated from kinetic analyses; in each case a Lineweaver-Burke plot of the data closely approximated a straight line. Values are the mean ± s.d. of two separate experiments.
FQFP does not mediate interactions with JNK MAP kinase
Elk-1 is a substrate for JNK (Minden and Karin 1997). To investigate whether FQFP mediates interactions with JNK, we used purified, recombinant, rat JNKβ and the wild-type and mutant forms of Elk-1 described above. Wild-type GST:Elk-1(307–428) had a Km of 5 μm (Fig. 7c-e). Disrupting the D box of wild-type Elk-1 or Elk-1 with a mutation of FQFP increased the Km 22-fold and 32-fold, respectively (Fig. 7e; lines 1,3 and 2,4). These data are consistent with previous findings that the D box mediates interactions with JNK (Yang et al. 1998b) and show that the D box functions independently of FQFP. By contrast, disrupting the FQFP motif in either wild-type Elk-1 or Elk-1 with a mutation in the D box did not affect Km significantly (Fig. 7e; lines 1,2 and 3,4). These data indicate that FQFP does not mediate interactions with JNK.
These results suggest that a protein that contains FXFP but lacks a D box will not function as a high-affinity substrate for JNK. To test this prediction, we analyzed JNKβ phosphorylation of Drosophila Ksr. GST: dKsr(467–532FNF) was a very poor substrate with a Km of 180 ± 20 μm (mean ± s.d. of two separate trials). GST: dKsr (467–532AAA) had a Km of 110 ± 10 μm. Thus, FXFP did not mediate an interaction between JNK and KSR.
Discussion
The results presented here lead to five major conclusions. First, FXFP is an evolutionarily conserved docking site that mediates high-affinity interactions between ERK and substrate proteins in at least two different protein families. Second, FXFP and the δ domain/D box, a different docking site, can function separately or together in substrate proteins to mediate binding to ERK. We refer to this as a modular system of docking sites. Third, the partially overlapping substrate specificities of ERK and JNK result from the ability of these enzymes to recognize shared and unique docking sites. Fourth, FXFP can be used to identify ERK substrates, such as KSR. Fifth, peptides containing FXFP can be used to inhibit ERK.
FXFP is a docking site for ERK
The evidence in support of this conclusion can be summarized as follows: (1) The Elk subfamily of ETS transcription factors includes some of the best documented physiological ERK substrates, since LIN-1, Aop/Yan, and Elk-1 were independently shown to be regulated directly by ERK in worms, flies, and vertebrates, respectively (O’Neill et al. 1994; Treisman 1994; Jacobs et al. 1998; Tan et al. 1998). The results presented here suggest that the KSR family of protein kinases are also physiological ERK substrates (described below). FXFP is evolutionarily conserved in both of these protein families, indicating that FXFP plays an important and perhaps similar role in these proteins. FXFP is positioned just carboxy-terminal to ERK phosphorylation sites in both of these protein families, consistent with it being involved in ERK phosphorylation. (2) FXFP was necessary for high-affinity interactions in vitro between ERK and C. elegans LIN-1 and KSR-1, Drosophila Aop/Yan and Ksr, and vertebrate Elk-1 and mKsr-1. The effects of mutating FXFP are not likely to be the result of nonspecific alterations of protein structure, because we observed a similar effect in all six proteins and mutating FXFP did not diminish phosphorylation of Elk-1 by JNK. (3) The addition of FXFP is sufficient to increase the affinity of protein and peptide substrates for ERK dramatically in vitro and to increase the efficacy of peptide inhibitors of ERK in vitro. Importantly, these peptide inhibitors do not require an S/TP site, demonstrating that FQFP-mediated binding to ERK can occur independently of S/TP-mediated binding to the active site of ERK. (4) Two different experiments suggest FXFP mediates interactions with ERK in vivo: Mutations that alter or eliminate FQFP cause C. elegans lin-1 to be unresponsive to RTK–Ras–ERK-mediated signaling in worms (Jacobs et al. 1998); peptides containing FXFP interfere with RTK–Ras–ERK-mediated signaling during Xenopus oocyte maturation. (5) Based on the six ETS and KSR proteins that were demonstrated to have a motif that mediates binding to ERK, we defined a consensus sequence: F-X-F-P (Fig. 1h). This consensus sequence is consistent with our findings that peptide substrates and inhibitors required both phenylalanine residues for high-affinity binding, whereas the residue in between was less significant. We speculate that ERK has a binding pocket that interacts with these bulky, hydrophobic residues. Additional experiments are necessary to define further the effect of particular residues at each position. We propose that this motif be named DEF (docking site for ERK, FXFP).
This conclusion is significant because it reveals a fundamental mechanism that enables ERK to interact with substrate proteins. This appears to be an ancient mechanism of substrate recognition, as it has been conserved during the evolution of worms, flies, and vertebrates. The demonstration that FXFP mediates interactions between ERK and two different protein families, and the finding that FXFP is present in additional demonstrated and candidate ERK substrates (described below), suggest that many different ERK substrates may share the same FXFP docking site.
Multiple docking sites on ERK substrate proteins form a modular recognition system
The δ domain of c-Jun is a docking site for JNK (Karin 1995). Yang et al. (1998a,b) extended the analysis of this motif by showing that the D box of Elk-1 and the δ domain have a similar sequence, are functionally interchangeable, and function as docking sites for both ERK and JNK. ERK-specific MAP kinase kinases are substrates for ERK (Matsuda et al. 1993), and the amino-terminal region of the ERK-specific MAP kinase kinase of S. cerevisiae, Ste7, mediates an interaction with ERK (Bardwell et al. 1996). Bardwell and Thorner (1996) noted that the amino termini of many ERK-specific MAP kinase kinases contain an evolutionarily conserved sequence that was proposed to mediate interactions with ERK and named a docking site. We noted that the sequence of the docking site is similar to the sequence of δ domain and the D box, as all three motifs are characterized by a cluster of basic residues amino-terminal to an L/I-X-L/I motif (Fig. 1g). Furthermore, we found a similar motif in a comparable position of JNK-specific MAP kinase kinases (Fig. 1e,g), consistent with the possibility that this motif interacts with JNK and ERK. We propose that the δ domain, D box, and docking site are versions of the same motif, and this motif functions as a docking site for ERK and JNK. Figure 1h shows two slightly different consensus motifs defined by these sequences. We propose that this motif be named DEJL (docking site for ERK and JNK, LXL). Our goal is to initiate a systematic and informative nomenclature for MAP kinase docking sites that can be used to name additional motifs described in the future.
Two types of evidence support the model that the DEF and DEJL form a modular system that mediates recognition by ERK (Fig. 8). First is our analysis of Elk-1. A mutant Elk-1 protein lacking both motifs did not interact with ERK significantly. Restoring the DEF or DEJL decreased the Km 12- and 50-fold, respectively, showing that these motifs can mediate interactions with ERK independently. Restoring both sequences decreased the Km 170-fold, showing that in combination these motifs function additively rather than redundantly or synergistically. These findings, together with the observation that the sequences of the DEF and DEJL are not similar, suggest that the DEF and DEJL may interact with separate binding pockets of ERK. If ERK has separate binding pockets, then ERK may be capable of simultaneously interacting with the DEF and DEJL. However, the results presented here do not test this possibility.
Figure 8.
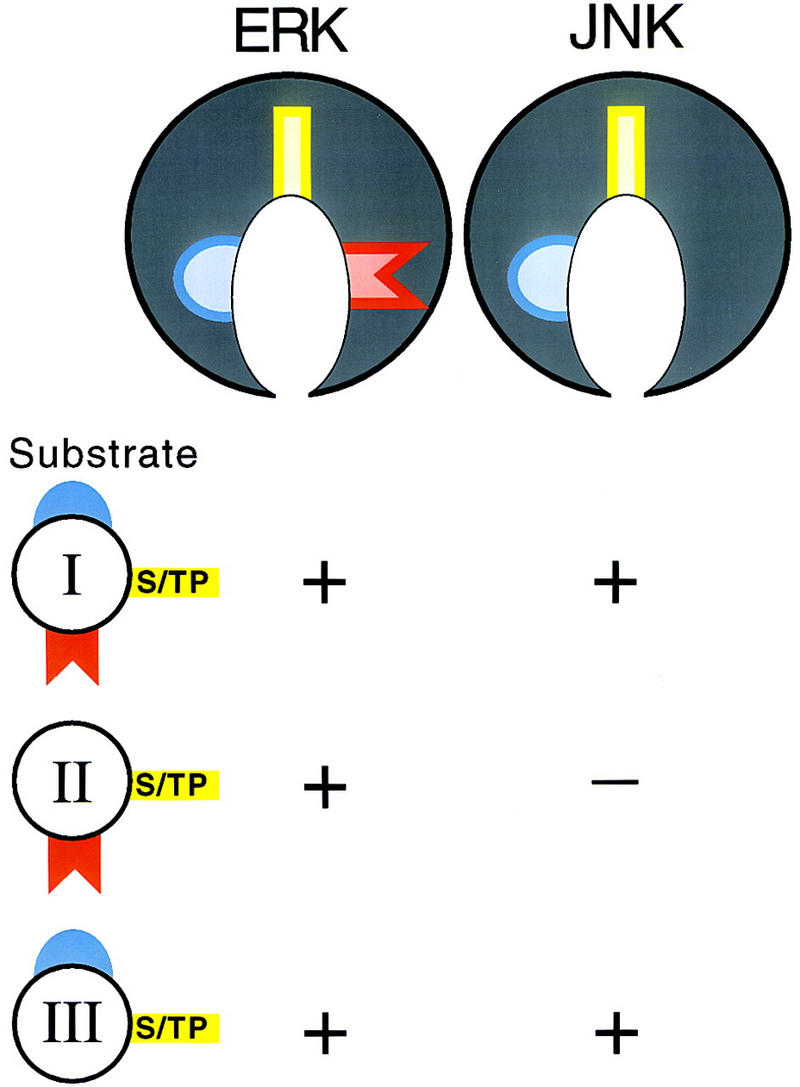
A model illustrating how a modular system of docking sites mediates substrate recognition by ERK and JNK. Class I substrates contain a DEF (red) and a DEJL (blue). Class II substrates contain only a DEF. Class III substrates contain only a DEJL. All substrates contain S/TP phosphoacceptor sites (yellow). We propose that ERK and JNK have similar active sites (yellow) and similar binding pockets for the DEJL (blue), whereas only ERK has a binding pocket for the DEF (red). (+) A substrate can be phosphorylated by the enzyme shown above.
Second, we analyzed the sequences of many proteins reported to be ERK substrates to determine if they contained a candidate DEF or DEJL. This analysis revealed four classes of substrate proteins (Fig. 8). Class I substrates contain both a DEF and DEJL. Examples include the Elk subfamily of ETS proteins (Fig. 1a) and S. cerevisiae Dig1 (K97RGRVPAPLNL; F412EFP; g1171410), a protein identified based on its ability to bind ERK in a two-hybrid experiment (Cook et al. 1996). In these proteins, the DEJL is amino-terminal to the DEF. Class II substrates contain only a DEF. Examples include GATA-2 (Fig. 1c) and rat c-Fos (F272LFP; g120473). Class III substrates contain only a DEJL. Examples include MAP kinase kinases (Fig. 1e,g) and c-Jun (Fig. 1f,g). Class IV substrates contain neither a DEF or DEJL. Examples include c-Myc, Sos, and pp90rsk. The existence of this class of reported substrates raises the possibility that ERK interacts with one or more yet-to-be-defined docking sites. Alternatively, these proteins may contain divergent DEF or DEJL motifs that do not match our current consensus sequences or they may not be physiological ERK substrates.
The conclusion that ERK uses a modular system of docking sites to recognize substrates is significant because it illuminates how ERK can appropriately phosphorylate such a wide spectrum of substrates. A modular recognition system offers several potential advantages. First, the number and arrangement of docking sites may modulate substrate affinity. Our analysis of Elk-1 supports this possibility. Furthermore, ETS proteins that contain a DEJL and a DEF had consistently higher affinities for ERK than KSR proteins that contain only a DEF (Fig. 2d). High affinity for ERK may be appropriate for substrates that are low in concentration or localized far from the site of ERK activation, such as transcription factors localized in the nucleus, whereas lower affinity may be appropriate for substrates that are high in concentration or localized at the plasma membrane close to the site of ERK activation, such as KSR, which is membrane localized (Xing et al. 1997). Second, the position of docking sites may determine which S/TP sites are phosphorylated. MAP kinases phosphorylate particular sites preferentially in substrates with multiple S/TP sequences. For example, positions 363, 368, 383, and 389 are preferred phosphorylation sites for ERK in Elk-1 (Marais et al. 1993). These sites in Elk-1 are the closest S/TP sequences amino-terminal to FQFP (Fig. 1a). The S/TP sites phosphorylated in KSR are also amino-terminal to FQFP. For proteins with multiple S/TP sequences, a combination of the position of S/TP sites relative to docking sites and residues such as proline immediately amino-terminal to S/TP sites may determine the order of phosphate addition and the stoichiometry of phosphorylation of different sites. Consistent with this possibility, the docking site of c-Jun appears to direct phosphorylation of particular S/TP sites (Kallunki et al. 1996). Third, a modular system may have made it easier for ERK to begin phosphorylating new substrates during evolution, as every substrate is not constrained to evolve the same docking site. By enabling ERK to phosphorylate many different proteins, this system probably contributed to the ability of ERK to mediate many different signals in many different cell types.
Although many enzymes act on multiple protein substrates, in most cases the features of the different substrates that mediate recognition are not well defined. One well-characterized case is UDP–GlcNAc:lysosomal enzyme N-acetylglucosamine 1-phosphotransferase, which catalyzes the transfer of N-acetylglucosamine-1-P to mannose residues in the oligosacccharides of many different lysosomal enzymes. In this case recognition does not appear to be mediated by a modular system, but instead is mediated by a lysine-rich epitope that appears to be similar in each substrate (for review, see Cuozzo et al. 1998). This epitope is separate from the oligosaccharides that are modified, and the lysines are not contiguous in the primary protein sequence but instead are juxtaposed by protein folding.
Partially overlapping substrate specificities of ERK and JNK result from recognition of shared and unique docking sites
This is illustrated by the model shown in Figure 8. We propose that ERK has two binding pockets, one for DEJL and one for DEF, whereas JNK has a binding pocket for DEJL but not for DEF. Because ERK and JNK are homologous, the binding pocket that interacts with DEJL may be similar in both enzymes. Kallunki et al. (1994) proposed that the region of JNK that interacts with the DEJL of c-Jun is a small β-strand-like region near the catalytic pocket. Both kinases can interact with proteins that contain a DEJL and a DEF or only a DEJL, but only ERK can interact with proteins that contain only a DEF, such as KSR. We speculate that JNK phosphorylates unique substrates by recognizing one or more docking sites that do not bind ERK. Furthermore, other MAP kinases such as p38 may also employ a modular system that includes shared and unique docking sites to interact with substrate proteins.
A DEF can be used to identify ERK substrates, such as KSR
Using visual and computer searches, we identified an evolutionarily conserved DEF positioned carboxy-terminal to multiple S/TP sites in several protein families including KSR, A-raf, GATA transcription factors, and dual-specificity protein phosphatases. To test the prediction that these proteins are ERK substrates, we focused on KSR. The model that KSR is a physiological substrate for ERK makes the following predictions: (1) KSR can be phosphorylated by ERK in vitro; (2) KSR phosphorylation correlates with ERK activity in vivo; (3) phosphorylation by ERK affects KSR function; and (4) KSR functions in an ERK-mediated process. The results presented here confirm the first prediction; KSR proteins from worms, flies, and vertebrates were high-affinity substrates for ERK, and the DEF was necessary for these high-affinity interactions. These findings are consistent with the observation that the amino-terminal region of mKsr-1 can interact with ERK-2 (Yu et al. 1997). The fourth prediction has been demonstrated previously, as KSR positively mediates RTK–Ras–ERK signaling in worms, flies and vertebrates (Kornfeld et al. 1995; Sundaram and Han 1995; Therrien et al. 1995; Xing et al. 1997). Although two predictions remain to be tested, these findings suggest that KSR is a physiological ERK substrate.
A-raf has two S/TP sites and FSFP positioned amino-terminal to the kinase domain (Fig. 1b). B-raf and Raf-1, two other vertebrate Raf proteins (for review, see Storm et al. 1990), do not appear to contain this domain. All Raf proteins share two regions of significant similarity with KSR proteins: The conserved region 1 (CR1) of Raf is similar to the CA3 region of KSR, and the kinase domain of Raf (CR3) is similar to the kinase domain of KSR (CA5) (Therrien et al. 1995). Interestingly, the FXFP motif of A-raf is in a comparable position to the CA4 region of KSR (Fig. 1b), suggesting that the common ancestral gene of Raf and KSR probably contained a CA4 region. Thus, A-raf and KSR may share a function (regulation by ERK) that is not shared by B-raf and Raf-1.
The GATA family of transcription factors regulates the development of multiple vertebrate tissues, such as hematopoeitic cells and the heart (for review, see Weiss and Orkin 1995; Evans 1997). Of the six characterized GATA subfamilies, GATA-2, GATA-3, and GATA-4 have multiple S/TP sites and an FXFP motif positioned in a region implicated in transcriptional regulation (Fig. 1c). GATA-1, GATA-5, and GATA-6 do not appear to contain this motif. Interestingly, GATA-2 has been reported to be a substrate for ERK (Towatari et al. 1995). We predict that GATA-3 and GATA-4 are also ERK substrates.
Dual specificity protein phosphatases can dephosphorylate both threonine and tyrosine residues. These enzymes appear to be physiological regulators of MAP kinases (for review, see Byon et al. 1997). MAP kinase phosphatase-1 (MKP-1) and dual-specificity protein phosphatase 4 (DUS4) contain multiple S/TP sites and FXFP at a similar position in the phosphatase domain (Fig. 1d). These findings suggest that FXFP may mediate an interaction between these phosphatases and ERK, which might facilitate phosphorylation of these phosphatases by ERK, dephosphorylation of ERK by these phosphatases, or both.
Three general approaches have been used previously to identify MAP kinase substrates. First, candidate substrates like Elk-1 have been identified based on circumstantial evidence such as involvement in a MAP kinase-mediated process or regulated phosphorylation (Treisman 1994). Second, biochemical purification of a kinase activity that can phosphorylate a particular protein, such as c-Jun, has resulted in the identification of MAP kinases, such as JNK (Karin 1995). Third, candidate substrates have been identified based on the ability to bind ERK, such as Dig1 (Cook et al. 1996). The identification of S/TP as a minimal consensus sequence for ERK phosphorylation was not sufficient to reliably predict ERK substrates, because ∼90% of all proteins contain one or more S/TP sequences (data not shown). A DEF is much less prevalent, and our findings demonstrate that combining these two motifs produces a relatively selective algorithm that can identify new ERK substrates. This is significant because an understanding of how ERK activity controls cell fates requires the identification of all ERK substrates, and it is likely that many ERK substrates have yet to be identified. Refinements of our search algorithm and the expansion of protein databases in the future should make it possible to identify many additional candidate substrates using this approach.
Peptide inhibitors of ERK
Peptides containing FQFP inhibited ERK activity in vitro. These peptides are likely to bind the enzyme and prevent protein substrates from docking. Furthermore, injecting these peptides into Xenopus oocytes reduced meiotic maturation and cdc2 kinase activity, events that are mediated by a RTK–Ras–ERK signaling pathway. These results are significant because specific ERK inhibitors are not available currently and many human tumors have constitutively active versions of proteins in the RTK–Ras–ERK pathway (Cantley et al. 1991). A specific ERK inhibitor might reduce such constitutive signaling activity and have clinical applications. Inhibiting ERK by targeting a substrate-docking site is an appealing strategy, as it is likely to be more specific than targeting the highly conserved active site. Furthermore, as multiple docking sites mediate substrate binding to ERK, it might be possible to inhibit phosphorylation of a subset of ERK substrates by targeting a particular docking site.
Materials and methods
Protein production and MAP kinase assays
Expression plasmids were derived from cloned cDNAs and pGEX vectors (Pharmacia) and modified by in vitro mutagenesis using standard techniques (Sambrook et al. 1989). Proteins were expressed in E. coli BL21, partially purified using glutathione–Sepharose (Pharmacia) essentially according to the manufacturer’s instructions, and dialyzed into kinase assay buffer. The amount of intact protein was estimated by comparison to known amounts of purified bovine serum albumin present in adjacent lanes of Coomassie blue-stained SDS-PAGE gels. Purified MBP was from GIBCO-BRL. Peptides were synthesized using standard 9-fluorenylmethoxycarbonyl chemistry and purified using HPLC by the Tufts Core Facility (Boston, MA).
Purified, recombinant, murine Erk2 (New England Biolabs) is fully active, because it is produced in E. coli containing constitutively active MEK. Assays were done according to Alessi et al. (1995). A 50-μl reaction contained 100 μm [32P]ATP (0.15 Ci/mmole) and 0.05 pmole Erk2 and was terminated after 15 min at 30°C, at which point 32P incorporation was linear with respect to time. Purified, recombinant, rat JNKβ (Stratagene) is partially active as a result of autophosphorylation. Assays were done as described for Erk2 except that [32P]ATP was 0.6 Ci/mmole. For each protein, we used SDS-PAGE and autoradiography to establish that intact fusion protein contained most or all of the incorporated 32P. To quantify phosphorylation, we measured radioactive protein bound to phosphocellulose paper (P81, Whatman) using a scintillation counter.
Oocyte microinjection
Stage VI fully grown immature oocytes were obtained from mature Xenopus laevis females and maintained in 1× modified Barth’s solution with HEPES, 1 mg/ml Ficoll 400, 1 mg/ml bovine serum albumin, and antibiotics (Muslin et al. 1993). Each oocyte was injected with 25–50 nl of water alone or 10 mm peptide dissolved in water and incubated at 19°C for 24 hr in the presence or absence of 8.25 μg/ml insulin. Using an estimate of 10 μl for the volume of an average oocyte, we calculate that the initial peptide concentration in the oocyte was ∼50 μm. GVBD was defined as the presence of a broad white spot on the animal pole of the oocyte. Oocytes were pooled and lysed as described by Muslin et al. (1993) and cdc2 kinase was assayed as described by Xing et al. (1997).
Binding assay
NIH 3T3 cells were grown in DMEM + 10% calf serum to ∼80% confluence and lysed using NP-40 lysis buffer (Xing et al. 1997). Following low-speed centrifugation, cell extracts were incubated for 1 hr at 4°C with ∼2 μg of GST:mKsr-1 fusion protein immobilized on glutathione–Sepharose and washed three times with NP-40 lysis buffer for 10 sec at 4°C. Samples of beads were boiled in SDS buffer, fractionated by SDS-PAGE, and transferred to a nitrocellulose membrane. Western blots were treated with rabbit, polyclonal, anti-ERK1, IgG antibody (C-16, Santa Cruz), anti-rabbit, secondary antibody conjugated to alkaline phosphatase (Santa Cruz) and NBT/BCIP reagent (Promega).
Acknowledgments
We thank Andrew Turk for constructing plasmids, Richard Maurer, Marc Therrien, Ilaria Rebay, and Gerry Rubin for cDNAs, Sean Eddy for computer assistance, and Jeff Gordon, Stuart Kornfeld, Tim Schedl, and Dwight Towler for advice. This research was supported by the Edward Mallinckrodt, Jr. Foundation (K.K.), a Howard Hughes Medical Institute grant (K.K.), the Barnes-Jewish Hospital Foundation (A.J.M.), and the National Institutes of Health (A.J.M.). K.K. is a recipient of the Burroughs Wellcome Fund New Investigator Award in the Basic Pharmacological Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kornfeld@pharmdec.wustl.edu; FAX (314) 362-7058.
References
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler V, Franklin CC, Kraft AS. Phorbol esters stimulate the phosphorylation of c-Jun but not v-Jun: Regulation by the N-terminal delta domain. Proc Natl Acad Sci. 1992;89:5341–5345. doi: 10.1073/pnas.89.12.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler V, Unlap T, Kraft AS. A peptide encoding the c-Jun δ domain inhibits the activity of a c-Jun amino-terminal protein kinase. J Biol Chem. 1994;269:11186–11191. [PubMed] [Google Scholar]
- Alessi DR, Cohen P, Ashworth A, Cowley S, Leevers SJ, Marshall CJ. Assay and expression of mitogen-activated protein kinase, MAP kinase kinase, and Raf. Methods Enzymol. 1995;255:279–290. doi: 10.1016/s0076-6879(95)55031-3. [DOI] [PubMed] [Google Scholar]
- Bardwell L, Thorner J. A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biol Sci. 1996;21:373–374. [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Signaling in the yeast pheromone response pathway: Specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel GJ, Tuck S, Greenwald I, Horvitz HR. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes & Dev. 1995;9:3149–3162. doi: 10.1101/gad.9.24.3149. [DOI] [PubMed] [Google Scholar]
- Byon JCH, Kenner KA, Kusari AB, Kusari J. Regulation of growth factor-induced signaling by protein-tyrosine-phosphatases. Proc Soc Exp Biol Med. 1997;216:1–20. doi: 10.3181/00379727-216-44153. [DOI] [PubMed] [Google Scholar]
- Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- Cantley LC, Auger KR, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Kron SJ, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes & Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- Cuozzo JW, Tao K, Cygler M, Mort JS, Sahagian GG. Lysine-based structure responsible for selective mannose phosphorylation of cathepsin D and cathepsin L defines a common structural motif for lysosomal enzyme targeting. J Biol Chem. 1998;273:21067–21076. doi: 10.1074/jbc.273.33.21067. [DOI] [PubMed] [Google Scholar]
- Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- Deshpande AK, Kung HF. Insulin induction of Xenopus laevis oocyte maturation is inhibited by monoclonal antibody against p21 ras proteins. Mol Cell Biol. 1987;7:1285–1288. doi: 10.1128/mcb.7.3.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T. Regulation of cardiac gene expression by GATA-4/5/6. Trends Cardiovasc Med. 1997;7:75–83. doi: 10.1016/S1050-1738(97)00010-8. [DOI] [PubMed] [Google Scholar]
- Ferrell JE. MAP kinases in mitogenesis and development. Curr Top Dev Biol. 1996;33:1–60. doi: 10.1016/s0070-2153(08)60336-1. [DOI] [PubMed] [Google Scholar]
- Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes & Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- Jacobs D, Beitel GJ, Clark SG, Horvitz HR, Kornfeld K. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics. 1998;149:1809–1822. doi: 10.1093/genetics/149.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T, Su B, Tsigelny I, Sluss HK, Derijard B, Moore G, Davis R, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes & Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- Korn LJ, Siebel CW, McCormick F, Roth RA. Ras p21 as a potential mediator of insulin action in Xenopus oocytes. Science. 1987;236:840–843. doi: 10.1126/science.3554510. [DOI] [PubMed] [Google Scholar]
- Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- Lopez M, Oettgen P, Akbarali Y, Dendorfer U, Libermann TA. ERP, a new member of the ets transcription factor/oncoprotein family: Cloning, characterization, and differential expression during B-lymphocyte development. Mol Cell Biol. 1994;14:3292–3309. doi: 10.1128/mcb.14.5.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr Opin Gen Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Gotoh Y, Nishida E. Phosphorylation of Xenopus mitogen-activated protein (MAP) kinase kinase by MAP kinase kinase kinase and MAP kinase. J Biol Chem. 1993;268:3277–3281. [PubMed] [Google Scholar]
- Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophys Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, MacNicol AM, Williams LT. Raf-1 protein kinase is important for progesterone-induced Xenopus oocyte maturation and acts downstream of mos. Mol Cell Biol. 1993;13:4197–4202. doi: 10.1128/mcb.13.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebreda AR, Porras A, Santos E. p21ras-induced meiotic maturation of Xenopus oocytes in the absence of protein synthesis: MPF activation is preceded by activation of MAP and S6 kinases. Oncogene. 1993;8:467–477. [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tijan R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Price MA, Rogers AE, Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET) EMBO J. 1995;14:2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Songyang Z, Lu KP, Kwon YT, Tsai L, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, DeMaggio AJ, Hoekstra MF, Blenis J, Hunter T, Cantley LC. A structural basis for substrate specificities of protein Ser/Thr kinases: Primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm SM, Brennscheidt U, Sithanandam G, Rapp UR. raf oncogenes in carcinogenesis. Crit Rev Oncogenesis. 1990;2:1–8. [PubMed] [Google Scholar]
- Sturgill TW, Wu J. Recent progress in characterization of protein kinase cascades for phosphorylation of ribosomal protein S6. Biochim Biophys Acta. 1991;1092:350–357. doi: 10.1016/s0167-4889(97)90012-4. [DOI] [PubMed] [Google Scholar]
- Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- Tan PB, Lackner MR, Kim SK. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell. 1998;93:569–580. doi: 10.1016/s0092-8674(00)81186-1. [DOI] [PubMed] [Google Scholar]
- Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- Towatari M, May GE, Marais R, Perkins GR, Marshall CJ, Cowley S, Enver T. Regulation of GATA-2 phosphorylation by mitogen-activated protein kinase and interleukin-3. J Biol Chem. 1995;270:4101–4107. doi: 10.1074/jbc.270.8.4101. [DOI] [PubMed] [Google Scholar]
- Treisman R. Ternary complex factors: Growth factor regulated transcriptional activators. Curr Opin Gen Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- ————— Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH. GATA transcription factors: Key regulators of hematopoiesis. Exp Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- Xing H, Kornfeld K, Muslin AJ. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- Yang S-H, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998a;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-H, Whitmarsh AJ, Davis RJ, Sharrocks AD. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998b;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Fantl WJ, Harrowe G, Williams LT. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol. 1997;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3A resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]