Abstract
An improved method has been developed for RNA interference in Cryptococcus neoformans, using opposing promoters to facilitate cloning and RNA interference targeting URA5 to allow selection of cells in which silencing is most effective. These advances significantly reduce the variability of silencing and the effort required for interference plasmid construction.
Keywords: Cryptococcus neoformans, RNA interference, pathogenic fungus
Cryptococcus neoformans is a basidiomycetous yeast that causes serious opportunistic infections, leading to over 600,000 deaths annually (Park et al., 2009). The fascinating biology of this organism and its major impact on world health have motivated intensive study of cryptococcal virulence factors and pathogenicity. One valuable molecular tool in this work is targeted gene silencing mediated by RNA interference (RNAi) (Liu et al., 2002; Panepinto et al., 2009). Unfortunately, the utility of this method has been limited by variability in RNAi penetrance and the labor required to generate silencing plasmids. We have now developed a double promoter construct for RNAi in C. neoformans to simplify cloning. We have also incorporated URA5 gene silencing to allow selection for cells that robustly perform RNAi.
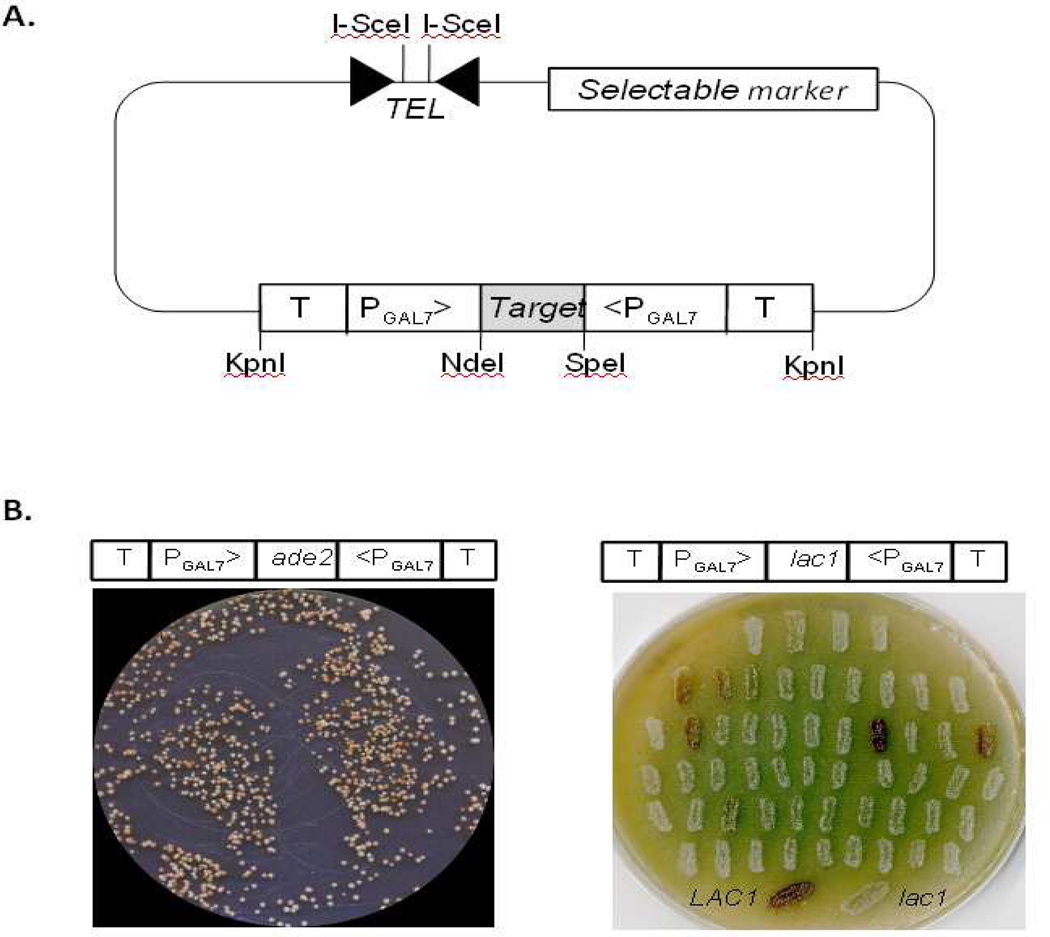
The plasmids we developed contain two cryptococcal promoters in opposing orientation, between which the sequence to be silenced can be inserted. Placing the target sequence between opposing promoters allows it to be transcribed from both directions simultaneously, producing complementary transcripts. These transcripts can hybridize to form the double-stranded RNA that activates the RNAi system. The opposing promoters of the plasmids are separated by restriction sites, allowing simple insertion of the gene segment(s) to be bidirectionally transcribed, and are flanked by terminators (Fig. 1A). This plasmid design eliminates the need to clone the target sequence twice in opposite orientations to yield a hairpin RNA, which is another way to produce double-stranded RNA and activate the RNAi pathway in fungi (Liu, 2002). The vectors also contain a selectable marker (ADE2, URA5, or neoR which confers geneticin-resistance) and telomeric (TEL) sequences (Edman, 1992); the latter become terminal when the vector is digested with I-SceI, thus stabilizing the plasmid (Fig. 1A).
Figure 1.
Opposing promoter constructs mediate RNAi. A. Schematic view of an RNAi plasmid with major elements shown; see text for details. TEL, telomeres; T, terminator; P, promoter; Target, sequence to be silenced. Restriction sites are indicated by vertical lines labeled with the abbreviated enzyme name. B. An opposing PGAL7 construct mediates silencing of ADE2 and LAC1. Left, A ura5 mutant (JEC43) was transformed with plasmid pIBB1, shown schematically, and grown on galactose medium lacking uracil (-ura/gal) for 2 days at 30°C. Right, a parallel study with plasmid pIBB29, showing a master plate with a random selection of transformants streaked onto niger seed agar with galactose (NSA/gal) and grown for 2 days at 30 °C. Controls streaked at the bottom of the plate are a wild-type strain that melanizes and appears brown (LAC1), and a deletion strain that does not melanize and therefore remains white (lac1).
We first tested URA5-marked plasmids with opposing copies of the constitutively active ACT1 promoter (Cox et al., 1995) (pIBB24), opposing copies of the regulatable GAL7 promoter (Wickes and Edman, 1995) (pIBB1), or one of each promoter (pIBB5). (Table 1 summarizes these plasmids and others discussed below.) As a target sequence each plasmid included a ~500 bp BglII fragment of ADE2 (Sudarshan et al., 1999), from the plasmid pADE2i (Liu et al., 2002). Successfully silencing ADE2 (which encodes phosphoribosylaminoimidazole carboxylase) yields pink colonies, due to accumulation of a purine biosynthetic intermediate. Each plasmid was electroporated into a ura5 serotype D strain (JEC43, Wickes et al., 1997) and transformants were selected on medium without uracil. The selection medium contained 2% galactose as the carbon source for plasmids with one or two copies of the GAL7 promoter and 2% glucose for other transformations. Transformation with pIBB24 and pIBB5 yielded uniformly white colonies (not shown), suggesting that ADE2 silencing was not effectively mediated by these constructs. In contrast, >80% of transformants carrying the pIBB1 plasmid were pink to red in color after 5 days of incubation at 30°C (Fig. 1B, left) demonstrating that the ADE2 gene had been effectively silenced by the plasmid with opposing GAL7 promoters.
Table 1.
| Plasmid | Forward promoter | Reverse promoter | Target gene | Marker gene |
|---|---|---|---|---|
| pIBB1 | GAL7 | GAL7 | 500 bp ADE2 | URA5 |
| pIBB5 | GAL7 | ACT1 | 500 bp ADE2 | URA5 |
| pIBB24 | ACT1 | ACT1 | 500 bp ADE2 | URA5 |
| pIBB29 | GAL7 | GAL7 | 650 bp LAC1 | URA5 |
| pIBB32 | GAL7 | GAL7 | 610 bp URA5 | ADE2 |
| pIBB69 | GAL7 | GAL7 | 610 bp URA5 + 650 bp LAC1 | ADE2 |
| pIBB75 | GAL7 | GAL7 | 250 bp URA5 | ADE2 |
| pIBB103 | GAL7 | GAL7 | 250 bp URA5 | neoR |
We observed wide variation in the colony phenotypes of cells transformed with pIBB1, with colors ranging from cream to dark red (Fig. 1B, left). Results were similar when we modified this plasmid to target LAC1 (pIBB29); this gene encodes a laccase responsible for melanin production that causes colonies to appear brown on appropriate media. While many transformants in this case appeared white, indicating successful silencing, a substantial minority displayed various levels of melanization (Fig. 1B, right). The apparent variability in RNAi efficacy might confound interpretation of studies employing RNAi. We therefore developed a system to eliminate those transformants in which silencing is inefficient.
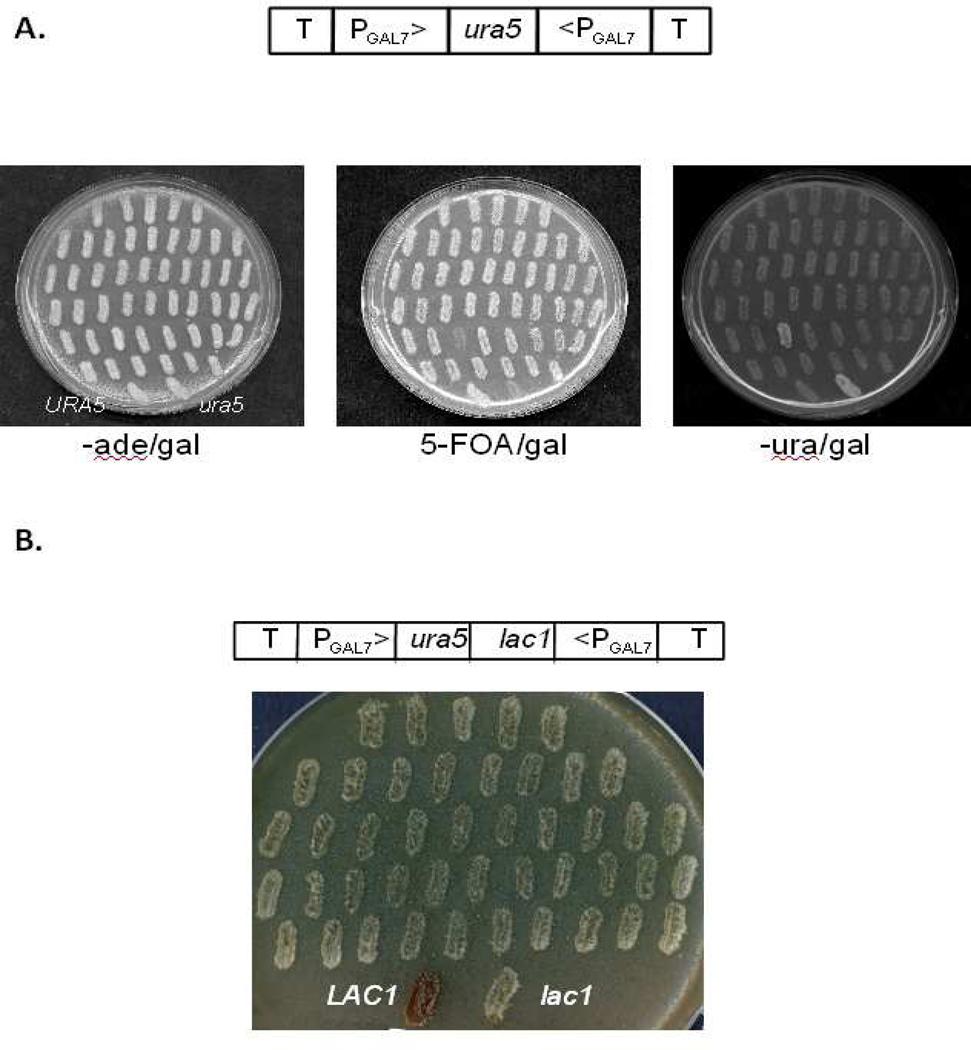
In our earlier studies of RNAi in C. neoformans, we proposed simultaneous silencing of ADE2 and a gene of interest, in order to use the red pigment that accumulates upon ADE2 silencing as an indicator of RNAi efficacy (Liu et al., 2002). GFP has also been used as an RNAi “sentinel” in Histoplasma capsulatum to identify individual transformants in which RNAi is most robust (Rappleye et al., 2004). Both of these indicators are useful, but we wished to develop a way to select transformants in which RNAi is most effective. We chose to target URA5 (Edman and Kwon-Chung, 1990) because silencing this gene would allow transformants to survive treatment with 5-fluoroorotic acid (5-FOA). (5-FOA is converted to 5-fluorouracil in strains bearing a functional URA5 gene; this product is toxic to yeast (Boeke et al., 1987).) To target URA5, we cloned a 610-bp SalI fragment of the gene between the GAL7 promoters of pIBB1 and replaced the existing URA5 marker with an ADE2 marker. We electroporated the resulting plasmid (pIBB32) into a serotype D ade2 mutant strain (JEC50) and grew the transformed cells for 3 h in medium lacking adenine (to select against untransformed cells) and containing galactose (to induce synthesis of double-stranded RNA corresponding to the URA5 sequence). We next treated the cells for 1 h with 1 mg/ml 5-FOA to select for cells in which interference was active and plated them on galactose medium lacking adenine. We restreaked the colonies onto the same medium, and then replica-plated onto galactose media either lacking uracil (so that cells with functioning RNAi cannot grow) or containing 1 mg/ml 5-FOA (so that wild-type cells cannot grow). Virtually all of the recovered colonies (100 of 101 tested) grew on 5-FOA (Fig. 2A and data not shown), confirming effective silencing of the URA5 gene. The telomeric vector remained episomal in these strains, as shown by plasmid loss upon plating on non-selective medium (data not shown). These results suggested that URA5 silencing with 5-FOA selection is an efficient way to isolate cells that are effectively performing RNAi. We confirmed this finding by using this strategy to target the LAC1 gene in tandem with URA5 (pIBB69). The colony color of transformants in this experiment matched that of a lac1 mutant (Fig. 2B) and was significantly (p<0.0002) more consistent (0% melanin-producing colonies, n=3) than in similar studies without the selection step (12 – 15% melanin-producing colonies, n=3) (Fig. 1B, right panel).
Figure 2.
Silencing URA5 allows 5-FOA selection of cells actively performing RNAi. A. Cells transformed with plasmid pIBB32 containing the RNAi cassette cartooned above were recovered and treated as described in the text before plating to medium with galactose but lacking adenine (left) and replica-plating to galactose media with 5-FOA (middle) or without uracil (right). Cells undergoing effective RNAi grow on 5-FOA and not on medium lacking uracil; this plate was chosen to show the single transformant (of over 100) that did not demonstrate this phenotype. The bottom two streaks on each plate are control strains with normal (left) or mutant (right) URA5 genes as indicated in the left panel. B. JEC50 cells were transformed with pIBB69 targeting LAC1 as cartooned above, pretreated with 5-FOA, and selected on –ade/gal medium as for the study in Fig. 2A. Transformants were restreaked on NSA/gal and grown for 2 days at 30°C, demonstrating effective silencing by white color matching that of control lac1 cells (bottom right of the plate). In contrast, the wild-type LAC1 strain is dark brown (bottom left).
In our initial studies we used 500- to 700-bp segments of chromosomal DNA to direct silencing of designated target genes (ADE2, URA5, and others not shown). Although we generally chose regions that were primarily exonic, the multiple short exons typical of C. neoformans genes (Tenney et al., 2004; Loftus et al., 2005) meant that the target regions often contained some intronic sequence as well. This raised the question of the optimum length of sequence for efficient silencing. To test this we amplified a series of fragments from genomic DNA using primer IB077 [5’-CCATTAATCAAGCTGGACAATTCCGAC-3’] as the reverse primer and forward primers designed to amplify a total of 100, 200, 300, 400, or 500 bp of the URA5 gene. We replaced the longer URA5 segment in pIBB32 with each fragment, transformed JEC50 with the resulting plasmids, and tested the transformants for resistance to 1 mg/ml 5-FOA as an assay of RNAi efficacy. In one representative experiment the fraction of colonies demonstrating robust RNAi was 0, 91, 95, 67, and 62% for plasmids containing target fragments of 100, 200, 300, 400, and 500 bp, respectively. Based on these and other similar results (not shown), we performed subsequent studies using the pIBB32-derived vector pIBB75 (Table 1), which bears a 250-bp URA5 target fragment amplified using primers IBO77 and IBO78 [5’-CCATTAATCAAGCTGGACAATTCCGAC-3’].
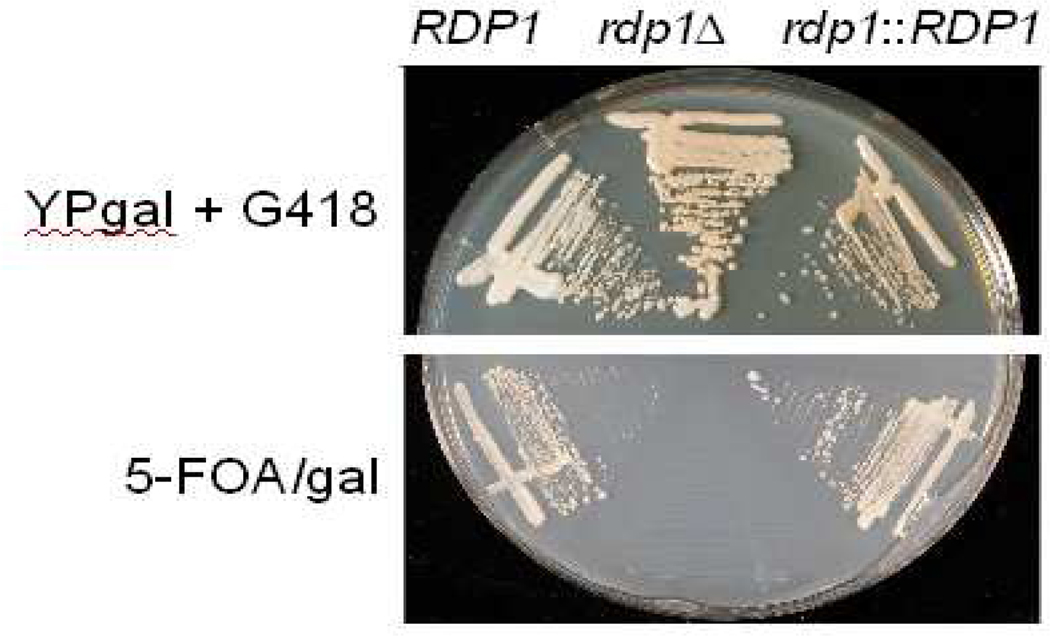
In all organisms studied so far, RNAi employs conserved proteins of the Dicer and Argonaute families; in many cases RNA-dependent RNA polymerases also play a major role in this phenomenon. We identified two orthologs of each of the Argonaute and Dicer genes and one RNA-dependent RNA polymerase (RdRP) gene in the JEC21 serotype D strain of C. neoformans, consistent with observations by Nakayashiki et al. (2006) and Janbon et al. (2010). To confirm the requirement for an RdRP in cryptococcal RNAi, we replaced the RDP1 gene (CNG01230) in serotype D strain JEC50 with a nourseothricin resistance cassette (McDade and Cox, 2001). We then transformed these and wild-type cells with pIBB103 (Genbank accession number HQ455038), a plasmid identical to pIBB75 but with a neomycin resistance marker that enables growth on medium containing G418 (Fig. 3, top). Wild-type transformants effectively silenced the URA5 gene and grew on media containing 5-FOA, while the rdp1Δ strain was not viable on 5-FOA-containing medium (Fig. 3, bottom). Complementing the rdp1Δ with wild-type RDP1 at the endogenous locus restored wild-type growth on 5-FOA (Fig. 3, bottom). RT-PCR of these strains confirmed that URA5 mRNA is reduced in the wild type and RDP1 complemented strains compared to the rdp1Δ strain (not shown). Together, our studies show that (1) the RDP1 gene is required for RNAi in C. neoformans, (2) opposing promoter constructs mediate RNAi, (3) 200–300 bp is the optimum length for target inserts, and (4) URA5 targeting is an effective strategy to select transformants that efficiently perform RNAi. We have now used the opposing promoter constructs described here to effectively silence multiple genes in serotype D C. neoformans (this report, Reilly (2010), and unpublished data from our laboratory), suggesting that the approach is target-independent. These findings should advance use of RNAi as a powerful tool in studies of an important pathogenic fungus.
Figure 3.
Deletion of RDP1 abolishes RNAi silencing. Wild-type RDP1, rdp1Δ, and rdp1::RDP1 complemented strains were electroporated with pIBB103 and transformants were grown on either complete media containing galactose and G418 (YPgal + G418) or 5-FOA/gal media at 30°C for 2 days.
Research Highlights.
-
--
An improved method has been developed for RNA interference in Cryptococcus neoformans
-
--
The method simplifies construction of interference plasmids
-
--
The method allows selection of cells in which RNAi is effective
-
--
These improvements reduce effort and improve results for RNAi in C. neoformans
Acknowledgments
This work was supported by National Institutes of Health grant GM071007 and an Award from the Edward J. Mallinckrodt, Jr. Foundation. The authors are grateful to Morgann C. Reilly for plasmid sequencing, Michael L. Skowyra for comments on the manuscript and sequence submission, and Doering lab members for helpful suggestions.
Abbreviations
- RNAi
RNA interference
- 5-FOA
5-fluoroorotic acid
- Ade
adenine
- Gal
galactose
- Ura
uracil
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Cox GM, Rude TH, Dykstra CC, Perfect JR. The actin gene from Cryptococcus neoformans: structure and phylogenetic analysis. J Med. Vet. Mycol. 1995;33:261–266. doi: 10.1080/02681219580000521. [DOI] [PubMed] [Google Scholar]
- Edman JC, Kwon-Chung KJ. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman JC. Isolation of telomeric sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol Cell Biol. 1992;12:2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Maeng S, Yang DH, Ko YJ, Jung KW, Moyrand F, Floyd A, Heitman J, Bahn YS. Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fumgal Genet. Biol. 2010;47:1070–1080. doi: 10.1016/j.fgb.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cottrell TR, Pierini LM, Goldman WE, Doering TL. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics. 2002;160:463–470. doi: 10.1093/genetics/160.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade HC, Cox GM. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol. 2001;39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H, Kadotani N, Mayama S. Evolution and diversification of RNA silencing proteins in fungi. J Mol Evol. 2006;63:127–135. doi: 10.1007/s00239-005-0257-2. [DOI] [PubMed] [Google Scholar]
- Panepinto J, Komperda K, Frases S, Park YD, Djordjevic JT, Casadevall A, Williamson PR. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol Microbiol. 2009;71:1165–1176. doi: 10.1111/j.1365-2958.2008.06588.x. [DOI] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Rappleye CA, Engle JT, Goldman WE. RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol Microbiol. 2004;53:153–165. doi: 10.1111/j.1365-2958.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- Reilly MC, Levery SB, Castle SA, Klutts JS, Doering TL. A novel xylosylphosphotransferase activity discovered in Cryptococcus neoformans. J Biol Chem. 2009;284:36118–36127. doi: 10.1074/jbc.M109.056226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarshan S, Davidson RC, Heitman J, Alspaugh JA. Molecular Analysis of the Cryptococcus neoformans ADE2 gene, a selectable marker for transformation and gene disruption. Fungal Genet Biol. 1999;27:36–48. doi: 10.1006/fgbi.1999.1126. [DOI] [PubMed] [Google Scholar]
- Tenney AE, Brown RH, Vaske C, Lodge JK, Doering TL, Brent MR. Gene prediction and verification in a compact genome with numerous small introns. Genome Res. 2004;14:2330–2335. doi: 10.1101/gr.2816704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Edman JC. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- Wickes BL, Edman U, Edman JC. The Cryptococcus neoformans STE12alpha gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]