Abstract
G-protein coupled receptors (GPCRs) belong to biologically important and functionally diverse and largest super family of membrane proteins. GPCRs retain a characteristic membrane topology of seven alpha helices with three intracellular, three extracellular loops and flanking N' and C' terminal residues. Subtle differences do exist in the helix boundaries (TM-domain), loop lengths, sequence features such as conserved motifs, and substituting amino acid patterns and their physiochemical properties amongst these sequences (clusters) at intra-genomic and inter-genomic level (please re-phrase into 2 statements for clarity). In the current study, we employ prediction of helix boundaries and scores derived from amino acid substitution exchange matrices to identify the conserved amino acid residues (motifs) as consensus in aligned set of homologous GPCR sequences. Co-clustered GPCRs from human and other genomes, organized as 32 clusters, were employed to study the amino acid conservation patterns and species-specific or cluster-specific motifs. Critical analysis on sequence composition and properties provide clues to connect functional relevance within and across genome for vast practical applications such as design of mutations and understanding of disease-causing genetic abnormalities.
Keywords: Transmembrane Helices, Membrane Topology, Amino acid conservation and substitutions, GPCR cluster association
Background
G-protein coupled receptors (GPCRs) possess seven transmembrane hydrophobic helices, with three extracellular loops and three intracellular loops alternating each other [1]. GPCRs retain a wide variety of functional domains [2] within and across species to activate G-proteins, bind with diverse ligands, participate in signaling pathways and oligomerization, and are also implicated in diseases [3, 4]. The relevance to various diseases has been the reason that GPCRs are primary targets (about 75% of drug targets) in the pharmaceutical industry [5, 6]. The conserved amino acid (AA) patterns i.e., motifs present in the helices and in the loop regions could be quite critical in preserving common function despite evolutionary pressures. It is equally interesting to observe the differences to explain the impact of amino acid substitutions (AAS) in functional diversity and genetic abnormalities due to single-residue mutations. For example, a single residue mutation (P23H) in rhodopsin gives rise to a severe genetic abnormality, Retinitis pigmentosa, affecting protein stability ultimately leading to blindness [7]. In another instance, of aspartate receptor with only two transmembrane helices, a single amino acid mutation (hydrophobic to another hydrophobic residue) was sufficient to impair its methylation function that is mediated by dimerization [8]. In the current study, starting from a cross-genome survey of H. sapiens and D. melanogaster GPCRs, leading to 32 clusters of eight major types as explained in our previous publication [9], we report the analysis of AAS and conserved motifs in all 32 clusters of GPCRs. This study was further extended to a cross-genome analysis of H. sapiens and C. elegans GPCRs.
Methodology
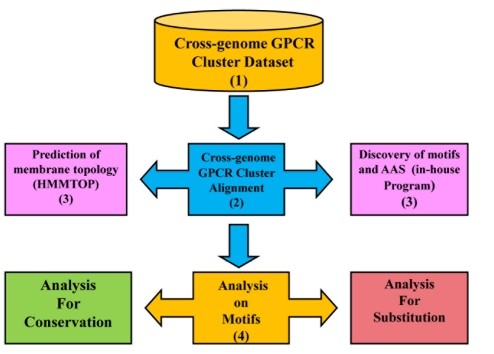
Figure 1 summarizes stepwise procedure for the identification of conserved AA (motifs) and residues exchanged at each position on MSA. This is split into four major steps:
Figure 1.
Flowchart depicting the methodology of the study
Step 1: GPCR cluster Dataset
A dataset of 32 clusters was created from our previous work [9] for selected H. sapiens and D. melanogaster (fruit fly) candidate GPCRs. The cluster association was established phylogenetically for eight major types like peptide receptors (PR), chemokine receptors (CMK), nucleotide and lipid receptors (N&L), biogenic amine receptors (BGAR), secretin receptors (SEC), cell adhesion receptors (CAR), glutamate receptors (GLU) and frizzled /smoothened (FRZ). The crossgenome GPCR cluster dataset was used in the current study for identifying key motifs and AA exchange patterns. (Please refer to Figure 1 for flow-chart).
Step 2: Alignment Procedure
Although the phylogenetically established GPCR cluster association was highly reliable in guiding the set of homologous sequences from the human and fruit fly genome, alignment tools play a crucial role in understanding sequence features, especially at remote homology. In the current study, CLUSTALW [10] was used for aligning sequences of human and fruit fly GPCR cluster dataset whereas MAFFT [18] was used to align human and C. elegans GPCRs for the 32 clusters. Alignments were manually examined and curated, where required, to retain equivalences of helices.
Step 3: Detection of Motifs and replacing amino acids
Cross-genome alignments for 32 clusters were taken as input to our in-house program to identify residue conservation and substitutions. AA conservation at an alignment position is simply an average of all possible pairwise sequences and the score is consulted from a normalized AA exchange matrix. A motif is defined by at least three consecutive conserved AAs with high amino acid conservation (more than 60% conservation score). The conservation of each residue in the set of aligned sequences was noted as ‘consensus’ and documented if the percentage conservation at a position is from 60 to 100%.
Step 4: Analysis of Identified Motifs
Once motifs were identified, the amino acids observed in the identified pattern were recorded and classified based on their property. The properties of substituting AA residues were denoted by a symbolic representation. The symbols @,*, +, , $ were used to represent the hydrophobic, aromatic, polar positive, polar negative and polar uncharged property of AA residues respectively. This symbolic representation at each position in the MSA helps to understand the extent of permitted amino acid exchanges and the proportion of AA conservation and replacement in the alignment. Separately, each sequence of the cross-genome alignment was annotated for membrane topology using HMMTOP 2.1 [11]. Incorporating the knowledge of predicted membrane topology and the identified motifs with AA substitutions in MSA enables us to understand the significant residue conservation and substitutions in TM helices and loop regions at cross-genome level.
Results & Discussion
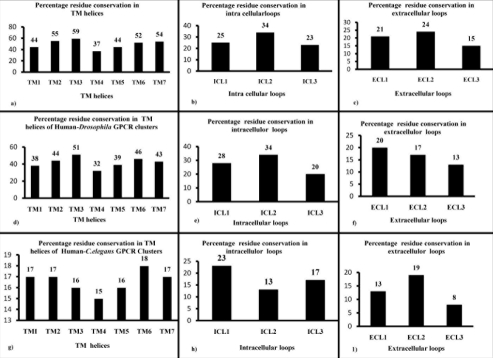
32 multiple sequence alignments from the GPCR cluster dataset were analyzed for the presence of motifs for human-Drosophila GPCRs and human-C. elegans GPCRs as described in Methods. ( http://caps.ncbs.res.in/download/crossgenomeGPCRs/align. zip provides full alignments for all 32 clusters). A total of 33 motifs were identified and 76% of them are within TM helices, predominantly in TM2 and TM7 (Table 1, see Table 1) in the human and Drosophila GPCR cluster dataset. Interestingly, peptide receptors retain 21 motifs and covers nearly 64% of the identified motifs, whereas other receptor types like chemokine receptors, nucleotide and lipid receptors and biogenic amine receptors contain 52%, 18% and 36% of motifs in the cross-genome cluster dataset. This could be due to the direct involvement of TM helices in ligand binding in the case of peptide receptors. In the current study, we have not included the N' and C' termini of the sequence and the study is focused only to selected set of sequences for the eight particular receptor types. The overall residue conservation is observed in the helices and the loop regions of human only, human- Drosophila and human-C. elegans GPCR clusters.(refer panels a - i in Figure 2).We found significant conservation in TM3 for the human-only and human-Drosophila GPCR clusters (refer panels a, d in Figure 2) and the ranking of conservation in the helices and loop regions are given in Table S1 Table S1 for human-only, human-Drosophila, human-C. elegans GPCR clusters. Notably, due to the occurrence of classical motif (E/DRY), significant motif conservation occurs in the intracellular loop (ICL2) of human GPCR clusters (refer panel b in Figure 2) and ECL2 retains high conservation in all the three cluster associations suggesting the crucial involvement and conservation of ligand binding residues in ECL2 loop (refer panels c, f and i in Figure 2). Aside from this, it is hard to obtain good-quality alignments of GPCRs from all three genomes simultaneously or to find motifs owing to poor sequence identity and high evolutionary divergence.
Figure 2.
Percentage residue conservation in TM helices and Loops in GPCR Clusters. Percentage residue conservation in the TM regions, intracellular loop, extra cellular loop of human GPCR clusters (shown in panels a, b, c), human-Drosophila GPCR clusters (shown in panels d, e, f), and human-C. elegans GPCR clusters (shown in panels g, h, i ).
Motifs observed in human-D. melanogaster cross-genome clusters
Motifs observed in transmembrane helices
We observed around 11 motifs occurring in single receptor type. Notably, VGL motif in transmembrane helix 1 (TM1), LGF motif in TM5 and NSC motif in TM7 are observed exclusively in peptide receptors (Table 1). Chemokine receptors exclusively possess YLLNLA motif in TM2 and HCC motif in TM7. On the other hand, the observed GNL motif in TM1, VMP motif in TM2, TASI motif in TM3, PFF motif in TM6, WLGY motif in TM7 are identified solely in biogenic amine type receptors. Further, the conservation of these motifs can be correlated to the cluster- or receptor-type specific properties at the sequence level.
We also observed around nine motifs occurring in two different types of receptors from our cluster dataset. SLA motif in TM2 is observed both in peptide and biogenic amine receptors. Interestingly, peptide and chemokine type receptors retain prominent conservation of motifs, with LFL, TLP and LPF motifs in TM2, AIA motif in TM3, LPL motif in TM5 and LYA in TM7 which explains the sequence conservation across two different receptor types and provide clues to common sequence properties (Table 1) among them. In a similar manner, IYL motif in TM2 and CIS motif in TM3 are observed not only in chemokine type receptors, but also in nucleotide and lipid type receptors. This emphasizes the utility of cross-genome clustering techniques, knowledge on receptor types for inferring the conservation of motifs across different receptor types at the cross-genome level. The significant occurrence of motifs in multi receptor type is also tabulated (Table 1). The NLA motif in TM2 occurs in three different receptor types like peptide, chemokine and nucleotide and lipid type receptors (Table 1). This motif has been observed for the maximum occurrence in our cluster dataset. The other motif DLL is also observed in TM2 helix in few clusters of peptide, chemokine, nucleotide, lipid and biogenic amine receptors. The same motif is also observed as ADL in TM2 in few clusters of all these four types of receptors (Table 1) and as ADLL motif in TM2 is observed in all three types of receptors, except peptide type receptors. The CWLP motif in TM6 is identified in peptide, chemokine, biogenic amine type receptors but not in nucleotide and lipid type receptors. In a broader sense, this significant conservation of motifs in TM2 explains the conservation of motifs not only with reference to the amino acid residues, but also with reference to their topology.
Motifs observed in loop regions
While observing motifs in the loop regions, eight different motifs were noted. The well-known E/DRY motif in ICL2 has the conservation as DRYLA in peptide (Cluster 3) and chemokine type receptors (Cluster 12) [17] and RYL in nucleotide and lipid type receptors (Cluster 15). ASG motif in ICL1 is conserved exclusively in glutamate receptors, whereas MRTVTN in ICL1 and LDR motif in ICL2 were conserved exclusively in peptide type receptors. Notably, WPFG and LCK motifs were found exclusively in ECL2 of peptide type receptors (Clusters 2 and 3 in Table 1, Table S2 Table S2). Interestingly, KLRN motif is observed in biogenic amine receptors (Cluster 21) and in secretin receptors (Cluster 26) in ICL1 (please see Table 1, Table S2 Table S2). Notably, Cluster 26 has a set of homologous sequences from Drosophila only GPCR clusters. However, Cluster 21 has GPCR sequences from both human and Drosophila genomes and we could identify common motifs observed across two taxa. This cluster can be a best illustration to emphasize the need of cross-genome phylogenetic analysis at sequence level even at distant relationships and during strong evolutionary drifts.
As prior studies [9] explain the important role of conserved AA in the ECL2 for the participation of ligand binding, this study reports around eight such motifs distributed in PR, N&L, BGAR, GLU, FRZ/SMT receptors. However, several motifs were identified in only one of the 32 cluster of receptors (Table S3 Table S3). For example, CLP motif from PR (Cluster 7) has AAS in the pattern as [C/P][L/F][P/C/S]. In the current study, there are 133 cluster-specific motifs observed in transmembrane helices and 59 cluster-specific motifs observed in the loop regions (Table S3 Table S3). The average sequence length of each of the TM - helices and loops were calculated from set of sequences based on the HMMTOP boundary predictions (Table S4 Table S4) and the average percentage of residue conservation in each TM helix and loop region was examined for the eight types of receptors (Table S5 Table S5). Interestingly, overall, the maximum amino acid conservation occurs as 42% and 46% in TM2 and TM3, respectively. Significant conservation of 55%, 80%, 61% occurs in TM1, TM2, TM3 within CMK receptors. Although the occurrence of motifs (consecutively preserved as three residues) is high in PR, it retains only 30-50% of conservation at TM2, TM6 and TM7. Generally, AA conservation is high at TM2 for BGAR, SEC, GLU, and FRZ type receptors. In most of the clusters, as expected, percentage residue conservation in ICL2 is higher than the other loop regions (Table S1 Table S1).
Motifs observed in human-C. elegans GPCR cross-genome clusters
Since the selected human-C. elegans GPCRs possess remote homology, the motifs are limited and are documented at the 30% – 100% conservation (refer Supplementary Tables S6 and S7). 295 motifs could be observed in the human and C. elegans GPCR clusters. This study will be further analyzed for comparative genome sequence analysis with other genome clusters in future.
Biological relevance of few previously observed GPCR motifs:
The detailed report on conserved motifs and substitutions in cross-genome GPCR cluster dataset for 32 clusters is given in Table S2 Table S2. However, to impart the feel for possible biological relevance, we will discuss few well-known motifs and substitutions.
Conserved E/DRY and NPXXY motifs in GPCR dataset
As cited earlier [12], the highly conserved characteristic E/DRY motif located at the boundary between transmembrane domain (TM3) and intracellular loop (ICL2) of Family A GPCRs play a pivotal role in regulating GPCR conformational states. The importance of DRY motif in connection with active MG4R in humans is well known [13]. Notably, in the cross-genome GPCR alignments, the preservation of characteristic DRY motif was observed in our current study (refer panel a in Figure 3). Tyr residue in this motif is highly conserved or retained an aromatic residue in most of the clusters in human GPCRs (example in chemokine receptors in Cluster 12, 13). However, in peptide receptors of Drosophila, there is a weak conservation of Tyrosine (refer panel a in Figure 3). Arg is conserved comparatively well and the substitution is of polar uncharged ($) or positively charged residue (+) of the same kind (for e.g. in biogenic amine receptors in Cluster 24) (Table S2 Table S2).
Figure 3.
Alignments showing conserved E/DRY, KLR/RLAR and PMNYM/PMSYM motifs in GPCR clusters (noted in the panel a, b, c respectively).
Conserved KLR/RLAR motif in Human Secretin receptor Dataset
Another highly conserved motif is seen within the third endoloop of the Family B Human Secretin receptor is KLR / RLAR motif [14]. Block deletion of KLRT and mutation of Lys323 (K323I) is known to reduce cAMP accumulation, and these mutations do not affect ligand interaction. Thus, KLRT region at the N-end of the third endoloop, particularly Lys323, is important for G protein coupling [14]. Also, it is noticed that for the RLAR motif, substitutions from Arg (R330) to Ala (342A), Glu (342E), or Ile (342I) as well as block deletion of the RLAR motif were all found to be defective in both secretinbinding and cAMP production (Chan et al., 2001) [14]. KLK/R and RLAR/K pattern is seen to be conserved in the two proteins GLR and GLP1 (refer panel b in Figure 3), which belong to the secretin family noted in Cluster 25 of our GPCR cluster dataset.
Conserved PMNYM / PMSYM motif in Human Adenosine receptor Dataset
The PMNYM / PMSYM pattern is conserved in the TM5 of GPCRs [7]. TM5 has been suggested to self associate and may be involved in the dimerization of the receptor A2aR (Human adenosine receptor). In adenosine A2b receptor, asparagine (N) residue is replaced by serine (S) generating the motif PMSYM, thus differentiating the two isoforms of receptors functionally (refer panel C in Figure 3). It is suggested that the motif PMNYM of A2aR and PMSYM of A2bR may be involved in TM assembly of the two isoforms of the receptors, respectively. Such information may provide an insight into the molecular mechanism of receptor-ligand interaction leading to design of tailored compounds. A careful observation of the alignment (please refer to Step 2 in Methods and Figure 1) reveals this important PMNYM/PMSYM motif in GPCR Cluster 23 albeit not identified at a score threshold of 60% (refer panel C in Figure 3).
Conclusion
Our approach for identifying conserved motifs and substituting AA residues are effective in recognizing functionally important residues in our GPCR cluster dataset. Along with the wellknown characteristic motifs (refer panel a, b, c in Figure 3), other preserved motif patterns in the MSA were also identified for their occurrence at 60-100% conservation. We have reported the residue conservation/identity, permitted AAS (based on their respective physiochemical property) at each position and cluster-specific motifs. This current approach can be applied to other membrane-bound receptors (such as olfactory receptors) and protein families to detect the conserved motifs. It will be interesting to map the identified motifs on predicted topology in MSA which may be helpful to perform evolutionary studies at the cross-genome level. Due to remote homology, there are chances of missing the key motifs in the generated MSA, especially in cross-genome GPCR alignments. Our approach (based on the recognition of motifs, derived from average AAS scores) is helpful in recognizing both classical and newer motifs, which have not been hitherto attributed any functional significance. Our approach of analyzing sequence properties in the set of aligned sequences can be applicable to compare with a reference sequence (of known 3D structure) to understand sequence similarity in the predicted topology and preserved motifs with AAS at each position. This method can be used as a guiding principle for 3-D modeling of GPCR sequences. Homology modeling, together with such motif analysis could uncover additional spatial clusters or ‘spatial motifs’, which may be critical for function.
Supplementary material
Footnotes
Citation:Nagarathnam et al, Bioinformation 7(1): 15-20 (2011)
References
- 1.K Palczewski, et al. Science. 2000;289:739. [Google Scholar]
- 2.R Fredriksson, et al. Mol Pharmacol. 2003;63:1256. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 3.SC Prinster, et al. Pharmacological Reviews. 2005;57:289. doi: 10.1124/pr.57.3.1. [DOI] [PubMed] [Google Scholar]
- 4.M Rocheville, et al. Science. 2000;288:154. [Google Scholar]
- 5.S Schlyer, R Horuk. Drug Discovery Today. 2006;11:481. doi: 10.1016/j.drudis.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 6.MJ Marinissen, JS Gutkind. Trends Pharmacol Sci. 2001;22:368. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 7.S Gleim, et al. Biochemistry. 2009;48:1793. doi: 10.1021/bi800968w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CJ Jeffery, DE Koshland. Jr. Biochemistry. 1994;33:3457. doi: 10.1021/bi00178a001. [DOI] [PubMed] [Google Scholar]
- 9.RP Metpally, R Sowdhamini. BMC Genomics. 2005;6:106. doi: 10.1186/1471-2164-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.JD Thompson, et al. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GE Tusnády, I Simon. Bioinformatics. 2001;17:849. [Google Scholar]
- 12.GE Rovati, et al. Mol pharmacol. 2007;71:959. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- 13.Y Yamano, et al. Biosci Biotechnol Biochem. 2004;68:1369. doi: 10.1271/bbb.68.1369. [DOI] [PubMed] [Google Scholar]
- 14.KY Chan, et al. Endocrinology. 2001;142:3926. doi: 10.1210/endo.142.9.8389. [DOI] [PubMed] [Google Scholar]
- 15.Pratibha Mehta Luthra, et al. PRIB. 2007;4774:41. [Google Scholar]
- 16.CM Westhoff, et al. Transfusion. 2000;40:321. [Google Scholar]
- 17.M Baggiolini, et al. Annu Rev Immunol. 1997;15:675. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 18.K Katoh, et al. Nucleic Acids Res. 2002;30:3059. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CJ Venglarik, et al. J Am Soc Nephrol. 2004;15:1168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.