Abstract
GAGA factor (GAF) is involved in both gene activation and gene repression and plays a role in the modulation of chromatin structure. In Drosophila, Trithroax like (Trl) gene encodes the DNA binding protein called GAGA factor (GAF). Trl-GAF binds to GAGA sites through its C2H2 zinc finger domain and has an N-terminal BTB/POZ domain. Identification of Trl-GAF homologue in mouse helps in deeper understanding of the mechanism and function. Conventional alignment tools such as BLAST and FASTA cannot identify homologues in mouse genome as their sequence identity is below 30%. In the present study, various sequence and structure analyses were followed for the detection of remote homologues of Drosophila GAGA FACTOR in mouse to identify as Zbtb3. Through homology modeling and docking approach, the zinc finger region of mouse Zbtb3 showed conserved residues and favorable DNA binding sites with GAGA sites similar to that of Drosophila GAGA FACTOR.
Keywords: Remote homologue, GAGA factor, BTB, Zinc finger, Zbtb3, molecular modeling
Background
In Drosophila, chromation factors such as Polycomb group (PcG) and trithroax group (trxG) proteins play a crucial role in the development of gene regulation. One of the trxG group gene is Trithorax like (Trl), also known as GAGA factor (GAF). It is identified as a sequence specific DNA binding protein, which could stimulate transcriptional activity [1]. GAGA promotes transcription by blocking repressive effects of histones. GAGA is required for proper expression of variety of development loci that contain GAGA binding sites in their upstream regulatory regions. Trl-GAF binds to GAGA sites through its C2H2 zinc finger domain and has an N-terminal BTB/PZ (broad complex tramtrack bric a brac/poxvirus zinc finger) protein-protein interaction domain. At N terminus of GAGA there is BTB/PZ domain required for oligomerization which is highly conserved and involves in protein-protein interaction. The C-terminal glutamine-rich region is required for transcriptional activation. Members of GAGA factor family bind to a 5'-GAGAG-3' DNA consensus binding site, and contain a Cys2-His2 zinc finger core as well as an N-terminal extension containing two highly basic regions. The zinc finger core binds in the DNA major groove and recognizes the first three GAG bases of the consensus in a manner similar to that seen in other classical zinc finger-DNA complexes [2]. It has been shown that GAGA factor has multiple roles in regulating genes during Drosophila development. A convenient way to study the function, structure and mechanism of a gene is to identify homologues (evolutionary relationships) in model organisms. It is interesting to identify homologues of GAGA factor in mouse. Dynamic programming based alignment tools such as BLAST and FASTA have been widely used to provide evidence for homology by matching a new sequence against a database of previously annotated sequences. However, these approaches can only detect homologous proteins that exhibit significant sequence similarity. But if sequence identity is below 30% it may be very difficult to detect homologue and they are called as remote homologue which share the same evolutionary ancestry. Accurate detection of homologue at low levels of sequence identity still remains a challenging problem. Recently, the vertebrate homologue of Drosophila GAGA factor was identified in mouse as c-Krox/Th-POK gene encoded by zbtb7b [3]. In this present study through insilico analysis another probable homologue of Drosophila GAGA factor in mouse is reported as Zbtb3 (Zinc finger and BTB domain-containing protein 3).
Materials and Methodology
Sequence retrieval
The sequence of Drosophila GAGA factor was retrieved from the UniprotKB (Q08605) and the domain boundaries were taken from Uniprot annotations. Drosophila GAGA factor contains BTB domain at N-terminus and zinc finger domain at Cterminus.
Remote Homology Detection Methodology
Similarity search
The retrieved sequence of GAGA factor (GAF) was searched against mouse genome database through Protein-Protein Blast (BlastP) of NCBI with default parameters.
PSI-Blast search
PSI-Blast makes use of iterated BLAST searches in order to extend the number of evolutionary relationships detected. Related proteins were searched using PSI-BLAST [4] with five iterations using the top twenty maximum hits.
Hidden Markow model and protein family search
The Drosophila GAGA factor sequence was used for searching databases of protein sequence using profile Hidden Markow Models (HMMs) for detection of remote homologs using the server HHpred [5]. Through this search about eight probable homologue clusters were obtained. The entire eight probable clusters were crosschecked with pfam database of protein families [ http://pfam.sanger.ac.uk/].
Multiple sequence alignment and phylogenetic analysis
From the eight probable homologue clusters, protein sequences were obtained and multiple alignment was constructed using ClustalW [6]. The phylogenetic tree was constructed with distance matrix method.
Homology modeling
The sequence of mouse zbtb3 was obtained from the Uniprot database (Q91X45) and the sequence was queried against the Protein DataBank (PDB) using PSI-BLAST. Through the PSIBLAST results, template, which has high sequence identity and fewer gaps were selected. Comparative modeling of both BTB domain and Zinc finger domain of Zbtb3 was performed by using Modeller software version 9v5 [7]. The resulting model was evaluated using Verify3D [8] and Procheck. The quality of models was assessed by Ramachandran plot analysis [9]. The structures were visualized using pymol.
Docking of Zbtb3 and GAGA-DNA complex
The homology modeled zinc finger domain of Zbtb3 was further studied to know how they interact with GAGA-DNA complex through docking approach. NMR structure of GAGA factor/DNA complex (PDB: 1YUI), which binds to the single zinc finger, was used. Zinc finger was removed and only GAGA factor/DNA complex was used for docking with homology modeled zinc finger of Zbtb3. The modeled zinc finger domain was docked with GAGA-DNA complex (1YUI) using Hex software [10].
Results and Discussion
Sequence analysis
Through BlastP search it was found that Drosophila GAGA factor had very less identity (below 20%) with mouse. Since sequence identity of the Drosophila GAGA factor with mouse was below the twilight zone, various rigorous methodologies were followed for the detection of remote homologues. PSIBlast search method groups the closely related protein sequences and reflects the information from a protein family better than pair wise sequence comparison method like Blast. PSI-Blast based search was carried out with default parameters against mouse genome with five iterations. Careful selection of the proteins containing both BTB and Zinc finger domain and removal of redundant proteins resulted in about twenty probable proteins. Profile Hidden Markov Model (HMMs) is one of the most powerful approaches for remote homolog identification. The mouse protein database was searched by profile Hidden Markow Models (HMMs) with HHPred and eight probable remote homologues were selected (Table 1, see Table 1). Crosschecking the entire eight clusters with pfam database of protein families [11] confirmed that they belong to the family of BTB and Zf-C2H2. Through pair wise sequence alignment, comparison of GAGA factor with the eight probable remote homologue clusters was carried out by EMBOSS using the Needleman-Wunsch alignment algorithm to find out the optimum alignment of two sequences along their entire length [12]. Zbtb3 was found to have maximum sequence identity in both domain wise and in whole sequence alignment (Table 1, see Table 1). Phylogenetic analysis with distance analysis suggests that zbtb3 was close to GAGA factor. Recently, it is reported that large number of transcription factors are mostly intrinsically disordered [13]. From the Disprot database [14], it is inferred that disordered residues were between regions 137-519 for Drosophila GAGA. The Zbtb3 predicted with consensus disorder prediction method [15] resulted in the disordered residues placed between regions 134-498 which agreed well with Drosophila GAGA factor. Intrinsic plasticity enables a disorder region to recognize and bind many biological targets with high specificity.
Structural analysis
Comparative modeling of Zbtb3 for both BTB domain and Zinc finger domain was carried out using Modeller program. For modeling of BTB domain, template of crystal structure of the Btb domain of human myoneurin from the PDB ID 2VPK was used. For modeling of Zinc finger domain, template of crystal structure of a designed zinc finger protein from the PDB ID 1Mey was used. The sequence alignment of target and template for both BTB domain and Zinc finger domain was about 40 42%. Individual alignments were given as input to Modeller to build 3D structures, and the resulting models were evaluated using Verify 3D and Procheck. This analysis has led to the conclusion that the models were more reliable. Homology model of zbtb3 zinc finger consists of three finger coordinate zinc ions with a combination of cysteine and histidine residues (Figure 1a). As Drosophila GAGA factor binds well with DNA, it is important to analyze and characterize the predicted homologue for affinity to DNA-binding sites. The Zbtb3 was predicted with DBS-PRED [16] and found that their binding affinity was about 34.7%. Solution and solid-state structural studies have revealed that alpha helix of Zinc finger makes contact with the major groove of DNA [17]. The homology modeled zinc finger domain of Zbtb3 was docked to GAGADNA, keeping DNA as a fixed molecule, using Hex. Hex employs spherical polar Fourier correlation and considers both shapes complementarily and with electrostatic effects. The binding energy score obtained was 3.14 kcal/mol and RMS value was about 1.00. From the docked model, it is inferred that it contains favorable binding sites like AGC sequence at N terminus, the presence of highly conserved glycine residues within alpha helix and TGEKP residues at the linker region necessary for promoting zinc finger to fit into the DNA major groove and also to strengthen the DNA binding (Figure 1b). This result agrees with the previous findings [18]. Zbtb3 was also found to be the homolog of lolla gene in Drosophila. Lolla gene functions with Trl for maintaining the repressed state of target genes and also binds to a DNA polycomb response element (PRE) at the bithroax complex [19]. As it is known that Trl binds with GAGA factor in Drosophila, it supports the hypothesis that Zbtb3 might play a similar function like Trl, binding with GAGA factor.
Figure 1.
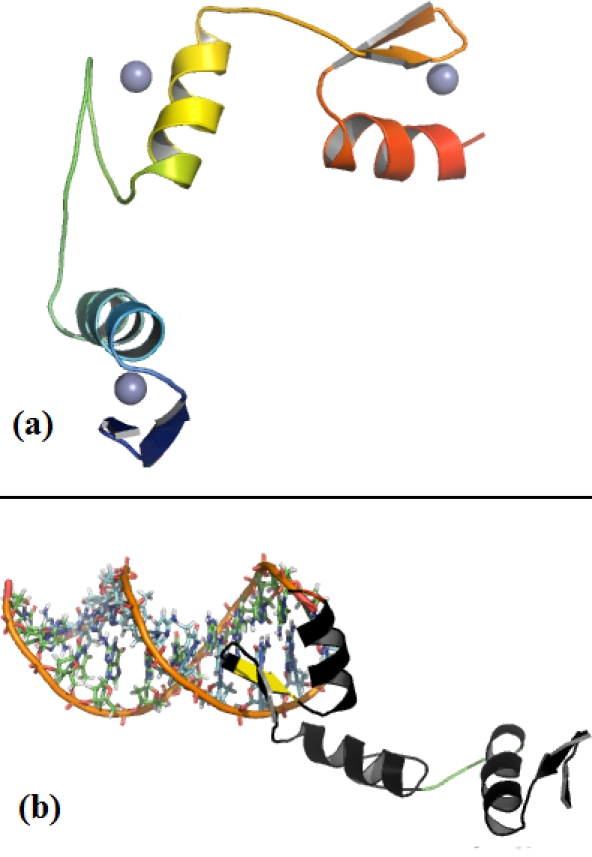
(a) Homology Model of zinc finger domain of Zbtb3 consisting of N and C terminus in the presence of zinc ions shown as spheres in cyan and (b) Docked zinc finger with DNA showing conserved TGEKP residues at linker region in green, glycine residue in red and AGC sequence is shown in yellow.
Conclusion
Drosophila GAGA factor plays a major role in developmental gene regulation. In this study, through computational analysis it is shown that mouse Zbtb3 is a remote homolog of Drosophila GAGA factor. The reported homolog exhibits strong similarity of highly conserved BTB domain as well as the zinc finger domain to the corresponding regions of Drosophila GAGA factor. The homology modeling and molecular docking was applied in this study to explore the binding mechanism of GAGA factor and Zbtb3. Through molecular modeling studies, it contains favorable DNA binding sites, which are conserved. These predictions confirm that Zbtb3 can bind to GAGA repeats.
Supplementary material
Footnotes
Citation:Kumar, Bioinformation 7(1): 29-32 (2011)
References
- 1.G Farkas, et al. Nature. 1994;371:806. [Google Scholar]
- 2.JS Omichinski, et al. Nat. Struct. Biol. 1997;4:122. doi: 10.1038/nsb0297-122. [DOI] [PubMed] [Google Scholar]
- 3.NK Matharu, et al. J Mol Biol. 2010;400:434. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 4.AA Schäffer, et al. Nucleic Acids Res. 2001;29:2994. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.J Söding. Bioinformatics. 2005;21:951. [Google Scholar]
- 6.R Chenna, et al. Nucleic Acids Res. 2003;31:3497. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A Sali, et al. Proteins. 1995;23:318. [Google Scholar]
- 8.JU Bowie, et al. Science. 1991;253:164. [Google Scholar]
- 9.GN Ramachandran, et al. J Mol Biol. 1963;7:95. [Google Scholar]
- 10.DW Ritchie, et al. Bioinformatics. 2008;24:1865. [Google Scholar]
- 11. http://pfam.sanger.ac.uk/
- 12. http://www.ebi.ac.uk/Tools/psa/emboss_needle/
- 13.J Liu, et al. Biochemistry. 2006;45:6873. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S Vucetic, et al. Bioinformatics. 2005;21:137. [Google Scholar]
- 15.S Kumar, O Carugo. Open Biochem J. 2008;2:1. doi: 10.2174/1874091X00802010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S Ahmad, et al. Bioinformatics. 2004;20:477-86. [Google Scholar]
- 17.ES Lander, et al. Nature. 2001;409:860. [Google Scholar]
- 18.SA Wolfe, et al. Annu Rev Biophys Biomol Struct. 2000;29:183. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 19.L Tracy, et al. Apoptosis. 2009;14:969. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


