Abstract
The accurate prediction of a comprehensive set of messenger putative antagomirs against microRNAs (miRNAs) remains an open problem. In particular, a set of putative antagomirs against human miRNA is predicted in this current version of database. We have developed Antagomir database, based on putative antagomirs-miRNA heterodimers. In this work, the human miRNA dataset was used as template to design putative antagomirs, using GC content and secondary structures as parameters. The algorithm used predicted the free energy of unbound antagomirs. Although in its infancy the development of antagomirs, that can target cell specific genes or families of genes, may pave the way forward for the generation of a new class of therapeutics, to treat complex inflammatory diseases. Future versions need to incorporate further sequences from other mammalian homologues for designing of antagomirs for aid in research.
Availability
The database is available for free at http://bioinfopresidencycollegekolkata.edu.in/antagomirs.html
Background
MicroRNAs (miRNAs) are a class of endogenous, small regulatory RNA averaging 22 nucleotides in length that mediate the post-transcriptional regulation of messenger RNAs. They bind to target messages in a sequence-specific manner, and induce translational repression or endonucleolytic cleavage. Recently, a novel class of chemically engineered oligonucleotides, termed antagomirs, is efficient and specific silencers of miRNAs. Moreover, the recent employment of synthetic analogues of these small RNA molecules termed ‘antagomirs’ has shown that microRNAs of interest can be specifically targeted [1, 2]. Functions have only been experimentally assigned to a small fraction of the experimentally designed antagomirs. These putative antagomirs are 20 nucleotides in length and complementary to the mature target human-miRNA. They specifically silenced miRNA expression (miR-122) up-regulating expression of hundreds of genes predicted to be repressed by miR [3]. Paradoxically, antagomir treatment also revealed a significant number of down-regulated genes that may be activated as opposed to repressed by miR [4, 5, 6]. Computational approaches are thus likely to remain an important means of studying antagomir targets for the foreseeable future, not least as a means of directing wet-lab experiments. Several algorithms have been used to predict antagomir targets in animal species; these are listed in Table 1 (see Table 1). We built an experimental antagomir in a way that it could adhere to several humiR, respectively. Interactions are viewed in a global context by predicting folds for the entire miRNA, rather than just its 3'- UTR or seed match. The stem-loop sequences of humiR have been used in our experiment.
The probability profile displays predicted accessible sites on the target RNA. Because an accessible site can be targeted by a number of antisense oligos, selection of the “optimal” one can be based on binding energy, together with other empirical rules such as GC content, avoidance of GGGG (or more stringent GGG) motifs, etc. Stronger binding is indicated by smaller binding energy (stacking energies are negatively valued). For example, an antisense oligo with a binding energy of 10 kcal/mol is more effective than an oligo with a binding energy of 5 kcal/mol. The antisense oligo binding energy is a weighted sum of the antagomir/miRNA stacking energies for the hybrid formed by the antagomir and the targeted sequence. For a base-pair stack, the weight for the sum is calculated by the probability of the unpaired dinucleotide in the target sequence that is involved in the stack. This weighting scheme accounts for the structural variation at the target site among the structures in the sample (Figure 1).
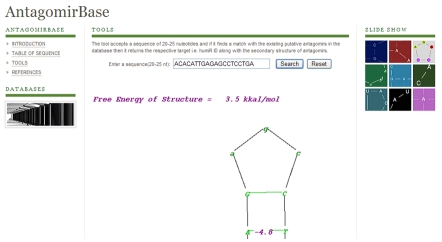
Figure 1.
Screenshot of the database main page
Implementation
Database
The antagomir database is based on the following assumptions. (1) Based on the humiR sequence the following details about the putative antagomirs are obtained with the help of Sfold. Column 1: target position (starting - ending) Column 2: target sequence (5'→3') Column 3: antisense oligo (5'→3') Column 4: GC content Column 5: oligo binding energy (kcal/mol) (2) The database also includes the secondary structure of antagomirs along with the other details mentioned above. The secondary structures have been designed with the help of mfold.
Tool
A tool has been integrated in this database. The tool accepts a sequence of 20-25 nuleotides and if it finds a match with the existing putative antagomirs in the database then it returns the respective target i.e. humiR ID along with the secondary structure of antagomirs.
Statistical analysis of predicted targets
Negative normalized free energy
The occurrence of favourable hybridizations of short antagomirs with long miRNAs can frequently be attributed to chance: the longer the miRNA, the more likely the incidence. In order to eliminate the effect of sequence length on our measure of free energy [7, 8], we define the negative normalized free energy where m is the length of the target sequence searched, and n is the length of the antagomir as shown in equation 1: gn = (g/log(mn)).
Extreme value statistics
Extreme value distributions (EVDs) are limiting distributions that describe the minimum or maximum of independent random variables [9]. If we consider the antagomir-miRNA duplex energy estimation to be essentially an optimization procedure that produces a minimum, the negative normalized free energy described above is a corresponding maximum, and can be described by an EVD having a distribution function of the form; P[G≤t]=D(t) =exp(exp(at/b)) (equation 2). A transformation then converts this distribution function into a straight line: Log(log(D))=(at) /b=(1/b)t + a/b (equation 3). By scanning for targets of random antagomir sequences in the miRNA sequences in the dataset, we obtain a set of negative normalized free energies, which we expect will follow an EVD. We then transform the distribution function of the empirical EVD into a straight line, as in Equation 3, and estimate the parameters of the EVD by a linear least squares fit to the line y = mx + c, obtaining b= 1/m (equation 4) and a=cb (equation 5). We can now compute, for each predicted antagomir-miRNA duplex, a p-value, the probability that the same or a more favorable free energy is observed due to chance: P[Z≥ gn]=1 exp(exp((a gn)/b)) (equation 6), where a and b are estimated EVD parameters, and gn is the negative normalized free energy from equation 1 [7].
Results
Antagomir database is the central online repository for antagomirs sequence data, annotation and target prediction. The current release (ver.1) contains 22 humiRNA from Homo sapiens, expressing 53 distinct putative antagomir sequences (Table 2, see Table 2). The humiRID and the secondary structure of antagomir are included in the database. The best four matches of the secondary structure with respect to free energy are given in Figure 2.
Figure 2.
Screenshot of the database showing the UID (User Interface Design
Conclusion
The prediction of antagomir targets is an open and difficult problem in spite of a few years of the existing research. Antagomir Database designed based on human miR targets, is shown to provide predictions characterized by favorable sensitivity, which comes at a price of an increased number of predictions. Our future work will concentrate in the inclusion of more miRNAs from different species as targets of antagomirs.
Supplementary material
Footnotes
Citation:Ganguli et al, Bioinformation 7(1): 41-43 (2011)
References
- 1.R Thadani, MT Tammi. BMC Bioinformatics. 2006;7(5):S20. [Google Scholar]
- 2.J Krützfeldt, et al. Nature. 2005;438:685. [Google Scholar]
- 3.AM Cheng, et al. Nucleic Acids Res. 2005;33:1290. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J Weiler, et al. Gene Ther. 2006;13:496. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 5.G Meister, et al. RNA. 2004;10:544. [Google Scholar]
- 6.J Mattes, et al. Am J Respir Cell Mol Biol. 2007;36:8. doi: 10.1165/rcmb.2006-0227TR. [DOI] [PubMed] [Google Scholar]
- 7.M Rehmsmeier, et al. RNA. 2004;10:1507. [Google Scholar]
- 8.S Karlin, SF Altschul. Proc Natl Acad Sci U S A. 1990;87:2264. [Google Scholar]
- 9.EJ Gumbel, et al. Statistics of Extremes. New York: Columbia University Press; 1958. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.