ABSTRACT
Cholera continues to be an important cause of human infections, and outbreaks are often observed after natural disasters, such as the one following the 2010 earthquake in Haiti. Once the cholera outbreak was confirmed, rumors spread that the disease was brought to Haiti by a battalion of Nepalese soldiers serving as United Nations peacekeepers. This possible connection has never been confirmed. We used whole-genome sequence typing (WGST), pulsed-field gel electrophoresis (PFGE), and antimicrobial susceptibility testing to characterize 24 recent Vibrio cholerae isolates from Nepal and evaluate the suggested epidemiological link with the Haitian outbreak. The isolates were obtained from 30 July to 1 November 2010 from five different districts in Nepal. We compared the 24 genomes to 10 previously sequenced V. cholerae isolates, including 3 from the Haitian outbreak (began July 2010). Antimicrobial susceptibility and PFGE patterns were consistent with an epidemiological link between the isolates from Nepal and Haiti. WGST showed that all 24 V. cholerae isolates from Nepal belonged to a single monophyletic group that also contained isolates from Bangladesh and Haiti. The Nepalese isolates were divided into four closely related clusters. One cluster contained three Nepalese isolates and three Haitian isolates that were almost identical, with only 1- or 2-bp differences. Results in this study are consistent with Nepal as the origin of the Haitian outbreak. This highlights how rapidly infectious diseases might be transmitted globally through international travel and how public health officials need advanced molecular tools along with standard epidemiological analyses to quickly determine the sources of outbreaks.
IMPORTANCE
Cholera is one of the ancient classical diseases and particularly prone to cause major outbreaks following major natural disasters, such as earthquakes and hurricanes, where the normal separation between sewage and drinking water is destroyed. This was the case following the 2010 earthquake in Haiti. Rumors spread that the disease was brought to Haiti by a battalion of Nepalese soldiers serving as United Nations peacekeepers. This possible connection has never been confirmed. Sequencing the genomes of bacteria can give detailed information on whether isolates from different sites share a common origin. We used this technology to sequence isolates of Vibrio cholerae from Nepal, identify single-nucleotide polymorphisms (SNPs), and compare these high-resolution genotypes to the complete genome sequences of isolates from the Haiti outbreak. We provide support for the hypothesis that the isolates were brought to Haiti from Nepal.
Introduction
Cholera is caused by the Gram-negative bacterium Vibrio cholerae, and the disease is usually transmitted through contaminated water (1). V. cholerae is normally present in coastal and brackish waters worldwide and has been found in countries where the disease is not found in humans. The bacterium can also be transmitted globally in the intestines of asymptomatic carriers. Thus, it is difficult to determine the origin of outbreaks associated with disaster situations where the normal water supply and hygiene measures are disrupted.
More than 200 serogroups of V. cholerae have been identified, but isolates belonging to serogroup O1 of the “classical” or El Tor biotype have been the most important human pathogen in the last century. Seven different cholera pandemics are believed to have occurred since 1817. The causative agents of the first five pandemics were not cultured, but the sixth pandemic (1899 to 1923) was caused by the classical biotype. El Tor strains were associated with sporadic cases during the sixth pandemic (2), but in 1961, this biotype was responsible for the seventh pandemic. El Tor and a number of variants have been implicated in numerous outbreaks worldwide and have become prevalent in some countries with limited access to clean water.
On 12 January 2010, a 7.0 MW earthquake hit Haiti. By 24 January, at least 52 aftershocks had been reported, and an estimated 316,000 people had died, 300,000 were injured and more than one million were homeless. This disaster destroyed the already fragile infrastructure and required international assistance in the form of food, water, and aid workers. On 21 October 2010, the Haitian public health authorities confirmed a cholera outbreak. By 7 July 2011, 386,429 cases, including 5,885 deaths have been reported (3). The outbreak has also spread to the neighboring Dominican Republic and to Florida and the United States (4) where sporadic cases have been observed. In the early days of the outbreak, rumors spread that the disease was brought to Haiti by a battalion of Nepalese soldiers serving as United Nations peacekeepers (2, 5–8). Though not proven definitively, the putative link to United Nations peacekeepers from Nepal gained global media attention and sparked riots in Haiti that disrupted relief efforts.
Conventional and molecular characterization of bacterial isolates is useful in determining the relationship between strains and can assist in identifying the sources. Traditionally, V. cholerae strains are classified into serogroups based on their outer membrane O antigen and further subdivided into biotypes based on biochemical testing; however, most outbreaks during the seventh pandemic have been caused by the same serogroup and biotype, El Tor, limiting the utility of these analyses for outbreak investigations. Molecular typing using pulsed-field gel electrophoresis (PFGE) is commonly used to characterize strains but does not always provide sufficient discriminatory power. Single-nucleotide polymorphisms (SNPs) and insertions/deletions have been used to further resolve global transmission of El Tor (9, 10). Whole-genome sequence typing (WGST) is a powerful tool providing an almost complete picture of genetic polymorphisms for evolutionary and epidemiological investigations (11–14).
A PFGE-based study by the U.S. Centers for Disease Control and Prevention indicated that the Haitian outbreak strain was related to contemporary strains circulating in South Asia and elsewhere (4). Another study using whole-genome sequencing has similarly shown that the Haitian outbreak strain is more closely related to recent strains from Bangladesh and Mozambique than to a strain from Peru (15); however, the Peruvian strain used in that study was more than 20 years old, which weakens their conclusions. So far, none of the published studies has included recent Nepalese V. cholerae isolates to evaluate their relatedness to the Haitian outbreak strain.
Cholera occurs in sporadic cases and outbreaks in Nepal each year. In 2010, a 1,400-case outbreak occurred in midwestern Nepal (http://www.irinnews.org/Report.aspx?ReportID=90231). The outbreak started around 28 July and was controlled by 13 or 14 August, just prior to the time the Nepalese soldiers left for Haiti. On the request by the public health authorities in Nepal and in our function as a World Health Organization Collaborating Centre, we conducted the current study to determine the genetic diversity of the most contemporary V. cholerae strains from Nepal. We then compared these data to the publicly available whole-genome sequences of isolates from the recent outbreak in Haiti, as well as those of other available strains.
RESULTS
All Nepalese isolates were susceptible to tetracycline but resistant to trimethoprim, sulfamethoxazole, and nalidixic acid and showed decreased susceptibility to ciprofloxacin. This susceptibility profile is consistent with that of isolates causing the Haitian outbreak (4). PFGE showed that the Nepalese isolates belonged to four clusters of indistinguishable patterns, including 2, 4, 4, and 14 isolates. One cluster containing four Nepalese isolates (isolates 12, 14, 25, and 26) was identical to a minor variant of the main pulsotype from Haiti, whereas another cluster of four Nepalese isolates (isolates 6, 15, 18, and 19) was indistinguishable from the most common pulsotype observed in Haiti, as determined by the U.S. Centers for Disease Control and Prevention. While the PFGE results show the great similarity of the Haitian to Nepalese isolates, the fine-scale affinities are discordant with WGST, perhaps due to convergent evolution by pulsotype of isolate 12.
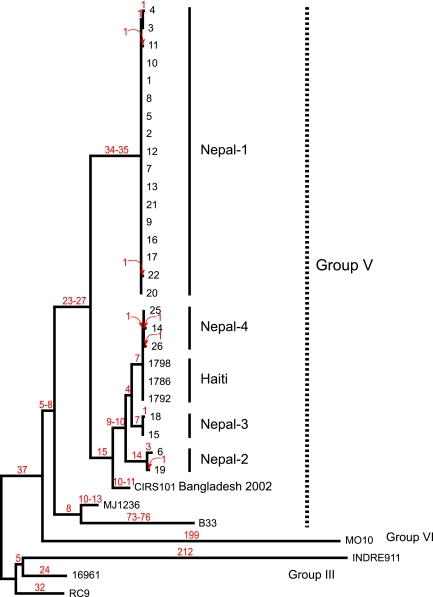
WGST and phylogenetic analysis showed that all 24 V. cholerae isolates from Nepal belong to a single well-supported monophyletic group that also contains isolates from Bangladesh and Haiti (Fig. 1). A single maximum parsimony tree was reconstructed using 752 SNPs from 34 whole-genome sequences. There were 184 parsimony-informative SNPs, of which 6 were homoplastic, resulting in a CI of 0.97 (excluding uninformative characters). The Nepalese isolates are subdivided into four closely related clusters, all within group V as defined by Lam et al. (16). One of the four Nepalese genotypic groups (Nepal-1), containing 17 out of the 24 isolates, is genetically distinct and highly homogeneous. There are 34 or 35 synapomorphic SNPs supporting its unique identity. (A synapomorphic SNP is a genome position that has mutated such that the new nucleotide is shared with all descendants.) The second group contains three Nepalese clusters along with a basal Bangladesh isolate (CIRS101 2002) and three Haitian isolates in a derived position. The three Nepalese isolates, isolates 14, 25, and 26 in cluster Nepal-4, and the three Haitian isolates, isolates 1786, 1792, and 1798, are extremely close and form their own monophyletic subclade supported by 7 synapomorphic SNPs, with no homoplasy. The lack of homoplasy is strong evidence of clonality in this population. Only a single synapomorphic SNP separates the Haitian isolates from isolates in cluster Nepal-4, although there are two autapomorphic SNPs within this cluster. (An autapomorphic SNP is a genome position that has mutated but is found only in a single descendant.)
FIG 1 .
Genetic relationships among V. cholerae isolates from Nepal and Haiti. A single maximum parsimony tree was reconstructed using 752 SNPs from 34 whole-genome sequences. There were 184 parsimony-informative SNPs, of which 6 were homoplastic, resulting in a CI of 0.97 (excluding uninformative characters). The branch lengths are labeled in red, and for branches affected by homoplasy, minimum and maximum branch lengths are designated. Members of SNP genotypic group V (16) are indicated. SNP differences among the three most closely related Nepali groups and the Haitian group are shown and characterized in Table S1 in the supplemental material.
Direct comparison between the three Haiti outbreak strains (strains 1786, 1792, and 1798) and the three most closely related strains from cluster Nepal-4 (strains 14, 25, and 26) showed that the 1- or 2-bp differences are nonsynonymous and give rise to amino acid differences (Table 1). The basal position of CIRS101 suggests a possible source for some of the Nepalese strains (clusters Nepal 2-3-4); its phylogenetic position among the clades argues for more than one infective focus for the Nepalese outbreak. The SNPs defining the Nepal-2,3,4 and Haitian cluster (branches A through K) appear to be under diversifying selection, as the nonsynonymous SNP (nSNP)/synonymous SNP (sSNP) ratio is 6.33, while the ratio for the entire data set is 1.08 (see Table S1 in the supplemental material). Of the six SNPs displaying homoplasy in Fig. 1, five were nSNPs for a ratio of 5.0 for this subset. Selective pressure (differentials, purifying, directional, etc.) in V. cholerae populations and in this outbreak deserves greater investigation.
TABLE 1 .
Different point mutations observed among the three sequenced isolates from the Haiti outbreak and the three most closely related isolates from Nepala
| Chromosome | Position | Nucleotide or amino acid in: |
||||||
|---|---|---|---|---|---|---|---|---|
| Reference strain | Haitian isolate |
Nepalese isolate |
||||||
| 1786 | 1792 | 1798 | 14 | 25 | 26 | |||
| I | 2787016 | C | C | C | C | T | T | T |
| Gly | Gly | Gly | Gly | Arg | Arg | Arg | ||
| I | 1090536 | T | T | T | T | T | T | G |
| Ile | Ile | Ile | Ile | Ile | Ile | Ser | ||
| II | 962762 | C | C | C | C | T | C | C |
| Ala | Ala | Ala | Ala | Ala | Ala | Ala | ||
DISCUSSION
Phylogenetic patterns indicate a close relationship between Haitian and Nepalese epidemic V. cholerae strains. Even with whole-genome sequencing, less than 100 SNPs were identified among these geographically disparate isolates; however, the few molecular characters that were available generated a robust and highly consistent phylogenetic topology with distinct subclade structure. The apparently identical Haitian genomes confirm the earlier findings that the Haitian outbreak originated from a single source (17). More importantly, one group that was well supported and had low diversity contained both Nepalese and Haitian isolates. In addition, the next two basal subclades were also Nepalese and more closely related to the Haitian outbreak strain than the Bangladeshi CIRS101 strain. Only a single SNP separates the Haitian and Nepalese isolates, providing strong evidence that the source of the Haitian epidemic was from this clonal group. This molecular phylogeny reinforces the previous epidemiological investigation (2) that pointed towards United Nations peacekeepers from Nepal as the source of the Haitian cholera epidemic. Given the implications of the epidemiological findings, it is imperative to use empirical laboratory data to support such findings. By using WGST to compare the entire genomes of available V. cholerae sequences, including the 24 added Nepalese strains, definitive basal/derived relationships have been established between and among these strains.
This study also showed that multiple clonal subclades were involved in the 2010 Nepalese outbreak, thus indicating that V. cholerae is prevalent in Nepal. Therefore, there is a general need for improved water hygiene and investment to reduce the occurrence of V. cholerae in Nepal.
Complete genomic analysis of pathogen populations is now a reality and is dramatically changing our approach to molecular epidemiology. With the cost and speed of new generation DNA sequencers improving exponentially, previously intractable problems can be resolved rapidly with modest expense. Outbreak pathogens will, almost by definition, have very little molecular diversity and may require comprehensive genomic analysis to differentiate and categorize isolates. In combination with evolutionary theory and advanced statistical methods, WGST represents the most powerful molecular approach imaginable and is setting a new standard for infectious disease epidemiology. While other descriptive and association-based epidemiological analyses (e.g., case control studies, geospatial analyses), along with limited-resolution molecular tools (e.g., PFGE), may leave room for interpretation on genetic linkage, WGST, as an empirical molecular epidemiological tool, does not (11, 13).
Infectious disease tracking requires global-scale information and cooperation. The current study was reliant upon genome analyses performed previously from other international studies. Future investigations will require high-quality genome databases that include representative isolates and metadata from geographically distributed samples, representing both historical and contemporary epidemics. Such databases will provide the contextual framework necessary to make definitive conclusions regarding infective sources and action plans for controlling epidemics. While we have precisely defined the Nepal-4 V. cholerae clade and the Haitian membership in it, its geographic distribution needs continued work. It is possible that this genetic group will be discovered in countries other than Nepal and Haiti. Attribution of outbreak sources based upon WGST alone requires comprehensive geographic strain collections. The current conclusion that Nepal is the source of the Haitian cholera outbreak can be reached only if both classical epidemiology and highly suggestive WGST are used together. Globally representative WGST databases will be available in the near future and increase our power to identify outbreak sources. It is now the charge of the world’s national health agencies and disease researchers to populate these databases with both sequences and rich metadata. Further, it must also be their mission to develop robust genomics and bioinformatics capabilities to rapidly generate and receive genomics-based data that can be turned into actionable public health knowledge.
Natural disasters such as the 2010 Haitian earthquake disrupt water and sanitation systems, adding to the vulnerability of affected populations. The United Nations, regional governments, and nongovernmental organizations respond rapidly to such disasters to bring aid and reduce suffering. The putative link between the Haitian and Nepalese cholera outbreaks underscores the speed at which infectious diseases can be transported globally and forces us to reconsider relief deployment strategies. In the current study, we used advanced molecular techniques to retrospectively characterize isolates from a devastating outbreak; in the future, we hope that rapid molecular diagnostics can be integrated into rapid screening programs for relief workers so their efforts will neither be delayed by ineffective diagnostics nor tainted by infectious diseases.
MATERIALS AND METHODS
Isolates.
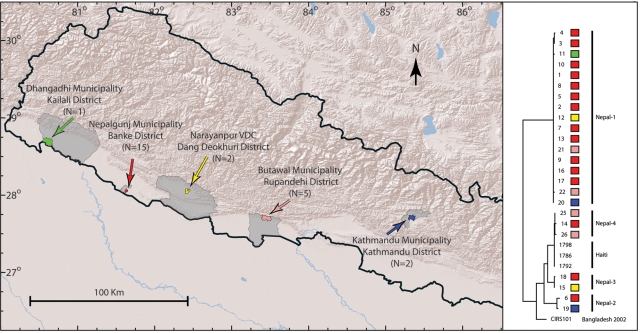
A total of 45 V. cholerae isolates were identified at the National Public Health Laboratory (NPHL), Kathmandu, Nepal, from Nepalese patients with diarrhea in 2010. Of these, 24 were available for analysis. The isolates were obtained from 30 July to 1 November 2010 and originated from five different districts in Nepal (Fig. 2; Table 2). All isolates with the exception of one from Kathmandu, Nepal, were obtained during the rainy season (June to August). Fifteen isolates, including the first laboratory confirmed case, were from a large outbreak in the municipality of Nepalgunj in Nepal that occurred in late July to mid-August. All isolates were identified as V. cholerae and serotyped at the NPHL. The isolates were shipped to the Technical University of Denmark (DTU) in February 2011.
FIG 2 .
Locations of the five districts in Nepal where the V. cholerae O1 Ogawa strains were isolated.
TABLE 2 .
Geographical, demographic, and clinical features of the laboratory-confirmed V. cholerae O1 Ogawa cases from Nepal during 2010
| Strain | Case ID | Location in Nepal | Collection date (day-mo-yr) | Gendera | Age (yr) |
|---|---|---|---|---|---|
| 1 | 31-OB NPHL | Banke district, Nepalgunj municipality | 30-7-2010 | M | |
| 2 | 32-OB NPHL | Banke district, Nepalgunj municipality | 30-7-2010 | M | |
| 10 | 44-OB NPHL | Banke district, Nepalgunj municipality, ward 12 | 1/8/2010 | F | |
| 4 | 36-OB NPHL | Banke district, Nepalgunj municipality, ward 4 | 1/8/2010 | M | |
| 6 | 38-OB NPHL | Banke district, Nepalgunj municipality, ward 4 | 1/8/2010 | F | |
| 9 | 41-OB NPHL | Banke district, Nepalgunj municipality, ward 4 | 1/8/2010 | F | |
| 3 | 35-OB NPHL | Banke district, Nepalgunj municipality, ward 5 | 1/8/2010 | M | |
| 5 | 37-OB NPHL | Banke district, Nepalgunj municipality, ward 5 | 1/8/2010 | F | |
| 8 | 40-OB NPHL | Banke district, Nepalgunj municipality, ward 5 | 1/8/2010 | M | |
| 14 | 49-OB NPHL | Banke district, Nepalgunj municipality, ward 5 | 1/8/2010 | M | |
| 7 | 39-OB NPHL | Banke district, Nepalgunj municipality, ward 6 | 1/8/2010 | F | |
| 13 | 47-OB NPHL | Banke district, Nepalgunj municipality, ward 8 | 1/8/2010 | F | |
| 12 | 46-OB NPHL | Dang Deokhuri district, Narayanpur VDC, ward 7 | 1/8/2010 | M | |
| 11 | 45-OB NPHL | Kailali district, Dhangadhi | 1/8/2010 | F | |
| 16 | 59-OB NPHL | Banke district, Nepalgunj municipality, ward 6 | 8/8/2010 | F | |
| 15 | 56-OB NPHL | Dang Deokhuri district, Narayanpur VDC, ward 5 | 8/8/2010 | F | |
| 19 | 508 | Kathmandu district, Kathmandu city | 15-8-2010 | F | 48 |
| 21 | LZH-11 | Rupandehi district, Butawal municipality | 30-8-2010 | M | 8 |
| 22 | LZH-21 | Rupandehi district, Butawal municipality | 30-8-2010 | F | 17 |
| 26 | LZH-23 | Rupandehi district, Butawal municipality | 30-8-2010 | F | 21 |
| 25 | LZH-24 | Rupandehi district, Butawal municipality | 30-8-2010 | M | |
| 17 | 65-OB NPHL | Banke district, Nepalgunj municipality | 31-8-2010 | M | |
| 18 | 66-OB NPHL | Banke district, Nepalgunj municipality | 31-8-2010 | M | |
| 20 | 526 | Kathmandu district, Kathmandu city | 1/11/2010 | M | 33 |
M, male; F, female.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility of the 24 V. cholerae isolates was determined utilizing MIC testing. The following antimicrobials were used: ampicillin, amoxicillin plus clavulanic acid, apramycin (veterinary approved aminoglycoside), cefotaxime, ceftiofur, chloramphenicol, ciprofloxacin, colistin, florfenicol, gentamicin, nalidixic acid, neomycin, spectinomycin, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim. Clinical and Laboratory Standards Institute guidelines and clinical breakpoints were utilized for the interpretation of the MIC values (18–20). Exceptions were made for interpretation of neomycin, where epidemiological cutoff values according to the EUCAST system were used (http://www.eucast.org/mic_distributions/). Due to the absence of interpretation guidelines, exceptions were made for the interpretation of apramycin and streptomycin which were interpreted according to research results from DTU. Quality control using Escherichia coli ATCC 25922 was conducted according to Clinical and Laboratory Standards Institute (CLSI) recommendations.
PFGE.
All of the V. cholerae isolates were analyzed for genetic relatedness by pulsed-field gel electrophoresis (PFGE) using the SfiI and NotI enzymes (Fermentas, Sankt Leon-Rot, Germany) according to the CDC PulseNet protocol (http://www.pulsenetinternational.org/protocols/Pages/default.aspx) (21). Electrophoresis was performed with a contour-clamped homogeneous electric field (CHEF) DR III System (Bio-Rad Laboratories, Hercules, CA) using 1% SeaKem gold agarose in 0.5× Tris-borate-EDTA. A two-block program was used consisting of block I with a pulse time of 2.0 to 10.0 s for 13 h and block II with a pulse time of 20.0 to 25.0 s for 6 h; the gels in both blocks were subjected to 6 V/cm on a 120° angle in 14°C TBE (Tris-borate-EDTA) buffer. A bundle file containing 14 pulsotypes of which 11 originated from the Haitian outbreak, including strain 201EL-1786, two from an early 1990s Latin American outbreak, and one from the U.S. Gulf Coast were sent to DTU by the United States CDC for comparison with the 24 isolates related to the Nepalese outbreak. The composite data set using both enzymes was evaluated by using Bionumerics software version 4.6 (Applied Maths, Sint-Martens-Latem, Belgium) where the average similarity of the experiments was used as settings for similarity, the enzymes were weighted equally, and unweighted-pair group method using average linkages (UPGMA) was used to generate a dendrogram.
Sequencing.
The DNA samples were prepared for multiplexed, paired-end sequencing on the Illumina GAIIx genome analyzer (Illumina, Inc., San Diego, CA). For each isolate, 1 to 5 µg of double-stranded DNA (dsDNA) in 200 µl was sheared in a 96-well plate with SonicMan (catalog no. SCM1000-3; Matrical BioScience, Spokane, WA) to a size range of 200 to 1,000 bp with the majority of material at ca. 600 bp using the following parameters: prechill at 0°C for 75 s, 20 cycles, sonication for 10 s, 100% power, lid chill at 0°C for 75 s, plate chill at 0°C for 10 s, and postchill at 0°C for 75 s. The sheared DNA was purified using the QIAquick PCR purification kit (catalog no. 28106; Qiagen, Valencia, CA). The enzymatic processing (end repair, phosphorylation, A-tailing, and adaptor ligation) of the DNA was done following the guidelines in the Illumina protocol (22). The enzymes for processing were obtained from New England Biolabs (catalog no. E6000L; New England Biolabs, Ipswich, MA), and the oligonucleotides and adaptors were obtained from Illumina (catalog no. PE-400-1001). After ligation of the adaptors, the DNA was run on a 2% agarose gel for 2 h, after which a gel slice containing 500- to 600-bp fragments of each DNA sample was isolated and purified using the QIAquick gel extraction kit (catalog no. 28706; Qiagen, Valencia, CA). Individual libraries were quantified with quantitative PCR (qPCR) on the ABI 7900HT (catalog no. 4329001; Life Technologies Corporation, Carlsbad, CA) in triplicate at two dilutions, 1:1,000 and 1:2,000, using the Kapa library quantification kit (catalog no. KK4832 or KK4835; Kapa Biosystems, Woburn, MA). Based on the individual library concentrations, equimolar pools of no more than 12 indexed V. cholerae libraries were prepared at a concentration of at least 1 nM using 10 mM Tris-HCl (pH 8.0) plus 0.05% Tween 20 as the diluent. To ensure accurate loading onto the flow cell, the same quantification method was used to quantify the final pools. The pooled, paired-end libraries were sequenced on the Illumina GAIIx to a read length of at least 76 base pairs. The average genome coverage for these 24 isolates was greater than 100× with a minimum of 75×. Over 97.6 of the genomes were at 10× cover or better. The Illumina genome sequencing data were deposited in the Short Read Archive at the National Center for Biotechnology Information (NCBI) under the accession no. SRA039806.1. The three Haitian genome sequences generated by the CDC were obtained from NCBI under the following accession numbers: strain 1786, SRX031665 (Illumina) and SRX031636 (454); strain 1792, SRX032204 (Illumina) and SRX032203 (454); and strain 1798, SRX032202 (Illumina) and SRX032201 (454).
Alignment.
Illumina WGS data sets were aligned against chromosomes I and II of the Vibrio cholerae O1 biovar El Tor strain N16961 (NC002505 and NC002506) using the short-read alignment component of the BWA alignment tool (23). 454 data for the publicly available Haitian genomes was aligned with BWA-SW (23). Where appropriate, isolates that were sequenced by both 454 and Illumina platforms were merged with Picard tools after the alignments were completed (http://picard.sourceforge.net). Reads containing insertions or deletions and those mapping to multiple locations in the reference were removed from the final alignments.
Identification of single-nucleotide polymorphism.
Each alignment was analyzed for SNPs using SolSNP (http://sourceforge.net/projects/solsnp/). SNPs were excluded if they did not meet a minimum coverage of 10× and if the variant was present in less than 90% of the base calls for that position. In parallel, publicly available genomes were aligned against both chromosomes of N16961 using MUMmer 3.22 (24). SNPs were extracted from the alignments using a custom script. Subsequently, regions found to be duplicated in the N16961 reference genome were identified using MUMmer version 3.22. SNPs residing within these repetitive regions were then removed. Loci that lacked reference sequence coverage data for one or more isolates were removed from the final analysis. This left us with a matrix of orthologous SNP loci shared across all genomes.
Phylogenetic analysis.
Phylogenetic reconstruction was performed using parsimony criteria and a heuristic search in PAUP 4.0 (25); 1,000 generations were run for bootstrap analysis. Reference genome mapping and read depth statistics were determined using the Genome Analysis Toolkit (26) and Lasergene’s SeqMan NGEN version 2.2 software (Lasergene, Madison, WI).
SUPPLEMENTAL MATERIAL
Additional phylogenetic analysis. Download Text S1, DOCX file, 0.01 MB.
Phylogenetic trees based on the results of maximum likelihood analysis (A) and Bayesian phylogenetic analysis (B). Download Figure S1, DOC file, 0.1 MB.
The phylogenetic tree from Fig. 1 annotated with branch names that correspond to particular SNPs in Table S1. Download Figure S2, PDF file, 0.1 MB.
Excel spreadsheet with all the SNP data represented in Fig. 1.
ACKNOWLEDGMENTS
We thank Christina AabySvendsen for technical assistance determining MICs and performing PFGE and Peter Gerner-Smidt for sharing the PFGE bundle file of the Haitian strains.
This study was supported by the Center for Genomic Epidemiology (09-067103/DSF) and the WHO Global Foodborne Infections Network (WHO GFN) (http://www.who.int/gfn/en/).
Footnotes
Citation Hendriksen RS, et al. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: Evidence on the Origin of the Haitian Outbreak. mBio 2(4):e00157-11. doi:10.1128/mBio.00157-11.
REFERENCES
- 1. Pollitzer R. 1959. Cholera. World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Piarroux R, et al. 2011. Understanding the cholera epidemic, Haiti. Emerg. Infect. Dis. 17:1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pan American Health Organization Monday, July 25, 2011,. Cholera and post-earth-quake response in Haiti. Health Cluster Bulletin 26 http://new.paho.org/blogs/haiti/?p=2043 [Google Scholar]
- 4. Centers for Disease Control and Prevention 2010. Update on cholera—Haiti, Dominican Republic, and Florida. MMWR Morb. Mortal. Wkly. Rep. 59:1637–1641 [PubMed] [Google Scholar]
- 5. Butler D. 2010. Cholera tightens grip on Haiti. Nature 468:2 [DOI] [PubMed] [Google Scholar]
- 6. Cyranoski D. 2011. Cholera vaccine plan splits experts. Nature 469:2. [DOI] [PubMed] [Google Scholar]
- 7. Enserink M. 2011. Epidemiology. Despite sensitivities, scientists seek to solve Haiti’s cholera riddle. Science 331:388–389 [DOI] [PubMed] [Google Scholar]
- 8. Enserink M. 2010. Infectious diseases. Haiti’s outbreak is latest in cholera’s new global assault. Science 330:738–739 [DOI] [PubMed] [Google Scholar]
- 9. Chun J, et al. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 106:15442–15447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grim CJ, et al. 2010. Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J. Bacteriol. 192:3524–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Engelthaler DM, et al. 2011. Next-generation sequencing of Coccidioides immitis isolated during cluster investigation. Emerg. Infect. Dis. 17:227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gardy JL, et al. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364:730–739 [DOI] [PubMed] [Google Scholar]
- 13. Harris SR, et al. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rasko DA, et al. 2011. Bacillus anthracis comparative genome analysis in support of the Amerithrax investigation. Proc. Natl. Acad. Sci. U. S. A. 108:5027–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chin CS, et al. 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam C, Octavia S, Reeves P, Wang L, Lan R. 2010. Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America. Emerg. Infect. Dis. 16:1130–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ali A, et al. 2011. Recent clonal origin of cholera in Haiti. Emerg. Infect. Dis. 17:699–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clinical and Laboratory Standards Institute 2006. M7-A7. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 19. Clinical and Laboratory Standards Institute 2008. M31-A3. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 20. Clinical and Laboratory Standards Institute 2010. M100-S20. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 21. Cooper KL, et al. 2006. Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog. Dis. 3:51–58 [DOI] [PubMed] [Google Scholar]
- 22. Illumina, Inc 2008. Preparing samples for multiplexed paired-end sequencing Part 1005361 Rev. B Illumina, Inc., San Diego, CA: . [Google Scholar]
- 23. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swofford DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4. Sinauer Associates, Sunderland, MA: [Google Scholar]
- 26. McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional phylogenetic analysis. Download Text S1, DOCX file, 0.01 MB.
Phylogenetic trees based on the results of maximum likelihood analysis (A) and Bayesian phylogenetic analysis (B). Download Figure S1, DOC file, 0.1 MB.
The phylogenetic tree from Fig. 1 annotated with branch names that correspond to particular SNPs in Table S1. Download Figure S2, PDF file, 0.1 MB.
Excel spreadsheet with all the SNP data represented in Fig. 1.