ABSTRACT
Novel pandemic influenza viruses enter the human population with some regularity and can cause disease that is severe and widespread. The emergence of novel viruses, historically, has often been coupled with the disappearance of existing seasonal virus strains. Here, we propose that the elimination of seasonal strains during virus pandemics is a process mediated, at the population level, by humoral immunity. Specifically, we suggest that infection with a novel virus strain, in people previously exposed to influenza viruses, can elicit a memory B cell response against conserved hemagglutinin stalk epitopes and/or neuraminidase epitopes. The anti-stalk and/or anti-neuraminidase antibodies then act to diminish the clinical severity of disease caused by novel influenza viruses and to eliminate seasonal virus strains.
EMERGENCE AND EXTINCTION OF INFLUENZA VIRUS SUBTYPES IN THE HUMAN POPULATION SINCE 1918
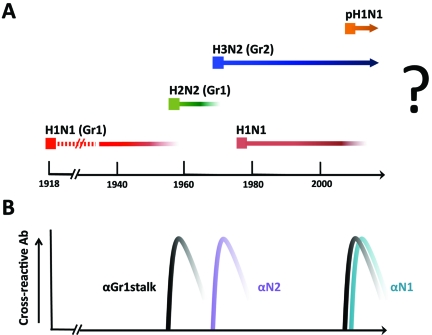
Historically, the emergence of novel influenza virus subtypes in the human population has been associated with worldwide epidemics (or pandemics). In recent history, the most devastating example of a new subtype emerging in the human population was the 1918 influenza virus, which expressed a subtype 1 hemagglutinin protein (H1) and a subtype 1 neuraminidase protein (N1). This H1N1 virus is estimated to have been responsible for 50 to 100 million deaths over a very short period of time. H1N1 variants then circulated for 39 years before being replaced by an H2N2 virus (H2 subtype and N2 subtype) in 1957. The H2N2 virus was prevalent for only 11 years until 1968, when it was replaced by an H3N2 virus (H3 subtype with retained N2 subtype). Curiously, in 1977, an H1N1 virus, which was actually the 1950 strain, reappeared and stayed on in parallel with the H3N2 seasonal virus until 2009. In April 2009, a novel pandemic H1N1 virus emerged in Mexico and proceeded to spread around the world. During the subsequent 2009-2010 and 2010-2011 winter seasons, most of the seasonal H1N1 viruses seem to have been replaced by this pandemic H1N1 strain (Fig. 1A) (1).
FIG 1 .
Influenza A viruses circulating in the human population and induction of cross-protective antibodies by pandemic viruses. (A) H1N1 indicates virus with hemagglutinin subtype 1 and neuraminidase subtype 1. H2N2 and H3N2 indicate viruses with hemagglutinin subtype 2 and neuraminidase subtype 2 and hemagglutinin subtype 3 and neuraminidase subtype 2, respectively. pH1N1 indicates the novel swine origin virus first isolated in 2009. (B) Antibody response in the human population, which we propose to have contributed to the elimination of existing seasonal influenza virus strains. Gr1, group 1 subtype; Gr2, group 2 subtype.
WHAT CAUSES THE EMERGENCE OF NOVEL INFLUENZA A VIRUS SUBTYPES?
Besides environmental climate, the following two independent elements appear to determine the ability of a new virus strain to take hold in the population: (i) factors present in the specific virus that enable transmission between humans and robust replication in human tissues and (ii) the immune status of the current human population.
In terms of the generation of novel virus strains, it is likely that all pandemic viruses (including the 1918 virus) result from a reassortment event following the coinfection of a host with two or more different influenza viruses. The genome of each influenza virus is made up of eight RNA segments, and during coinfection of a single cell, the parental virus segments can mix, causing the generation of new virus strains which may express novel combinations of hemagglutinin and neuraminidase subtypes. It is a complex stochastic event that results in the emergence of a successful virus strain from all of the 254 possible gene combinations that can occur during reassortment of any two parent viruses. The production of a new human strain by reassortment is also limited by the host species in which the mixing event occurs. Thus, the emergence of a reassortant virus that can cause pandemic human disease is a rare event, and the specific properties of such a virus are difficult to predict.
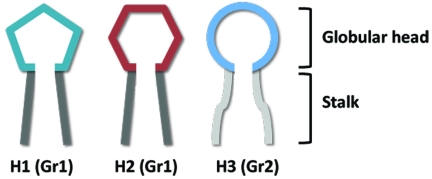
Seasonal influenza virus strains are continually changing in response to the existing herd immunity in the human population. This phenomenon, called antigenic drift, results in structural changes within the globular head of the hemagglutinin protein, while the hemagglutinin stalks are largely conserved within each of the following two phylogenetic groups: the group 1 subtype (e.g., H1 and H2) and the group 2 subtype (e.g., H3) (Fig. 2). The immune status of any population against influenza viruses is largely defined by the presence or absence of neutralizing antibodies. Two basic types of neutralizing antibodies have been described: the highly potent, virus-specific globular head antibodies and the less potent, cross-reactive anti-stalk antibodies. The broadly neutralizing stalk-specific antibodies have been only recently described, and it is not clear what role they play in the protection of humans from influenza viruses. We propose that broadly neutralizing anti-stalk antibodies serve to diminish the clinical severity of influenza disease and, importantly, that they can act in the elimination of seasonal virus strains during influenza pandemics. We further propose that antibodies against a second surface glycoprotein, viral neuraminidase, can also lessen the burden of influenza disease and act in the elimination of old seasonal strains. Neuraminidase-specific antibodies do not prevent virus infection; however, studies have demonstrated that neuraminidase-specific antibodies are generated during virus infection and that these specificities can mediate protection against disease (2–4).
FIG 2 .
Representation of the major antigenic differences between hemagglutinin subtypes. Group 1 subtypes, such as H1 and H2, share a conserved stalk domain (dark gray); group 2 subtypes, such as H3, have a stalk domain that is structurally different (light gray) from the group 1 stalk. The globular heads of the different hemagglutinin subtypes are structurally distinct (green/red/blue).
When a new pandemic strain enters the human population, it predominantly induces an immune response against the hemagglutinin protein, but the viral neuraminidase is also an immunogenic protein. In each year following the disease pandemic, novel escape mutations occur (mostly in the hemagglutinin) in response to the existing immunity present in the human population; many of these mutations get fixed in the viral genome and are likely to cause reduced viral fitness. This may be reflected by the increased mortality rate during pandemic influenza seasons in the largely naive age group of children under the age of 4 years. This age group is less affected by herd immunity; thus, differential disease rates between pandemic and typical seasons likely reflect true differences in the virulence of pandemic versus seasonal virus strains (5–8).
WHAT MAY HAVE CAUSED THE DYING OUT OF THE H1N1 SUBTYPE STRAIN IN 1957?
In 1957, the H2N2 virus emerged, which had a new hemagglutinin subtype (H2 versus H1). Although infection with an H2 virus would have induced a distinct set of antibody specificities, we hypothesize that people who were previously infected with H1N1 viruses would also have generated a memory response specific for conserved group 1 stalk epitopes (9, 10). While exposure to the antigenic head of the H2 hemagglutinin would have caused an immunological priming event, exposure to the group 1 stalk would have lead to a high-titer, class-switched, affinity-matured memory response (Fig. 2). Members of the population who had experienced multiple seasonal exposures to previously circulating H1 viruses would have had memory cells capable of responding to conserved group 1 stalk epitopes present in the 1957 pandemic virus; thus, this older population was largely protected (or may have experienced subclinical infection) in 1957—it was the people aged 20 years and younger who were seriously affected, with disease rates of over 50% (11, 12). With the novel 1957 virus causing a sudden boost in herd immunity against group 1 viruses, and because it was spreading within the susceptible population at rates much higher than the existing seasonal H1 strain, the older H1 virus was quickly extinguished. Thus, we suggest that the induction of cross-neutralizing antibodies directed against the stalk of the H1 hemagglutinin following infection with the related group 1 virus (H2N2) played a significant role in the protection of older segments of the population from disease in 1957 and in the elimination of the existing seasonal H1N1 virus (Fig. 1A and B).
HOW DID THE H3N2 VIRUS SUCCESSFULLY ELIMINATE THE H2N2 VIRUS IN 1968?
The H3N2 virus expressed a group 2 hemagglutinin, so it was less likely to have induced cross-reactive anti-hemagglutinin antibodies that mediated the elimination of the existing H2N2 virus. However, in this case, the neuraminidase between the H2N2 and the H3N2 viruses was the same. It has been suggested that the low rate of disease in older segments of the population during the 1968 pandemic was a result of anti-N2 antibodies present due to prior exposure to H2N2 virus strains (11). We postulate that the sudden generation of herd immunity against the N2 protein in the younger population through infection with the H3N2 virus along with the existing immunity in the older segments of the population caused the elimination of the less-fit H2N2 seasonal virus strain. The ability of neuraminidase-specific antibodies to limit virus replication and to mediate protection in animal models against heterosubtypic virus strains that share the same neuraminidase subtype has been established (2–4, 13–15). Thus, in 1968, an anamnestic immune response against N2 likely decreased disease rates in the older population, but we postulate that anti-N2 antibodies also had a dampening effect on the seasonal H2N2 virus, resulting in its extinction.
WHY DID INFECTION WITH AN H1N1 VIRUS IN 1977 RESULT IN COEXISTENCE WITH H3N2 VIRUSES?
In 1977, a 1950-like H1N1 strain took hold in the population. We hypothesize that the H1N1 virus stayed on because both the hemagglutinin and the neuraminidase of the new strain were very different from those of the H3N2 virus and because a substantial portion of the population had not been exposed to antigenically similar viruses. The hemagglutinins of the two strains belong to different phylogenetic groups, and preexisting immunity against the 1977 strain did not exist in most of the (younger) population.
WHY DID PANDEMIC H1N1 REPLACE SEASONAL H1N1?
A new pandemic H1N1 strain (pH1N1) appeared in 2009. It contained the hemagglutinin and the neuraminidase from influenza viruses circulating in the pig population. The antigenic character of the hemagglutinin was close to those of early H1N1 viruses but was very different from those of the circulating seasonal H1N1 viruses. Some of the population may have been protected by existing cross-neutralizing antibodies, particularly older individuals who were exposed to early strains similar to the pH1N1 virus (16). Recent data suggest that broadly neutralizing antibodies may have significantly diminished disease severity while not protecting against infection with the 2009 pandemic virus (17). Although the H1N1 hemagglutinins were sufficiently different to allow the seeding of the new pH1 strain within susceptible populations, the pH1 stalk region (which is highly conserved in the two H1N1 strains) induced a sudden large-scale cross-protective immune response (17) that likely contributed to the elimination of the seasonal H1N1 virus over the 2009-2010 and 2010-2011 seasons. Antibodies against the highly similar neuraminidase proteins in the seasonal and pandemic H1N1 strains may have further reduced the spread of the less virulent seasonal H1N1 strain.
WHAT CAN WE PREDICT FOR THE FUTURE?
Based on the hypothesis that related viruses induce cross-protective immunity via their conserved hemagglutinin stalks and/or their conserved neuraminidases, it is likely that pH1N1 will completely displace the seasonal H1N1 viruses. Thus, the pH1N1, H3N2, and B viruses will continue to circulate until a new pandemic influenza virus strain emerges. If a new pandemic strain induces cross-reactive stalk antibodies, it will likely eliminate one or both of the circulating A strains—the emergence of a pandemic H2N2 virus strain would likely result in the elimination of both seasonal strains due to a sudden boost in titer, at a population level, of both anti-group 1 hemagglutinin and anti-N2 protective antibodies. If, however, the new pandemic strain is sufficiently different in both its hemagglutinin and neuraminidase, it may actually cocirculate with the present strains without causing the H3N2 or pH1N1 strains to die out. Which particular hemagglutinin subtype such a new pandemic strain will possess and when such an event will occur cannot be predicted.
In summary, the present discussion suggests that the induction of a large-scale humoral immune response against conserved hemagglutinin stalk epitopes and/or against the neuraminidase protein results in the clearance of old seasonal influenza virus strains. If the newly introduced pandemic strain has a very divergent hemagglutinin (belonging to a different hemagglutinin group) and does not share the neuraminidase subtype, it is likely to coexist with the prepandemic seasonal influenza virus strain.
ACKNOWLEDGMENTS
We thank Rafi Ahmed and Patrick Wilson for sharing unpublished results and for important discussions.
We also thank Diane Post for her unwavering support of our CRIP/CEIRS contract HHSN266200700010C. Partial support was also provided by NIH grants U54 AI057158-06, U01 AI070469-02, AI058113-06, and 1RC1 AI086061-01. T.T.W. was supported by the Mount Sinai Medical Scientists training grant T32 GM007280.
Footnotes
Citation Palese P, Wang TT. 2011. Why do influenza virus subtypes die out? A hypothesis. mBio 2(5):e00150-11. doi:10.1128/mBio.00150-11.
REFERENCES
- 1. Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p. 1647–1689 In Knipe DM. et al. (ed), Fields virology, vol. 2, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Chen Z, et al. 2000. Cross-protection against a lethal influenza virus infection by DNA vaccine to neuraminidase. Vaccine 18:3214–3222 [DOI] [PubMed] [Google Scholar]
- 3. Sandbulte MR, et al. 2007. Cross-reactive neuraminidase antibodies afford partial protection against H5N1 in mice and are present in unexposed humans. PLoS Med. 4:e59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kilbourne ED, et al. 2004. Protection of mice with recombinant influenza virus neuraminidase. J. Infect. Dis. 189:459–461 [DOI] [PubMed] [Google Scholar]
- 5. Mullooly JP, Barker WH. 1982. Impact of type A influenza on children: a retrospective study. Am. J. Public Health 72:1008–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glezen WP. 1996. Emerging infections: pandemic influenza. Epidemiol. Rev. 18:64–76 [DOI] [PubMed] [Google Scholar]
- 7. Bodewes R, et al. 2011. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin. Vaccine Immunol. 18:469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karageorgopoulos DE, Vouloumanou EK, Korbila IP, Kapaskelis A, Falagas ME. 2011. Age distribution of cases of 2009 (H1N1) pandemic influenza in comparison with seasonal influenza. PLoS One 6:e21690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ekiert DC, et al. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sui J, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cox NJ, Subbarao K. 2000. Global epidemiology of influenza: past and present. Annu. Rev. Med. 51:407–421 [DOI] [PubMed] [Google Scholar]
- 12. Epstein SL. 2006. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J. Infect. Dis. 193:49–53 [DOI] [PubMed] [Google Scholar]
- 13. Van Reeth K, et al. 2009. Prior infection with an H1N1 swine influenza virus partially protects pigs against a low pathogenic H5N1 avian influenza virus. Vaccine 27:6330–6339 [DOI] [PubMed] [Google Scholar]
- 14. Kilbourne ED, Christenson WN, Sande M. 1968. Antibody response in man to influenza virus neuraminidase following influenza. J. Virol. 2:761–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murphy BR, Kasel JA, Chanock RM. 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N. Engl. J. Med. 286:1329–1332 [DOI] [PubMed] [Google Scholar]
- 16. Xu R, et al. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328:357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wrammert J, et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208:181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]