Abstract
MicroRNAs (miRNAs) influence many biological processes, including cancer. They do so by posttranscriptionally repressing target mRNAs to which they have sequence complementarity. Although it has been postulated that miRNAs can regulate other miRNAs, this has never been shown experimentally to our knowledge. Here, we demonstrate that miR-107 negatively regulates the tumor suppressor miRNA let-7 via a direct interaction. miR-107 was found to be highly expressed in malignant tissue from patients with advanced breast cancer, and its expression was inversely correlated with let-7 expression in tumors and in cancer cell lines. Ectopic expression of miR-107 in human cancer cell lines led to destabilization of mature let-7, increased expression of let-7 targets, and increased malignant phenotypes. In contrast, depletion of endogenous miR-107 dramatically increased the stability of mature let-7 and led to downregulation of let-7 targets. Accordingly, miR-107 expression increased the tumorigenic and metastatic potential of a human breast cancer cell line in mice via inhibition of let-7 and upregulation of let-7 targets. By mutating individual sites within miR-107 and let-7, we found that miR-107 directly interacts with let-7 and that the internal loop of the let-7/miR-107 duplex is critical for repression of let-7 expression. Altogether, we have identified an oncogenic role for miR-107 and provide evidence of a transregulational interaction among miRNAs in human cancer development.
Introduction
MicroRNAs (miRNAs) are involved in diverse biological processes including embryonic development, metabolism, viral infections, and human malignancies. These small noncoding RNAs belong to a novel class of gene regulators and control gene expression by binding to complementary sequences in the 3′ UTRs of target mRNAs (1, 2). let-7, the first identified tumor-suppressing miRNA, plays an important role in cancer pathogenesis as a regulator of Ras and high mobility group AT-hook 2 (HMGA2) expression (3–5). Regulation of miRNAs occurs at both the transcriptional and posttranscriptional levels. For example, transcription of the let-7 gene is directly suppressed by c-Myc, a nuclear protein that regulates cell proliferation and differentiation (6). As cells undergo differentiation, the pluripotency factor Lin28 posttranscriptionally inhibits the biogenesis of let-7 via binding to its primary and precursor transcripts (7). In general, miRNAs inhibit translation of target mRNAs via base-pairing interactions with their 3′ UTRs (1). Interestingly, in Drosophila it has been postulated that miRNAs may target other miRNAs as well (8). This possibility prompted us to hypothesize that miRNAs may regulate non-mRNA targets, including other miRNAs.
In this study, we show that, as an independent predictor of clinical outcome in human breast cancer, miR-107 acts as an endogenous suppressor of let-7, which plays a major role in determining metastatic progression. Furthermore, we provide what we believe is a novel mechanistic explanation of the interaction among miRNAs. This evidence sheds light on the current understanding of miRNA regulation and the function of miRNA during tumor progression.
Results
miR-107 suppresses let-7 function.
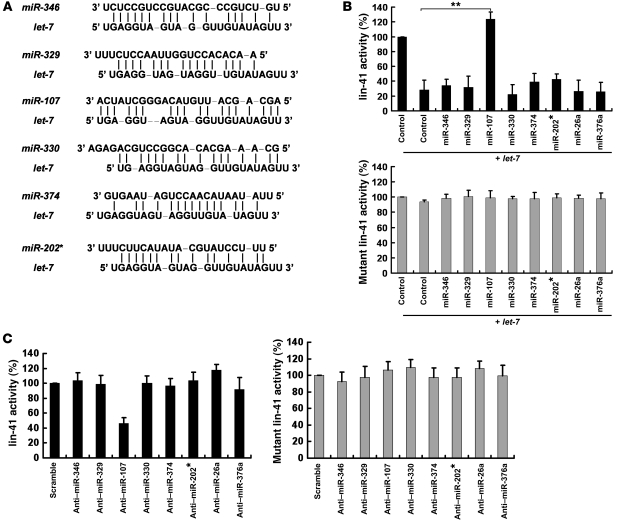
We employed RNAhybrid and miRanda algorithms (9, 10) to search for miRNAs that potentially interacted with let-7 members (Figure 1A; let-7a as a representative member of the let-7 family). To evaluate whether the identified miRNAs could interfere with let-7’s function in regulation of gene expression, we utilized the lin-41 reporter construct containing 2 let-7 binding sites in its 3′ UTR. When overexpressing these miRNAs (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI45390DS1), the let-7–mediated inhibition of lin-41 expression was abolished by miR-107 but not by the other candidate miRNAs or nonspecific miRNAs; this finding was statistically significant (Figure 1B). Moreover, expression of miR-107 had no effect on the inhibition of lin-41 when the let-7–binding sites were mutated (Figure 1B). In addition, to confirm this effect under endogenous conditions, we depleted miR-107 from MDA-MB-231 cells and observed that the depletion of miR-107 enhanced the repression of lin-41 activity by let-7, indicating that endogenous miR-107 antagonized endogenous let-7 (Figure 1C).
Figure 1. miR-107 suppresses let-7 function.
(A) Potential duplexes formed between let-7a and candidate miRNAs. (B) A549 cells were cotransfected with lin-41 and the indicated antagomirs. Firefly luciferase reporter activity was normalized to renilla luciferase. MiR-26a and miR-376a were included as nonspecific miRNAs. Control cells in lane 1 carried pcDNA6.2-EmGFP empty vectors and were transfected with the lin-41 reporter. (C) Depletion of endogenous miR-107 results in enhanced let-7–suppressed lin-41 expression. MDA231 cells were cotransfected with lin-41 and the miRNA antagomirs indicated. Firefly luciferase reporter activity was normalized to renilla luciferase. MiR-26a and miR-376a were included as nonspecific miRNAs. Scramble oligo-transfected cells were used as the control group. *P < 0.05; **P < 0.01. Data are presented as mean ± SD.
miR-107 directly interacts with let-7.
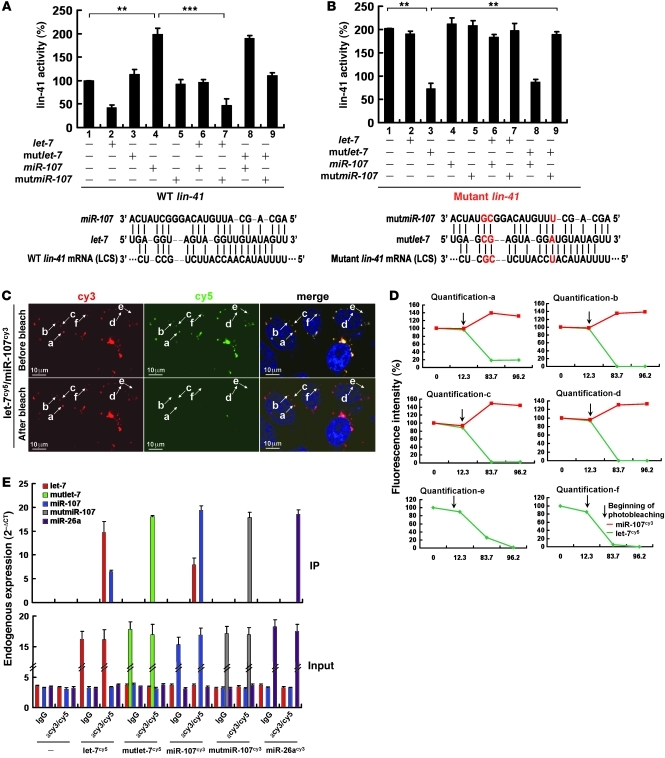
To determine whether miR-107 regulated lin-41 in a let-7–dependent manner, we generated compensatory mutations in the 3′ UTR of lin-41 (mutlin-41), in let-7 (mutlet-7), and in miR-107 (mutmiR-107) and evaluated their regulation using a lin-41 luciferase reporter assay. The mutant forms were designed to maintain the hypothetical base pairings between let-7 and lin-41 and between miR-107 and let-7 (Figure 2A). As anticipated, wild-type, but not mutant, miR-107 restored lin-41 reporter expression (Figure 2B). However, only mutlet-7, but not wild-type let-7, could suppress mutant lin-41 expression, and this repressive effect was abolished by expression of mutmiR-107 (Figure 2B), which emphasized that miR-107 modulated let-7 activity via base-pairing interactions. Taken together, these results indicate that miR-107 interacted with let-7 and suppressed let-7–mediated gene regulation.
Figure 2. Direct interaction between miR-107 and let-7.
(A) Design of constructs containing wild-type (A) and mutant (B) miR-107, let-7, and lin-41. The 3′ UTR of lin-41 was appended to the luciferase open reading frame (Luc). The let-7 complementary sites are indicated. miR-107, let-7, lin-41, and their mutants were transfected into H1299 cells and assayed for luciferase activity. **P < 0.01; ***P < 0.001. (C) In vivo cellular localization of miR-107 and let-7a in A549 cells after photobleaching. Cells with colocalization of miR-107/let-7a observed by confocal microscopy (Supplemental Figure 2B) were bleached repeatedly by λ= 633 nm, and the fluorescence intensity of cy3-labeled miR-107 was subsequently monitored at λ= 575–615 nm before and after photobleaching. (D) The FRET efficiency in each location shown in C. a–d: colocalized complexes; e, f: complexes without colocalization. (E) The interaction between miR-107 and let-7a as assessed by immunoprecipitation. A549 cells expressing cy3–miR-107 and cy5–let-7a were subjected to immunoprecipitation using anti-cy3/cy5 antibodies and then assayed by qRT-PCR. Data are presented as mean ± SD.
To monitor the miR-107–let-7 interaction in vivo, we synthesized duplex precursors that had 5′ phosphorylation and a cy3 or cy5 3′ modification on their functional strands. cy3-labeled miR-107 or nonspecific miR-26a was coexpressed with cy5-labeled let-7 mRNA in A549 cells. Immunoprecipitation with anti-cy3/cy5 antibodies was performed to determine whether these miRNAs could recruit Ago1, a key component of the RNA-induced silencing complex. We found that Ago1 was present in all immunoprecipitates (Supplemental Figure 2A). By using confocal microscopy, we found that 70% of miR-107 colocalized with let-7 in the cytoplasm (Supplemental Figure 2B). However, mutmiR-107 did not colocalize with let-7, but mutations in let-7 (mutlet-7) restored its interaction with mutmiR-107 (Supplemental Figure 2B).
To further confirm the interaction between let-7 and miR-107 in vivo, we used an acceptor photobleaching method to evaluate fluorescence resonance energy transfer (FRET) between the cy3-labeled miR-107 and cy5-labeled let-7. After repeated photobleaching, real-time quantitative analysis showed an inverse correlation between the fluorescence intensity of let-7 and miR-107 in miR-107–let-7 colocalized foci (Figure 2, C and D), indicating that the direct interaction between miR-107 and let-7 enabled energy transfer between their respective fluorophores. Similar results were also found in mutlet-7–mutmiR-107 foci, as compensatory mutations restored their interaction (Supplemental Figure 3). We further immunoprecipitated miR-107 with an anti-cy3/cy5 antibody and found that miR-107 immunoprecipitant contained a significant amount of let-7; we also observed the presence of miR-107 within the let-7 immunoprecipitant (Figure 2E). We did not detect let-7 in the miR-26a or mutmiR-107 fractions and did not find miR-26a in the let-7 immunoprecipitant (Figure 2E). These observations support the direct interaction between miR-107 and let-7.
miR-107 promotes let-7 degradation and antagonizes let-7–suppressed AIG in human cancer cell lines.
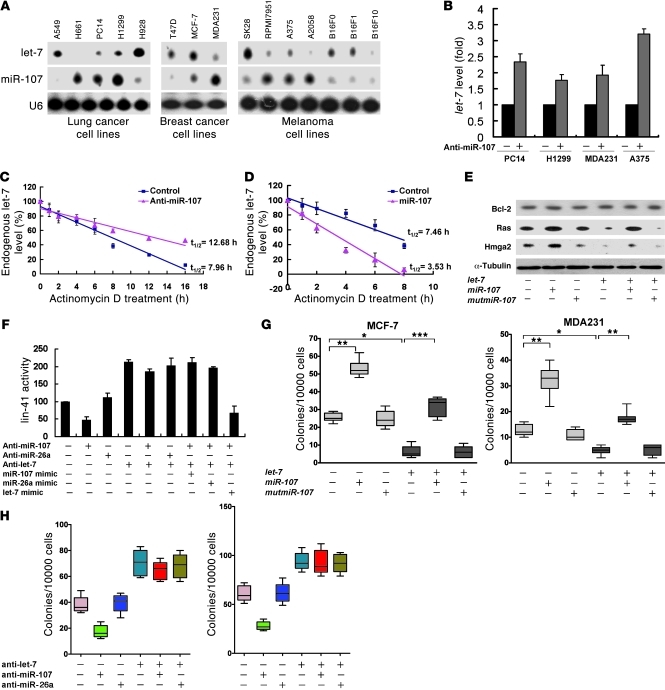
When the expression levels of let-7a and miR-107 in human lung, breast, and melanoma cancer cell lines were examined, we noticed an inverse expressing pattern between these 2 miRNAs. The let-7a–overexpressing cell lines (A549, H928, T47D, MCF-7, SK28, B16F0, and B16F1 cells) exhibited low levels of miR-107 (Figure 3A), whereas the miR-107–overexpressing cell lines (H661, PC14, H1299, MDA231, RPMI7951, A375, and A2068 cells) exhibited low, if any, expression of let-7a (Figure 3A). We also found similar phenomenon in public data sets from the NCI-60 cancer cell panel (Supplemental Figure 4; data set from EMBL EBI: ID No. E-MTAB-327), in which miR-107 was inversely correlated with let-7 family members such as let-7a,b,c,f,g,i, and miR-98. These results (Figure 3A and Supplemental Figure 4) confirmed the inversed correlations in multiple types of cancer cell lines. Furthermore, when clustering the expression data by tumor type, we found stronger negative correlations in some cancer types, such as CNS, breast, and kidney (Supplemental Figure 4), suggesting that the reciprocal regulation between miR-107 and let-7 may be tumor-type specific. In addition, miR-107 depletion via antagomir significantly increased the level of let-7a in different cancer cell lines (Figure 3B). However, depletion of miR-107 had no effect on the expression of the let-7 primary and precursor transcripts (pri–let-7 and pre–let-7) (Supplemental Figure 5).
Figure 3. miR-107 promotes let-7 degradation and antagonizes let-7–suppressed AIG in human cancer cell lines.
(A) Expression of miR-107 and let-7 in human cancer cell lines. Total RNA (20 μg) was isolated and assayed by Northern blotting with specific probes for let-7a and miR-107. (B) Effect of miR-107 on let-7a expression. Total RNA was isolated from cells transfected with miR-107 antagomir and then assayed by qRT-PCR. U6 was used as an internal control, and the fold change of the let-7a level was normalized with the paired untreated group. (C) An miR-107 antagomir was introduced into H1299 cells in the presence of 1 μg/ml of actinomycin D. Total RNA was isolated and assayed by qRT-PCR for mature let-7a. (D) miR-107 was introduced into A549 cells in the presence of 1 μg/ml of actinomycin D. Total RNA was isolated and mature let-7a was assayed for by qRT-PCR. (E) Forced expression of miR-107 induced the expression of both Ras and HMGA2. H1299 cells were transfected with indicated miRNAs, and Ras and HMGA2 levels were assayed by Western blotting. (F) Dual-luciferase assays were performed to determine the lin-41 expression in the presence of miR-107, let-7a mimics, or antagomirs. (G and H) MiRNA mimics or antagomirs were introduced into MCF-7 or MDA231 cells and effects of miR-107 on colony formation were determined. The indicated miRNAs or antagomirs were transfected into MCF-7 (G, H), MDA-MB-231 (G), and H1299 (H) cells. Soft agar assays were performed over 14 days. *P < 0.05; **P < 0.01; ***P < 0.001. Data are presented as mean ± SD.
These results prompted us to examine whether targeting let-7 by miR-107 could result in let-7a degradation. We found that depletion of endogenous miR-107 slowed let-7 degradation (Figure 3C) without affecting its precursors (Supplemental Figure 6, A and B), whereas the degradation rate of mature let-7 was dramatically accelerated upon miR-107 overexpression (Figure 3D), supporting the idea that the interaction between these miRNAs only occurred between the mature miRNA molecules. Furthermore, the degradation rate of miR-26a remained unchanged following depletion of endogenous miR-107 (Supplemental Figure 6C), indicating that miR-107–mediated let-7 degradation was a specific event and did not affect all miRNAs.
Next, we sought to determine whether miR-107 could regulate the expression of let-7 targets, including HMGA2 and Ras. Overexpression of miR-107 significantly enhanced the expression level of HMGA2 and Ras proteins but not that of Bcl-2, a known target of miR-15/16, in the absence or presence of ectopic let-7 (Figure 3E). These results indicated that miR-107 antagonized the inhibitory effect of let-7 on gene expression. To corroborate the above findings, we also examined whether endogenous miR-107 could regulate let-7 activity using antagomirs to deplete miR-107. Expression of lin-41 and HMGA2 was suppressed by miR-107 depletion but not by miR-26a depletion (Figure 3F and Supplemental Figure 7). On the other hand, when let-7 was depleted, the levels of lin-41 and HMGA2 were significantly elevated and miR-107 depletion did not efficiently reduce reporter expression, unless exogenous let-7 was expressed (Figure 3F). These results indicated that miR-107 modulated the stability of let-7 and therefore had an impact on its ability to regulate gene expression.
To investigate whether miR-107 could affect anchorage-independent growth (AIG), we transfected miR-107 and let-7 into MCF-7 and MDA-MB-231 cells. Expression of miR-107 abolished the let-7–mediated suppression of AIG in both cell lines (Figure 3G). Such effects were not observed when let-7 was depleted (Figure 3H). Moreover, miR-107 had no effect on AIG in let-7–deficient H661 cells (Supplemental Figure 8), consistent with the above data and indicating that miR-107–mediated regulation of AIG was dependent on let-7.
miR-107–mediated induction on the let-7–suppressed AIG requires direct base pairing of miR-107:let-7:HMGA2.
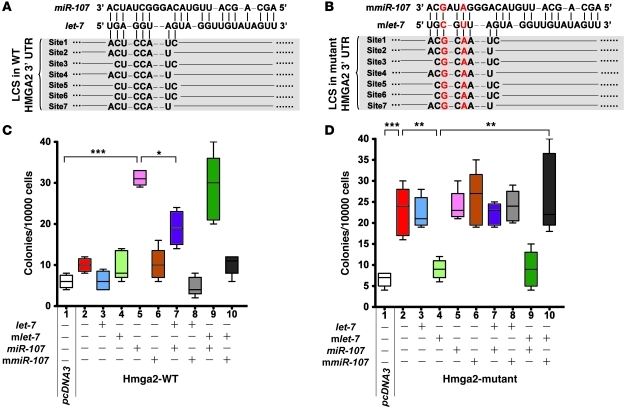
HMGA2 is an oncogenic factor that can modulate cell colony formation, and its expression is regulated by let-7 (11). Consistent with previous findings (11), expression of miR-107 (but not a mutant miR-107) promoted AIG in cells that expressed wild-type HMGA2, but such activity was abolished by expression of let-7 (Figure 4, A and C). To further confirm that miR-107 could modulate let-7 activity, we established mutant let-7 (mlet-7) and mutant miR-107 (mmiR-107), which contained compensatory point mutations that restored the pairing between mlet-7 and the 3′ UTR of mutant HMGA2 and between mmiR-107 and mlet-7 (Figure 4B). We observed that expression of mutant HMGA2 was insensitive to wild-type let-7–mediated control and demonstrated greatly enhanced colony formation (Figure 4D). Moreover, the AIG conferred by mutant HMGA2 was completely abolished by expression of mlet-7 (Figure 4D) and was again restored by expression of mmiR-107 (Figure 4D), but not by wild-type miR-107 (Figure 4D). These results recapitulated the importance of miR-107 in regulation of let-7–dependent AIG.
Figure 4. miR-107–mediated induction of the let-7–suppressed tumorigenicity requires direct base-pairing of miR-10:let-7:HMGA2.
(A and B) HMGA2 constructs used in the AIG assays. The let-7 complementary and mutated sites are indicated. The mutated HMGA2 3′ UTR site had 2 point substitutions that disrupted pairing to let-7 but allowed pairing to mlet-7. LCS, let-7 complementary site. (C and D) The requirement of pairing between miR-107 and let-7 on colony formation. The indicated miRNA-expressing constructs were transfected into wild-type or mutated HMGA2-expressing MCF-7 cells. Cells were then assayed for colony formation as described above. *P < 0.05; **P < 0.01; ***P < 0.001. Data are presented as mean ± SD.
In addition to suppressing tumorigenesis, let-7 has also been implicated in the regulation of self renewal in breast tumor–initiating cells (5). To elucidate whether let-7 could affect mammosphere formation, we cultured mammospheres from 4T1 mouse mammary tumor cells (Supplemental Figure 9). RT-PCR and Western blot analyses confirmed that the levels of 2 breast cancer stem cell markers, oct-4 and cd44, were significantly elevated in mammosphere-forming 4T1 cells (Supplemental Figure 9A). Accordingly, immunofluorescence microscopy demonstrated nuclear localization of oct-4 in such cells (Supplemental Figure 9B). Ectopic expression of miR-107 significantly restored the mammosphere-forming ability of the 4T1 cells, even in the presence of let-7 (Supplemental Figure 9, C and D). Collectively, our findings indicate that miR-107 antagonized the suppressive effect of let-7 on tumorigenicity and self renewal in cancer cells.
The internal loop of the miR-107–let-7 duplex is crucial for the interaction.
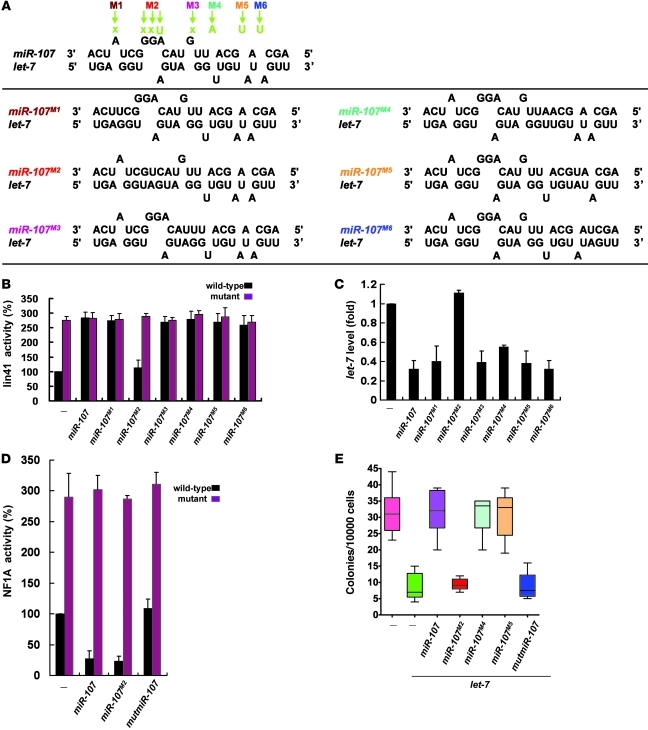
Several studies have indicated that asymmetric internal loops and bulged-A residues are critical for miRNA-mRNA interactions (12–14). To test whether these structures were important for the miR-107–let-7 interaction, we mutated nucleotides in miR-107 to eliminate the individual mismatch/loop structures of the miR-107–let-7 duplex (Figure 5A). Except for the mutant miR-107M2, which was missing the putative internal GAA loop, all other miR-107 mutants retained the ability to promote lin-41 and let-7 expression (Figure 5, B and C). In contrast, miR-107M2 could still suppress the expression of NF1A, a known mRNA target of miR-107 (Figure 5D). Moreover, colony formation analysis revealed that miR-107M2 cannot recover let-7–mediated AIG suppression (Figure 5E). Together, these data indicate that the internal loop structure of the miR-107–let-7 duplex was essential for miR-107’s activity in let-7–mediated gene expression.
Figure 5. The internal loop of the miR-107–let-7 duplex is crucial for their activities.
(A) Duplex structures formed between mutant forms of miR-107 and let-7a (bottom). Design of mutated miR-107 constructs. The top panel shows the wild-type miR-107–let-7a duplex structure, which exhibits several mismatched regions; mutations in miR-107 eliminating those mismatches are indicated (M1–M6). (B and C) The internal loop is required for miR-107 to antagonize let-7 function. The effects of structure-based miR-107 mutants on lin-41 activity (B) and let-7a levels (C) were measured in T47D cells. Luciferase activity of wild-type lin-41 in the control group served as the reference (100%). (D) Effect of the mutant forms of miR-107 on NF1A suppression. The activity of NF1A, a known mRNA target of miR-107, was measured using a luciferase reporter assay. The wild type and mutant refer to the NF1A constructs used in the reporter assay. (E) Requirement of miR-107’s internal loop for in vitro tumorigenesis. The effect of miR-107M2 mutants on colony formation, as measured by AIG assays. Data are presented as mean ± SD.
Effects of miR-107 on let-7–suppressed tumorigenesis and metastasis in a mouse model.
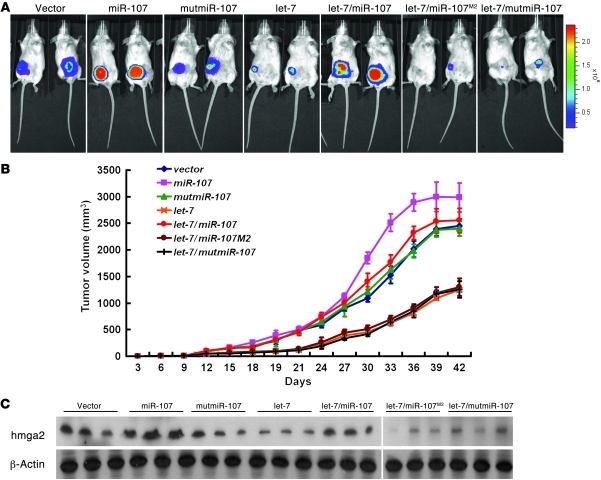
Using an in vivo luciferase imaging assay, we observed that expression of let-7 exhibited a suppressive effect on tumor growth (Figure 6, A and B), which was in agreement with our in vitro results. To investigate whether miR-107 could counteract this tumor suppression activity of let-7, we injected 4T1/luciferase vector (4T1/vector), 4T1/miR-107, and 4T1/mutmiR-107–transfected cells orthotopically into female Balb/c mice. The 4T1/miR-107 tumors grew more rapidly than the 4T1/vector tumors (Figure 6B), and they also had a higher photon intensity than the 4T1/vector tumors, as measured using an in vivo luciferase image assay (Figure 6A). In contrast, the 4T1/mutmiR-107 tumors behaved similarly to the vector control for both tumor growth and photon intensity (Figure 6, A and B). Coexpression of let-7 and miR-107, but not mutmiR-107 or miR-107M2, completely abolished the let-7–mediated tumor suppression (Figure 6, A and B), suggesting a causal role for miR-107 in the regulation of let-7–dependent tumor suppression. In support of these findings, expression of let-7 inhibited HMGA2 expression in 4T1-bearing tumors, which was restored by coexpression of miR-107 (Figure 6C).
Figure 6. Effects of miR-107, let-7, and their mutants on tumorigenesis and metastasis in an animal model.
(A and B) The effects of expression of miR-107, let-7a, and their mutants on orthotropic tumor growth were assessed by in vivo image detection. (A) The tumor burden was measured and calculated directly every 3 days, and the results were plotted as tumor volume over time (B). (C) Expression of HMGA2 in orthotropic tumor tissues was measured by Western blotting. Blot lanes were run on the same gel but were noncontiguous (white line). Data are presented as mean ± SD.
To determine whether miR-107 had any effect on the metastatic capability of breast cancer cells, we examined metastatic lung colonies in 4T1/vector, 4T1/miR-107, and 4T1/mutmiR-107 tumor-bearing mice. All of these tumors had high metastatic ability, and we did not observe a significant difference in metastases between them (Supplemental Table 1). However, tumors expressing let-7 did not exhibit any macroscopically visible lung nodules (Supplemental Table 1). Coexpression of miR-107 partially rescued the metastatic ability of 4T1/let-7–bearing tumors, whereas coexpression of either miR-107M2 or mutmiR-107 did not affect this ability (Supplemental Table 1). Together, these data indicate that miR-107 targeted and inhibited let-7, thereby promoting tumorigenesis and metastasis in this animal model. In total, the results of these animal and clinical experiments further confirmed the role of miR-107 in the let-7–mediated regulatory circuit in vivo.
Regulation of let-7 family members by miR-107.
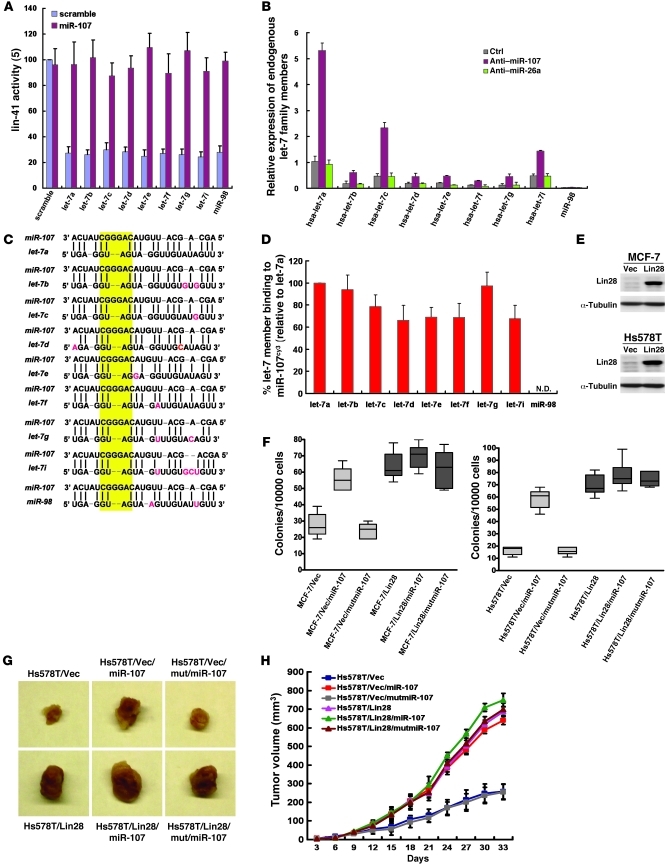
In addition to let-7a, we also wondered whether the effect of miR-107 could be extended to other let-7 family members. To this end, we tested the effect of miR-107 on lin-41 reporter activity in the presence of specific let-7 family members. As shown in Figure 7A, expression of miR-107 abolished the inhibitory effects mediated by specific let-7 members on lin-41 activity (Figure 7A), indicating that miR-107 could widely regulate let-7 members instead of let-7a alone. To investigate whether the expression of let-7 members could also be regulated by miR-107, we also tested the effect of miR-107 on regulating let-7 members and found that their expression levels were dramatically elevated after depleting endogenous miR-107 (Figure 7B). In addition, we found that the duplex structures have no obvious change between different let-7 members, especially within the highly conserved regions of internal loop (Figure 7C), even though there are variations in the sequence of let-7 family members. In support of this observation, we also precipitated miR-107cy3 to identify whether it could directly bind with endogenous let-7 family members. Data from specific quantitative RT-PCR (qRT-PCR) indicated that, to a different extent, miR-107 directly binds to let-7 family members with similar affinities (Figure 7D). Together with the above information, this suggests that miR-107 could directly bind and suppress all let-7 family members.
Figure 7. Regulation of let-7 family members by miR-107.
(A) Effect of miR-107 on lin-41 expression suppressed by different let-7 members. H661 cells were cotransfected with lin-41 and miR-107 in the presence or absence of let-7 member overexpression. Firefly luciferase reporter activity was normalized to renilla luciferase. (B) Expression of let-7 family members in miR-107 knockdown cells. The indicated antagomirs were transfected into MCF-7 cells and then assayed by real-time RT-PCR. (C) Duplex between miR-107 and let-7 family members. The yellow box represents the conserved sequence within the regions of internal loop. Red, residues differ from let-7a. (D) Binding between miR-107 and let-7 family members. miR-107cy3 was transfected into MCF-7 cells. The binding between miR-107cy3 and endogenous let-7 family members was determined by real-time RT-PCR. Percentage of let-7 members binding to miR-107cy3 was normalized with control. (E) Successful overexpression of Lin28 in MCF-7 and Hs578T cells. MCF-7 and Hs578T cells were stably transfected with pBabe-hLin28B. Lysates were collected and assayed by Western blot. (F) Effect of miR-107 on anchorage-independent growth in Lin28-overexpressed (let-7 knockdown) cells. MCF-7 and Hs578T cells were transfected with indicated miRNAs in the presence or absence of Lin28 overexpression. Soft agar assays were performed for 14 days. (G and H) Effect of miR-107 on in vivo tumor growth in Lin28-overexpressed cells. Hs578T/Vec and Hs578T/Lin28 cells were transfected with indicated miRNAs in the presence or absence of Lin28 overexpression. The tumor burden was measured and calculated every 3 days, and the results were plotted as tumor volume over time. Data are presented as mean ± SD.
Furthermore, we stably overexpressed Lin28 to suppress the expression of all let-7 members in MCF-7 and Hs578T cells (Figure 7E) and verified the effects of miR-107 on AIG (Figure 7F) and in vivo tumor growth (Figure 7, G and H). Similar to the results from H661 cells (Supplemental Figure 8), miR-107 has no effect on AIG in Lin28-overexpressed MCF-7 and Hs578T cells (Figure 7F), confirming that miR-107 acts through let-7 to enhance AIG in cancer cells. Furthermore, we also confirmed the requirement of let-7 in miR-107–enhanced tumor growth in vivo, in which its oncogenic effect was significantly reduced in the circumstance of let-7 knockdown (Figure 7, G and H). These results lead to the conclusion that miR-107 acts mainly through let-7 during tumor formation. However, the neoplastic growth in an orthotropic animal model was further enhanced by miR-107 under the condition of Lin28 overexpression (Figure 7, G and H), suggesting the existence of a let-7–independent mechanism, such as canonical miRNA-mediated gene repression, in tumor growth. These results conclude that miR-107 acts mainly through the let-7 family to promote tumorigenesis and may also control other let-7–independent mechanisms during tumor formation.
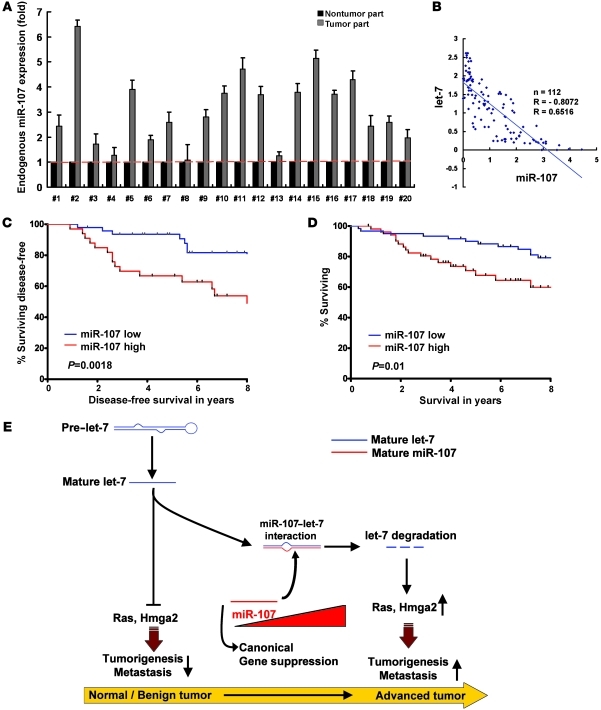
miR-107 is overexpressed in human breast cancer tissues and is related to poor prognosis.
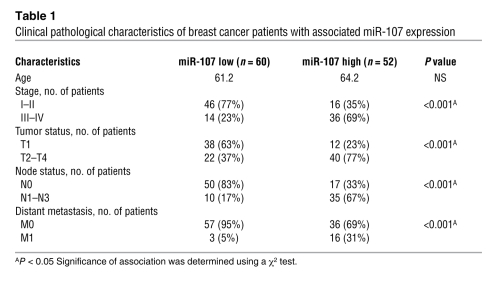
In contrast with the downregulation of let-7a in human cancer tissues (Supplemental Figure 10), we noticed a significant overexpression of miR-107 in most cancerous specimens (20 breast tumors) compared with their paired nonneoplastic counterparts (Figure 8A), suggesting that miR-107 may have a promoting effect during tumorigenesis. Furthermore, to determine whether miR-107 acts as a regulator of let-7, we analyzed the levels of miR-107 and let-7a in human breast (Figure 8B) and lung (Supplemental Figure 11) cancer tissues by qRT-PCR. There was a strong inverse correlation between miR-107 and let-7a levels in the 112 human breast (Figure 8B) and 38 lung (Supplemental Figure 11) cancer tissues. We next asked whether the expression of miR-107 correlated with the clinical outcome in patients with breast cancer. log-rank analysis indicated that disease-free survival was significantly worse in patients with higher (n = 52) miR-107 expression in their tumor tissues (Figure 8C), whereas higher let-7a levels were observed in patients with better disease-free survival (Supplemental Figure 12A). Importantly, patients with breast cancer who had higher levels of miR-107 or lower levels of let-7a had a poor overall survival (Figure 8D and Supplemental Figure 12B). To further ascertain whether expression of miR-107 correlated with the clinicopathological characteristics of advanced stage breast cancers, we used qRT-PCR to analyze miR-107 expression in 112 human breast cancer specimens. Patients whose tumors expressed higher levels of miR-107 demonstrated more advanced tumor status, increased lymph node metastases, and increased metastasis in distant organs (Table 1), whereas let-7a was downregulated in these advanced tumors (Supplemental Table 2). In addition to clarifying the overexpression of miR-107 in advanced human cancers, these results confirmed the role of miR-107 in let-7 regulation in human breast tumors.
Figure 8. miR-107 is overexpressed in human breast cancer tissues and is related to poor prognosis.
(A) Expression of miR-107 in normal/tumor portions of breast cancer tissues. We isolated RNAs from paired tumor and nontumor samples. Expression of miR-107 was detected by qRT-PCR. The level of miR-107 in nontumor tissue was used as a reference (= 1), and the relative fold expression in the tumor tissue was determined. (B) The correlation between miR-107 and let-7a levels in human breast cancer tissues. qRT-PCR was used to detect expression of miR-107 and let-7a. Each point on the graph corresponds to the relative expression levels from an individual patient. We normalized let-7a and miR-107 levels with U6 to calculate their relative expression levels. We used the averaged expression level of all patients as cutoff value. (C and D) miR-107 level predicts poor clinical outcome in patients with breast cancer. The log-rank test (2-sided) was used to compare differences between groups. The Kaplan-Meier curves show analyses of disease-free (C) and overall (D) survival in patients with breast cancer. (E) Working model of the miR-107–let-7 interaction during cancer progression. Data are presented as mean ± SD.
Table 1 .
Clinical pathological characteristics of breast cancer patients with associated miR-107 expression
Taken together, our results show that increased expression of miR-107 leads to cancer progression, including tumor growth and metastasis. Interestingly, like an endogenous antagomir of the let-7 miRNA, miR-107 plays a role in the let-7 degradation regulatory circuit and results in the upregulation of let-7–suppressed oncoproteins. We demonstrated for what we believe is the first time that miRNAs can be regulated by way of miRNA-miRNA interactions. Furthermore, manipulation of an miRNA by its antagonist miRNA may provide a potential strategy for treating cancers (Figure 8E).
Discussion
Regulation of miRNA biogenesis and turnover is crucial, as alteration of miRNA expression has been linked to human diseases including several cancers. Maintaining accurate levels of miRNAs is important for physiological function, and miRNA regulation occurs both transcriptionally and posttranscriptionally (15–18). For example, the transcriptional factors MyoD, Mef2, and SRF activate heart-specific expression of miR-1 via recognition of cis-regulatory elements within the miR-1 promoter (19). It is also known that the protooncogene c-Myc activates the expression of the miR-17-92 cluster in a variety of cancers through transcriptional regulation (20). Posttranscriptional regulation of miRNAs occurs at various steps, including processing of primary and precursor miRNAs, miRNA editing, and regulation of miRNA maturation, stabilization, and degradation (16). Our current study suggests that expression of an miRNA can be regulated by other miRNAs at the posttranscriptional level and that this regulation is critical for cancer progression.
Binding activity of miRNAs to mRNAs and structural features of these complexes are crucial for gene silencing. The patterns of known miRNA-mRNA duplexes indicate the requirement of a mismatch/internal loop within the central region (12, 21). Herein, we postulate that the internal loop structure in the miR-107–let-7 hybrid plays an important role in let-7 stability and in target mRNA recognition and/or degradation. However, its underlying mechanism requires further investigation. According to our study, the characteristics of the internal loop structure may provide a starting point for prediction of miRNA-miRNA interactions. In addition, future resolution of the 3D structure of this miRNA-miRNA duplex may provide insight into the structural aspects of the interaction among other miRNAs.
Although there is only 1 nucleotide that is different between miR-107 and a closely related miRNA, miR-103, in their 3′ end, we found that miR-103 has only a marginal effect on let-7a, as a mere change in let-7 activity was observed upon the manipulation of miR-103 (Supplemental Figure 13). To consider this discrepancy, we investigated the potential formation of a duplex between miR-103 and let-7, and found that the differences in nucleotide sequence between miR-107 and miR-103 at the 3′ end weaken the binding affinity and stability of the interaction with let-7. Also consistent with this finding, we did not observe a similar inverse correlation between miR-103 and let-7a in the same specimens used in Figure 8B (Supplemental Figure 14). This evidence indicates that miR-103 cannot regulate let-7 as miR-107 does.
Stability of individual mature miRNAs can be regulated by protein factors. For example, GLD2 specifically stabilizes and monoadenylates miR-122 in liver cells (22). Exoribonucleases have been reported to degrade mature miRNAs to limit their accumulation in plants and Caenorhabditis elegans (23, 24). RNA-induced silencing complex (RISC) has also been reported to play an important role in miRNA processing and targeting of mRNAs. As targeting let-7 by miR-107 causes miRNA degradation, whether these factors or other proteins participate in miRNA-miRNA duplex formation and subsequent miRNA degradation is an important subject for further study.
miR-107 is overexpressed in several tumor types such as colon, pancreas, and stomach cancers (25). Subsequently, recent study has also shown that breast cancer patients with higher miR-107 levels display a significantly lower probability of metastasis-free survival (26). Our findings indicate that downregulation of the tumor-suppressing miRNA let-7 is important for miR-107–mediated tumorigenesis. Interaction with and degradation of let-7 may provide a possible mechanism for how miR-107 accelerates tumor progression. let-7 has been reported to prevent the maintenance of breast cancer–initiating cells through inhibition of HMGA2 and Ras (5). In our current study, we found that miR-107 is highly expressed in breast cancer–initiating cells and helps maintain their self-renewal ability, which may result from the miR-107–let-7 interaction. Moreover, in contrast with the downregulation of let-7, increased levels of miR-107 have been found during early embryonic stages (27). Therefore, we propose that through suppression of let-7, miR-107 regulates multipotency by enhancing self renewal and promoting differentiation in both normal development and cancer. In addition, we have further examined the effect of let-7 family members on the expression of miR-107. After blocking endogenous let-7 family members, the level of miR-107 was significantly upregulated (Supplemental Figure 15), suggesting that let-7 family members could suppress the expression of miR-107. These results conclude that interaction of 2 miRNAs may result in the reciprocal suppression to each other. The reciprocal expression between miR-107 and let-7 suggests a balanced regulation that may be controlled by miRNA interactions in different physiological contexts. In support of this idea, we have also established miR-107 knockout mice and found a significant increase in the expression of let-7 family members in the tissues of liver and kidney of mice (Supplemental Figure 16).
Our study indicates that miRNA expression is posttranscriptionally controlled by interactions with other miRNAs. At present, how the Ago-containing RNA-induced silencing complex participates in miRNA duplex formation and regulates let-7 stability is yet unclear. The question of whether this heterodimeric miRNP contains specific proteins for strand selection and degradation remains a topic of further interest. The identification of mechanisms that regulate global or individual miRNA biogenesis and turnover represents an important challenge for future studies. Nevertheless, our study provides what we believe is a novel mechanism for regulation of individual miRNA expression. In particular, miR-107–mediated stability control of let-7 appears to be important for cancer progression and metastasis and also holds a promising therapeutic potential for cancer treatment.
Methods
Cell culture, plasmids, antibodies, and reagents.
All human cancer cell lines were obtained from ATCC. These cell lines were maintained in minimum essential medium (MEM) supplemented with 10% FBS (Gibco BRL; Invitrogen), 2 mM l-glutamine (Life Technologies), 100 μg/ml streptomycin, and 100 U/ml penicillin in a humidified 5% CO2 atmosphere. The miRNA inhibitors and miRNA mimics, including cy3-labeled miR-107, miR-26a, and cy5-labeled let-7, were purchased from Dharmacon. The lin-41 (pISlin-41w; ref. 28), mutant lin-41 (pISlin-41m; ref. 28), HMGA2 (pcDNA3.1 Hmga2 wild type; ref. 11), mutant HMGA2 (pcDNA3.1 Hmga2 m7; ref. 12), and Lin28B (p-Babe-hLin28B; ref. 29) plasmids were provided by Addgene. The Ras antibody (RB-1627-PCS) was purchased from Lab Vision, the HMGA2 antibody (ab41878) was purchased from Abcam, and the Cy-3/Cy-5 antibody (c3117) was purchased from Sigma-Aldrich. We established miRNA-expressing constructs by using the BLOCK-iT pol II miR RNAi Expression Vector Kit with EmGFP from Invitrogen (K4936-00). The mature miRNA sequences were obtained from the Sanger Center miRNA Registry ( http://microrna.sanger.ac.uk/sequences/), and the human miRNA gene was designed according to the manufacturer’s instructions (30, 31). These miRNA gene double strands were ligated with the Block-iT Pol II miR RNAi Expression Vector, pcDNA6.2-GW/EmGFP-miR, to generate constructs expressing specific miRNAs.
Detection of miRNA.
Total RNA was isolated using Trizol (Invitrogen) according to the manufacturer’s instructions. miRNA-specific TaqMan RT primers (Applied Biosystems) were used to perform reverse transcription. Real-time PCR was performed using an Applied Biosystems 7900 Fast Real-Time PCR system with miRNA-specific primers and TaqMan Universal PCR Master Mix (Applied Biosystems). The results were calculated and normalized to an endogenous reference control gene (RNU-6B). Detection of mutant forms of miRNA was performed using DNA primers and SYBR green dye (Applied Biosystems) as described previously (32). To detect precursor (pri- and pre-) miRNAs, we used SYBR green qRT-PCR as previously established (33, 34).
Transfection of miRNA mimics, antagomirs, and cy3-labeled miR-107, miR-26a, and cy5-labeled let-7a.
All the oligos were transfected by using Lipofectamine 2000 according to the manufacturer’s instructions. Briefly, 200 nM (100 μl) of mimics, antagomirs, and dye-labeled miRNAs were mixed and incubated with 10 μl of Lipofectamine 2000 reagent for 30 minutes. Eight hours after transfection, medium was replaced for loner incubation.
Fluorescence resonance energy transfer.
We synthesized duplex precursors that had 5′ phosphorylation and 3′ modification with cy3 or cy5 on their functional strands (Dharmacon). Cy3-labeled miR-107 or miR-26a was cotransfected into A549 cells with cy5-labeled let-7. The cells were fixed with 4% paraformaldehyde at 24 hours after transfection, and the images were captured using a confocal microscope. Quantification of colocalization was performed in triplicate by assaying confocal images in which total and colocalized foci of miR-107 and let-7 were counted randomly in a total of 20 cells. To further determine the miR-107–let-7 interaction in vivo, we used a real-time FRET assay in which the transfer of energy from fluorescent donor molecules to fluorescent acceptor molecules, which occurs by a nonradioactive dipole-dipole interaction, was detected. The FRET efficiency between miR-107 and let-7 was measured using the acceptor photobleaching method. Cells cotransfected with miR-107/miR-26a (acceptors) and let-7 (donor) were fixed and imaged using a LSM510 confocal microscope. Selective areas of the cells were repeatedly photobleached at λ = 633 nm, and the fluorescence intensity of cy3-labeled miR-107 was subsequently monitored at λ = 575–615 nm.
Orthotropic tumor model.
4T1 cells were transfected with the indicated plasmids, and stable colonies were selected using blasticidin. These cells were maintained in DMEM medium containing 5 μg/ml of blasticidin prior to injection. We injected 5 × 105 cells orthotropically into the fat pads of each group (9 mice/group). Animals were analyzed 35 days after cell injection using an in vivo luciferase imaging system. Animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of National Taiwan University College of Medicine.
RNA immunoprecipitation.
The procedure for RNA immunoprecipitation was modified from that of a previous study (35). Briefly, cells were treated with formaldehyde to generate RNA-RNA cross-links between molecules that were in close proximity in vivo. miRNAs that cross-link with another miRNA were isolated by immunoprecipitation using the anti-cy3/cy5 antibody (Sigma-Aldrich), and reversal of the formaldehyde cross-linking permitted recovery and quantitative analysis of the immunoprecipitated miRNA by qPCR.
Statistics.
Data are presented as mean ± SD. The 1-tailed Student’s t test was used to compare data between groups. The correlations exhibited in Table 1 and Supplemental Table 2 were calculated by using Pearson’s χ2 test. Statistical analyses of clinicopathological data were performed as described previously (36). Tissue preparation and analysis were approved by the Institutional Review Board (IRB) of National Taiwan University Hospital. Statistical significance was assumed at P < 0.05.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Council, Taiwan (NSC97-2320-B-002-036-MY3, NSC97-2323-B-002-011, NSC96-2314-B-002-123-MY3, and 96-3114-P-002-002-Y), and the Ministry of Education (97R0066-08). This work was also partially supported by National Science Council grants NSC 96-2320-B-004-MY2, NSC 97-2320-B-039-039-MY3, NSC 98-2815-C-039-082-B, and NSC 99-2314-B-039-002-MY3 from the National Science Council of Taiwan; National Health Research Institutes grants from Taiwan (NHRI-EX100-10033BI, NHRI-EX98-9712BC); Department of Health, Executive Yuan grants from Taiwan (DOH97-TD-G111-024, DOH97-TD-G111-011); grants from China Medical University (CMU96-220, CMU96-189, CMU97-253, CMU97-277 and CMU99-TC-22), Taiwan Department of Health Clinical Trial and Research Center of Excellence (DOH100-TD-B-111-004), and an Odyssey Scholarship to J-L. Su. We thank Mien-Chie Hung and Gunnar Johansson for their help in editing this paper.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(9):3442–3455. doi:10.1172/JCI45390.
References
- 1.Bartel DP. MiRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 spec no 1:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Johnson SM, et al. RAS is regulated by the let-7 miRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Takamizawa J, et al. Reduced expression of the let-7 miRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 5.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 6.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Lai EC, Wiel C, Rubin GM. Complementary miRNA pairs suggest a regulatory role for miRNA:miRNA duplexes. RNA. 2004;10(2):171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cevec M, Thibaudeau C, Plavec J. Solution structure of a let-7 miRNA:lin-41 mRNA complex from C. elegans. Nucleic Acids Res. 2008;36(7):2330–2337. doi: 10.1093/nar/gkn088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem Biol. 2004;11(12):1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004;18(2):132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 16.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11(12):1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 18.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38(3):323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 20.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe Y, Yachie N, Numata K, Saito R, Kanai A, Tomita M. Computational analysis of microRNA targets in Caenorhabditis elegans. Gene. 2006;365:2–10. doi: 10.1016/j.gene.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 22.Katoh T, et al. Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23(4):433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321(5895):1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461(7263):546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 25.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 27.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1(1):47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 28.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 29.Viswanathan SR, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41(7):843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaughlin J, et al. Sustained suppression of Bcr-Abl-driven lymphoid leukemia by microRNA mimics. Proc Natl Acad Sci U S A. 2007;104(51):20501–20506. doi: 10.1073/pnas.0710532105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su JL, et al. Downregulation of microRNA miR-520h by E1A contributes to anticancer activity. Cancer Res. 2010;70(12):5096–5108. doi: 10.1158/0008-5472.CAN-09-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11(11):1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33(17):5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmittgen TD, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44(1):31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selth LA, Gilbert C, Svejstrup JQ. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb Protoc. 2009;2009(6):pdb.prot5234. doi: 10.1101/pdb.prot5234. [DOI] [PubMed] [Google Scholar]
- 36.Shih JY, et al. Collapsin response mediator protein-1 and the invasion and metastasis of cancer cells. . J Natl Cancer Inst. 2001;93(18):1392–1400. doi: 10.1093/jnci/93.18.1392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.