Abstract
The ubiquitin-proteasome system degrades most intracellular proteins, including misfolded proteins. Proteasome functional insufficiency (PFI) has been observed in proteinopathies, such as desmin-related cardiomyopathy, and implicated in many common diseases, including dilated cardiomyopathy and ischemic heart disease. However, the pathogenic role of PFI has not been established. Here we created inducible Tg mice with cardiomyocyte-restricted overexpression of proteasome 28 subunit α (CR-PA28αOE) to investigate whether upregulation of the 11S proteasome enhances the proteolytic function of the proteasome in mice and, if so, whether the enhancement can rescue a bona fide proteinopathy and protect against ischemia/reperfusion (I/R) injury. We found that CR-PA28αOE did not alter the homeostasis of normal proteins and cardiac function, but did facilitate the degradation of a surrogate misfolded protein in the heart. By breeding mice with CR-PA28αOE with mice representing a well-established model of desmin-related cardiomyopathy, we demonstrated that CR-PA28αOE markedly reduced aberrant protein aggregation. Cardiac hypertrophy was decreased, and the lifespan of the animals was increased. Furthermore, PA28α knockdown promoted, whereas PA28α overexpression attenuated, accumulation of the mutant protein associated with desmin-related cardiomyopathy in cultured cardiomyocytes. Moreover, CR-PA28αOE limited infarct size and prevented postreperfusion cardiac dysfunction in mice with myocardial I/R injury. We therefore conclude that benign enhancement of cardiac proteasome proteolytic function can be achieved by CR-PA28αOE and that PFI plays a major pathogenic role in cardiac proteinopathy and myocardial I/R injury.
Introduction
The ubiquitin-proteasome system (UPS) mediates the targeted degradation of abnormal and most normal intracellular proteins in the cell and generally includes 2 main steps: ubiquitination of a specific protein molecule and subsequent degradation of the ubiquitinated protein by the proteasome (1–3). Ubiquitinated proteins usually accumulate in the cell during proteasome functional insufficiency (PFI). Increases in steady-state ubiquitinated proteins were observed in the myocardium of patients with end-stage heart failure resulting from a variety of heart diseases, such as dilated cardiomyopathy and ischemic heart disease (4), which suggests that PFI is a common phenomenon of cardiac pathogenesis. PFI occurs when the proteasome is impaired and/or when the demand for proteasome function surpasses the functional capacity of proteasomes (4).
As exemplified by neural degenerative diseases, proteinopathies are diseases caused by protein misfolding and are characterized by aberrant protein aggregation (4). Desmin-related cardiomyopathy (DRC) is the cardiac manifestation of desmin-related myopathy (5), representing the best-studied example of cardiac proteinopathy. The presence of desmin-positive protein aggregates in cardiomyocytes is characteristic of DRC. Mutations in the desmin, αB-crystallin (CryAB), and myotilin genes have been linked to human DRC (5). A recent experimental study suggests that the more commonly seen pressure-overloaded cardiomyopathy may also display characteristics of proteinopathy as well (6).
Terminally misfolded proteins are degraded mainly by the UPS. Misfolded proteins form aberrant aggregates when escaping from the vigilance of the UPS. Hence, PFI allows more misfolded proteins to aggregate, and the aberrant protein aggregation can impair proteasome function (7). Consequently, PFI and protein aggregation form a vicious cycle, which is believed to result in proteinopathy (4). Characteristics of PFI, which include increases in ubiquitinated proteins and formation of pre-amyloid-like oligomers (8, 9), were reported in explanted human hearts with end-stage heart failure, suggestive of the involvement of PFI in at least a subset of human cardiomyopathies (1). However, the necessity of PFI in the genesis of proteinopathies or any disease has not been proven.
Detection of PFI in experimental animals was facilitated by the development of proteasome function reporter mice in which an easily detected biologically inert surrogate substrate for the UPS was expressed (10). We had previously created and validated a Tg mouse model that expresses a modified GFP with carboxyl fusion of the degron CL1 (GFPdgn) (10). Degron CL1 signals for ubiquitination via the surface-exposed hydrophobic structure of its predicted amphipathic helix (11), a signature structure that is shared by misfolded proteins and constitutes a signal for their ubiquitination (12). Therefore, GFPdgn is considered a surrogate of misfolded proteins (4). Using the GFPdgn reporter mice, we were able to reveal PFI in the heart of DRC mice (13, 14), produced by cardiac overexpression of a missense mutation of CryAB (CryABR120G) or a 7–amino acid (R172–E178) deletion mutation of the desmin gene (15, 16), both linked to human DRC (5). Further interrogation proved that aberrant protein aggregation is required for CryABR120G- or mutant desmin–induced PFI in cardiomyocytes (14, 17). Notably, PFI has also been observed or suggested in animal models of several other types of heart disease (18, 19), including myocardial ischemia/reperfusion (I/R) injury and pressure overload–induced cardiomyopathy (4, 20, 21). Proteasome dysfunction has been recently reported in human hearts with hypertrophic or dilated cardiomyopathy (19, 22). Therefore, PFI was hypothesized to play a major role in the progression of various heart diseases, especially cardiac proteinopathies, to congestive heart failure. However, this hypothesis has been difficult to test because a relatively benign method to enhance proteasome function has not been reported until very recently (23, 24).
In the multisubunit proteasome functional complex, the proteolytic activities reside in its 20S core particle, a barrel-shaped structure consisting of 28 protein subunits. To degrade a target protein molecule, the 20S requires the assistance of proteasome activators (PAs) that attach to one or both ends of the 20S (2). In mammalian cells, the PA mainly includes the 19S (also known as PA700) and the 11S (also known as PA28 or REG) subcomplexes (25). Although 19S-associated 20S is perhaps the predominant proteasome form, 11S-associated 20S proteasomes are found in many cell types, including cardiomyocytes (1). The 11S can be formed by PA28α and PA28β in heteroheptamers (α3β4 or α4β3) or by PA28γ in homoheptamers (γ7) (26, 27). The association of the 11S with the 20S may play a role in antigen processing by modulating peptide cleavage in the 20S (28, 29), but it appears that the 11S may play a greater role in intracellular protein degradation than in antigen processing (30).
Using adenoviral-mediated PA28α overexpression (PA28αOE) in cultured cardiomyocytes, we recently found that PA28αOE upregulates 11S proteasomes by interacting and stabilizing PA28β, leading to significant enhancement of proteasome proteolytic function in the cell, as evidenced by increased degradation of a validated surrogate substrate of the proteasome. Furthermore, the induced proteasome functional enhancement significantly attenuated oxidative stress–induced accumulation of oxidized proteins and cell death in the cultured cardiomyocytes (23).
In the present study, we investigated whether upregulation of the 11S proteasome, via cardiomyocyte-restricted PA28αOE (CR-PA28αOE), enhances the proteolytic function of the proteasome in intact mice and, if so, whether the enhancement can rescue a bona fide proteinopathy and protect against I/R injury. We found that CR-PA28αOE significantly enhanced UPS-mediated degradation of a surrogate misfolded protein in mouse hearts without showing adverse effects on intracellular proteostasis and cardiac function, establishing a Tg mouse model of benign enhancement of cardiac proteasomal function that we believe to be novel. Using this model, we demonstrated that enhancing cardiac proteasome function protected against cardiac proteinopathy and myocardial I/R injury in intact mice.
Results
Establishment of a Tg mouse model of cardiomyocyte-restricted overexpression of 11S proteasomes.
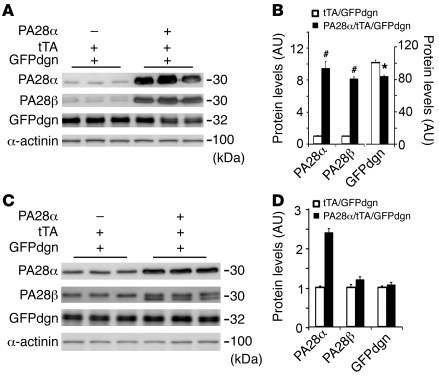
A genetic model of proteasome functional enhancement has not been reported. Our previous discovery that PA28αOE is sufficient to upregulate 11S proteasomes and, more interestingly, destabilize a surrogate UPS substrate in cultured cells prompted us to generate stable Tg mouse lines in which CR-PA28αOE could be reversibly activated with the tetracycline (Tet) analog doxycycline (Dox). The cardiac-specific Tet-suppressible binary Tg system (commonly known as Tet-Off system) was used (31). Mouse Psme1 cDNA was inserted behind the modified mouse Mhc6 promoter in the responder vector and used for creating the responder lines via microinjections of fertilized eggs. To achieve Tet-suppressible PA28αOE, the responder mice were cross-bred with the previously described cardiac-specific Tet-controlled trans-activator (tTA) Tg mice (31). For characterization of the initial cohort, no Dox was administered, so that PA28α Tg expression occurred upon activation of the α-MHC promoter. No embryonic or postnatal lethality was observed. In the absence of tTA, the baseline myocardial PA28α protein levels of line 1 and line 2 responder Tg mice were slightly higher than those of non-Tg littermates, indicating a slightly leaky expression of Tg PA28α, but this was not the case in line 3 (Figure 1, A and B). When coupled with Tg tTA, the abundance of PA28α markedly increased by 9.8-, 8.3-, and 5.0-fold in lines 1, 2, and 3, respectively, compared with non-Tg littermates (Figure 1, A and B). Consistent with our cell culture data (23), upregulation of PA28α was accompanied by proportional increases in PA28β protein levels (Figure 1, A and B), but PA28β transcript levels were not affected (Figure 1, D–F).
Figure 1. PA28α and PA28β expression in hearts of mice with CR-PA28αOE.
(A) 3 Tg responder lines carrying Tg PA28α were crossbred with the tTA Tg mice. No Dox was given to the breeding pairs or pups. Western blot analyses of 2-month-old mouse heart samples show that CR-PA28αOE resulted in a proportional increase of PA28β protein in the PA28α/tTA double-Tg mice. (B) Quantitative densitometry analyses of PA28α and PA28β protein levels. n = 3, *P < 0.001 vs. non-Tg (NTG); #P < 0.001 vs. tTA single-Tg. (C and D) Representative Northern blot analyses for PA28α (C) and PA28β (D). (E) Quantitative analysis of PA28β mRNA levels. CTL, control. n = 4. (F) RNA dot blot analyses of PA28α and PA28β transcript levels. GAPDH and ribosome 18S RNA served as loading controls. (G) Reciprocal IP with PA28α and PA28β antibodies showed increased PA28α-associated PA28β and PA28β-associated PA28α in myocardium with PA28αOE.
Co-IP (Figure 1G and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI45709DS1) and gel filtration (Supplemental Figure 2A) further confirmed the increased abundance of PA28α-associated PA28β in PA28α/tTA double-Tg mouse hearts compared with littermate controls. The native PA28α/PA28β complex has a molecular weight of approximately 200 kDa (Supplemental Figure 2A), similar to the size of the reported heteroheptamer (α3β4 or α4β3) 11S proteasome subcomplex (26).
PA28αOE increases the abundance of the 11S-associated 20S.
Given that the 20S can be capped by either 19S or 11S, we then examined whether upregulation of the 11S affects the dynamics of the interaction between 19S and 20S proteasome subcomplexes. The size distribution of native proteasome subcomplexes was assessed by gel filtration and compared between PA28α/tTA double-Tg and tTA single-Tg control hearts (Supplemental Figure 2A). The presence of 20S particles (marked by proteasome subunit β5 [PSMB5]) in a smaller protein complex (fractions 29–35) increased, whereas the presence of 19S (marked by RPN2) decreased, in fractions 23–32. Furthermore, the 11S proteasome, as represented by PA28α and PA28β, was increased in fractions 23–29. Consistently, IP using an Ab against the α3 subunit of 20S, which effectively immunoprecipitates the 20S proteasome (32), showed that more PA28α, but less Rpt6, was co-immunoprecipitated with the 20S from the myocardial protein extracts with PA28αOE compared with those lacking PA28αOE (Supplemental Figure 2B). These findings indicate that the overexpressed 11S subcomplexes might compete with the 19S particle for binding to 20S and result in more 11S-20S-19S (hybrid) and 11S-20S-11S proteasome complexes.
To determine the impact of PA28αOE on 19S and 20S abundance, representative subunits of 19S and 20S in ventricular tissues were analyzed using IB (Supplemental Figure 2, C and D). Compared with the respective controls, PA28αOE did not cause significant changes in RPN2 and RPT6 or in the abundance of the core subunits of the 20S, which indicates that abundance of 19S and 20S is not altered by PA28αOE.
CR-PA28αOE enhances cardiac UPS proteolytic function.
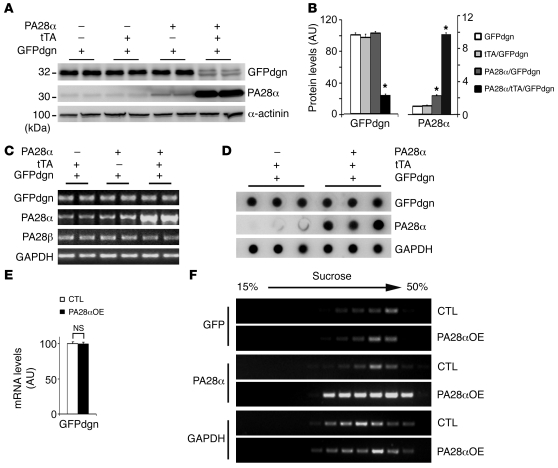
A previously validated UPS activity reporter (GFPdgn) Tg mouse model was used to probe the effect of 11S upregulation on cardiac UPS proteolytic function. In the absence of changes in synthetic rates, alterations of GFPdgn protein levels in a cell or tissue inversely correlate to UPS proteolytic function (10). Mice harboring homozygous GFPdgn and hemizygous tTA were cross-bred with mice carrying hemizygous PA28α responder Tg. The resultant offspring mice were examined for GFPdgn protein levels in the heart at 2 months. Western blot analysis revealed that cardiac GFPdgn levels of tTA/GFPdgn double-Tg mice were not significantly different from those of GFPdgn single-Tg mice (Figure 2, A and B). However, CR-PA28αOE significantly reduced GFPdgn protein levels in the PA28α/tTA/GFPdgn triple-Tg mouse heart compared with littermate controls; an 80% reduction of GFPdgn protein was observed upon a 9-fold increase in PA28α levels (Figure 2, A and B). Both quantitative RT-PCR and RNA dot blot analyses showed that GFPdgn transcript levels were unchanged by CR-PA28αOE compared with those of tTA/GFPdgn double-Tg controls (Figure 2, C–E). Furthermore, we assessed the ribosomal distribution of GFPdgn mRNA in the heart and found no apparent difference between PA28α/tTA/GFPdgn triple-Tg and control tTA/GFPdgn double-Tg hearts (Figure 2F), which indicates that PA28αOE did not alter the translational efficiency of GFPdgn mRNA in the heart. Therefore, the decreases in GFPdgn protein levels in the PA28α/tTA/GFPdgn triple-Tg hearts indicate that CR-PA28αOE enhances cardiac UPS proteolytic function.
Figure 2. Effect of CR-PA28αOE on UPS proteolytic function in the heart.
Mice were generated via cross-breeding between those harboring homozygous GFPdgn and hemizygous tTA and those carrying hemizygous PA28α responder Tg. (A and B) Western blot analyses of PA28α and GFPdgn protein levels (n = 4 mice/group). *P < 0.001 vs. all other groups. (C–F) Semiquantitative RT-PCR (C) and RNA dot blot analyses (D and E) of steady-state GFPdgn transcript levels, and assessment of GFPdgn mRNA polysomal distribution (F), in the ventricles of PA28α/tTA/GFPdgn triple-Tg (PA28αOE) and tTA/GFPdgn double-Tg control littermates. (C and D) GAPDH was analyzed for the loading control. (F) Polysomes were isolated from ventricular myocardium using sucrose gradients (see Methods). RNAs were extracted from the gradient fractions and used for RT-PCR to detect the distribution of GFPdgn mRNA. PA28α and GAPDH were probed as positive and negative controls, respectively.
Effectiveness of the inducible system on CR-PA28αOE and proteasomal proteolytic function.
We next tested whether CR-PA28αOE can be initiated in adult mice with the newly created PA28α responder mice and if so, whether CR-PA28αOE initiated in adult mice can also enhance cardiac proteasome function in a separate cohort of PA28α/tTA/GFPdgn triple-Tg and tTA/GFPdgn double-Tg mice. Dox administration was started when breeding was initiated and continued until the offspring mice reached 8 weeks of age, when Dox was withdrawn. For these mice at 8 weeks of age, changes in the protein levels of PA28α, PA28β, and GFPdgn in PA28α/tTA/GFPdgn triple-Tg mice were blocked by Dox (data not shown), with levels comparable to those in PA28α/GFPdgn double-Tg mice that had not received Dox treatment (Figure 2). PA28αOE in tTA/PA28α/GFPdgn mice was successfully induced 8 weeks after Dox was removed (Figure 3, A and B). The resultant protein levels of PA28α and PA28β were comparable to those achieved in PA28α/tTA/GFPdgn mice that never received Dox (Figure 1A). Upon CR-PA28sOE, there was a significant decrease in GFPdgn in PA28α/tTA/GFPdgn compared with tTA/GFPdgn mouse hearts (P < 0.05; Figure 3, A and B), which demonstrates that PA28αOE initiated in adult mice is also able to increase GFPdgn protein degradation. For those receiving Dox during the entire 16-week period after birth, the tTA-driven myocardial PA28αOE in PA28α/tTA/GFPdgn triple-Tg mice was completely blocked; as a result, the myocardial GFPdgn protein levels in these mice were identical to those of tTA/GFPdgn double-Tg mice (P > 0.05; Figure 3, C and D).
Figure 3. Inducible activation of UPS proteolytic function.
(A and B) Western blot analysis of PA28α, PA28β, and GFPdgn. Dox administration was started from the breeding pairs and withdrawn when the offspring reached 8 weeks of age. Cardiac tissue was collected 8 weeks after Dox withdrawal for the analyses. (C and D) For age-matched mice that received Dox treatment throughout, induced expression of PA28α was blocked, and GFPdgn protein levels were not significantly altered. Each lane was from an individual mouse. #P < 0.01, *P <0.05 vs. tTA/GFPdgn.
Effects of CR-PA28αOE on baseline cardiac growth and heart function.
Given that upregulation of 11S proteasomes enhanced UPS proteolytic function, we sought to determine the effect on cardiac structure and function. Pathological cardiac remodeling is often associated with reactivation of the fetal gene program. The transcript levels of the fetal gene panel in ventricular myocardium were examined in 1-year-old PA28α/tTA double-Tg and tTA single-Tg mice using RNA dot blot analysis (Supplemental Figure 3). PA28αOE did not change the transcript levels of any of these genes. Serial echocardiography analyses up to 1 year revealed no detectable alterations in cardiac geometry and function (Supplemental Table 1). The absence of any pathology or hypertrophy was further demonstrated by myocardial histopathological analysis (Supplemental Figure 4) and gravimetric measurements (Supplemental Table 2).
CR-PA28αOE attenuates cardiac hypertrophy and delays premature death of DRC mice.
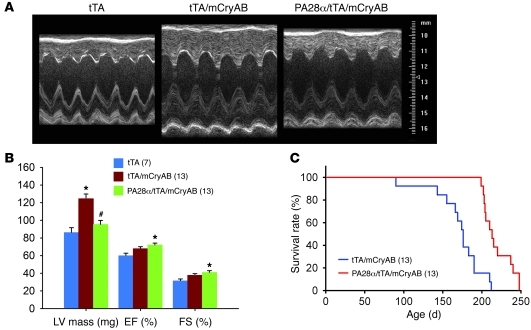
Since CR-PA28αOE can enhance cardiac proteasome proteolytic function in a manner that appears to be benign, and PFI is observed in CryABR120G DRC mouse hearts, we crossed the 2 Tg mouse models to test whether CR-PA28αOE is able to rescue the DRC. Previous studies have characterized that DRC in line 134 CryABR120G Tg mice progresses in a well-defined time course. The animals do not show overt pathology at 1 month, but develop concentric cardiac hypertrophy at 3 months and congestive heart failure at 6 months, dying shortly thereafter with a median lifespan of approximately 25 weeks (33). Therefore, this Tg line provides an excellent cardiac proteinopathy model for pathogenesis and experimental intervention studies. We obtained a cohort of mice with a genotype of either tTA/CryABR120G double-Tg or PA28α/tTA/CryABR120G triple-Tg (i.e., DRC without or with forced PA28αOE, respectively) and carried out a Kaplan-Meier survival analysis. Transthoracic echocardiography was performed at 12 weeks of age along with a group of age- and sex- matched tTA single-Tg mice of the same FVB/N inbred strain (Figure 4, A and B, and Supplemental Table 3). Compared with tTA single-Tg mice, which are known to display no phenotype within their 6–8 months of life, double-Tg mice developed concentric hypertrophy, as revealed by increases in LV posterior and anterior wall thickness at the end of both diastole and systole as well as in calculated LV mass, whereas LV internal dimensions at the end of both diastole and systole, ejection fraction (EF), and fractional shortening (FS) were not significantly altered in the tTA/CryABR120G double-Tg mice (Figure 4, A and B). However, these abnormal changes were significantly attenuated in the PA28α/tTA/CryABR120G triple-Tg group compared with the tTA/CryABR120G double-Tg group (P < 0.05, P < 0.01; Figure 4B and Supplemental Table 3). Compared with the tTA single-Tg group, the average EF and FS were increased in both the double-Tg and the triple-Tg group, but only the increase in the triple-Tg group reached statistical significance (P < 0.05; Figure 4B). These findings suggest that CR-PA28αOE prevents and/or attenuates cardiac hypertrophy and preserves cardiac function in DRC mouse hearts. Because cardiac hypertrophy represents a secondary adaptive response of the heart to the primary lesion caused by mutant CryAB proteins, attenuating hypertrophy without compromising heart function is consistent with the hypothesis that CR-PA28αOE attenuates the primary lesion in the DRC heart, thereby being protective.
Figure 4. PA28αOE attenuates cardiac hypertrophy and delays premature death of mice with CryABR120G-based cardiomyopathy.
(A and B) Effect of CR-PA28αOE on LV mass, EF, and FS, assessed by echocardiography at 12 weeks (see Supplemental Table 3 for other parameters). mCryAB, CryABR120G. *P < 0.05 vs. tTA; #P < 0.05 vs. tTA/CryABR120G. (C) Survival rate of a cohort of mixed-sex littermate tTA/CryABR120G double-Tg or PA28α/tTA/CryABR120G triple-Tg mice was monitored daily, and survival data were used for Kaplan-Meier analysis. P < 0.01, log-rank test. In B and C, n is shown in parentheses.
Consistent with the above observation, Kaplan-Meier survival analysis of the cohort revealed that CR-PA28αOE significantly delayed the premature death of the DRC mice. By 200 days of age, 75% (9 of 12) of the double-Tg mice had died, whereas more than 90% (10 of 11) of the triple-Tg mice were still alive (P < 0.01; Figure 4C).
CR-PA28αOE reduces aberrant protein aggregation in DRC mouse hearts.
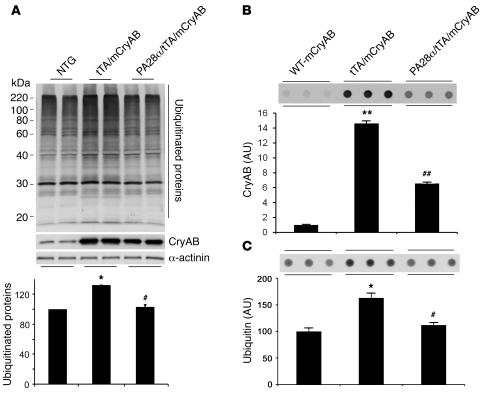
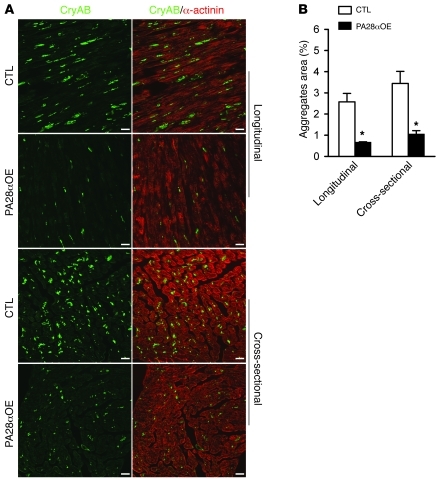
Improving proteasome function in a PFI setting should prevent and/or attenuate aberrant protein aggregation associated with the PFI. To test whether CR-PA28αOE attenuates aberrant protein aggregation in DRC mouse hearts, we used a cohort of mice with the same combinations of genotypes as those for the Kaplan-Meier survival analysis. Western blot analyses show that the total ubiquitinated proteins in ventricular myocardium were significantly increased in tTA/CryABR120G double-Tg mice (P < 0.05), but this increase was significantly attenuated by forced CR-PA28αOE (P < 0.05; Figure 5A). Given that detergent-resistant soluble oligomers are likely the toxic species of aggregation-prone proteins in causing proteinopathy (1), we performed filter trapping assays to quantify the amount of detergent-resistant CryAB- or ubiquitin-positive soluble oligomers in heart muscle tissues (Figure 5, B and C). Compared with myocardium from mice with Tg overexpression of an equivalent or even a higher level of WT CryAB, the amount of filter-trappable CryAB proteins in the tTA/CryABR120G double-Tg mice was markedly increased (P < 0.01). This increase was significantly attenuated by CR-PA28αOE (P < 0.01; Figure 5B). When ubiquitin was probed in the filter trapping assay, similar results were obtained (Figure 5C). Moreover, CryAB aggregates were assessed by immunostaining for CryAB in myocardial sections. In agreement with previous data, CryAB-positive protein aggregates were only detectable in the CryABR120G Tg mouse hearts, not in tTA or PA28α Tg hearts (Supplemental Figure 5). Microscopically visible CryAB-positive protein aggregates in CryABR120G Tg hearts were significantly reduced by CR-PA28αOE (Figure 6). These findings compellingly demonstrated that CR-PA28αOE reduces aberrant protein aggregation in DRC mouse hearts.
Figure 5. PA28αOE reduces CryABR120G-induced aberrant protein aggregation.
(A) Western blot analyses of total ubiquitinated proteins in mouse ventricular myocardium. Representative images and pooled densitometry data (n = 4 mice/group) are shown. Total CryAB protein levels were probed to verify CryABR120G overexpression in the PA28α/tTA/CryABR120G triple-Tg mice. α-actinin was probed as a loading control. *P < 0.05 vs. non-Tg; #P < 0.05 vs. tTA/CryABR120G. (B and C) Ventricular myocardium from WT CryAB Tg, tTA/CryABR120G double-Tg, or PA28α/tTA/CryABR120G triple-Tg mice was processed for the filter trapping assay (see Methods). The proteins retained on the filter were immunoprobed for CryAB (B) or ubiquitin (C). Summarized densitometry data are also shown. *P < 0.05, **P < 0.01 vs. WT CryAB; #P < 0.05, ##P < 0.01 vs. tTA/CryABR120G.
Figure 6. Confocal microscopic analysis of immunofluorescence-stained CryAB aggregates in DRC mouse hearts.
Cryosections of perfusion-fixed ventricular myocardium from PA28α/tTA/CryABR120G triple-Tg mice and tTA/CryABR120G double-Tg controls were used for immunofluorescence staining for CryAB (green) and α-actinin (red). (A) Representative images of longitudinal sections and cross-sections. Scale bars: 20 μm. (B) Morphometric quantification of CryAB-positive protein aggregates in myocardial sections from CryABR120G Tg hearts. *P < 0.05 vs. control.
PA28α knockdown increases, whereas PA28αOE decreases, the stability of CryABR120G overexpressed in cultured cardiomyocytes.
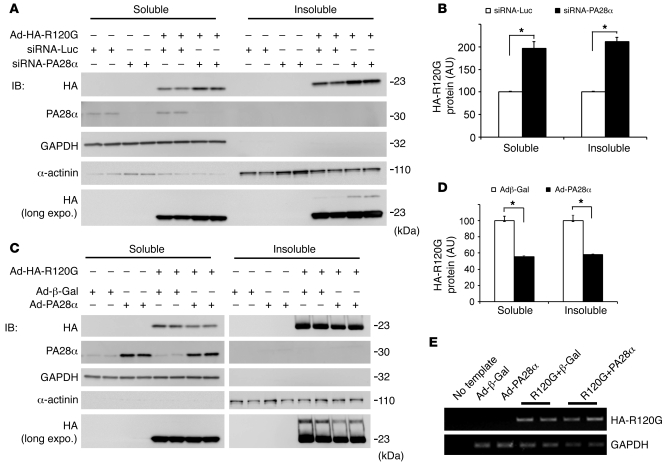
To further test whether the protection of PA28αOE against aberrant protein aggregation in CryABR120G Tg mouse hearts is mediated by enhancing the degradation of misfolded CryABR120G, we examined the effects of genetically induced PA28α loss or gain of function on the protein stability of CryABR120G protein overexpressed in cultured neonatal rat cardiomyocytes (NRCMs). In this in vitro system, the potential feedback from changes in heart function on individual cardiomyocytes — which would occur in intact mice — is avoided; hence, the direct impact of altered PA28α expression can be better evaluated. PA28α loss of function was induced by PA28α knockdown using specific siRNA, whereas PA28α gain of function was achieved by recombinant adenovirus-mediated PA28αOE. Compared with control siRNA transfection, PA28α knockdown significantly increased the steady-state CryABR120G protein level in both the soluble and the insoluble fractions of cultured NRCMs, without changing CryABR120G mRNA expression. Reciprocally, a comparable level of CryABR120G mRNA overexpression resulted in a significantly lower CryABR120G protein level in PA28αOE NRCMs than in control adenovirus–infected (Ad–β-gal) cells (Figure 7). Results of these in vitro experiments further demonstrate that PA28α in cardiomyocytes is capable of negatively regulating the stability of CryABR120G, a bona fide misfolded protein.
Figure 7. Effects of genetic manipulation of PA28α on the stability of a bona fide misfolded protein in cultured NRCMs.
(A and B) PA28α knockdown was achieved via 2 consecutive transfections of siRNA against rat PA28α; siRNA for luciferase (siRNA-Luc) was used as control. (C and D) PA28α or HA-tagged CryABR120G overexpression was achieved by infection of Ad-PA28α and Ad–HA-CryABR120G (Ad-HA-R120G), respectively; Ad–β-gal was used as control. Manipulations of PA28α were performed 24 hours before initiation of HA-CryABR120G overexpression. Cells were collected for protein and RNA extractions 4 days after Ad–HA-CryABR120G infection. (A and C) Representative Western blot (IB) images of the indicated proteins. A longer exposure (long expo.) of IB for HA-tag illustrates that a higher–molecular weight species of HA-CryABR120G in the insoluble fraction was also altered by changing PA28α expression. (B and D) Changes in HA-CryABR120G protein levels by PA28α knockdown (B) or PA28αOE (D). Shown are HA-CryABR120G protein levels normalized with the corresponding in-lane loading control, GAPDH or α-actinin. *P < 0.05. (E) Representative RT-PCR images for HA-CryABR120G in NRCMs. PA28αOE did not alter HA-CryABR120G mRNA levels compared with controls.
Effects of CR-PA28αOE on infarct size and cardiac function of myocardial I/R–injured mice.
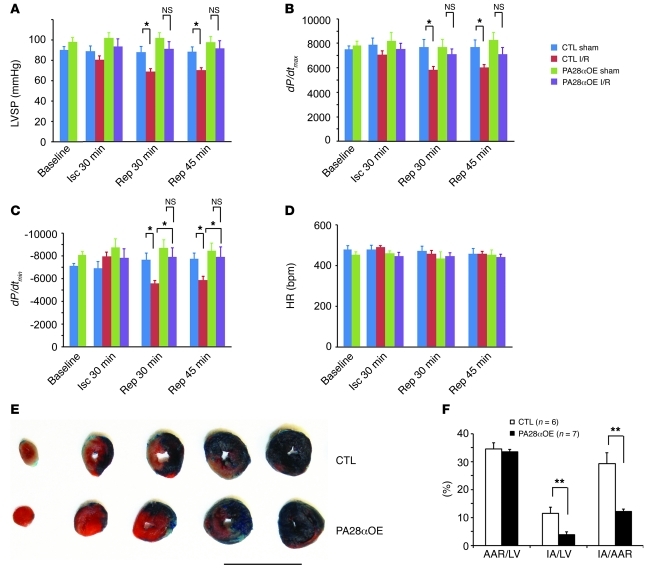
Previous reports have shown impaired proteasome function in hearts subjected to I/R injury (20, 21, 34). Increased oxidative stress is a main known cause of I/R injury (35). Our recent study demonstrated that PA28αOE protects against oxidative stress in cultured cardiomyocytes (23). To test whether CR-PA28αOE affects acute myocardial I/R injury in vivo, we created I/R by ligating the left anterior descending coronary artery for 30 minutes and releasing the ligation for 24 hours in tTA single-Tg control and PA28α/tTA double-Tg mice. We determined infarct size at the terminal experiment and assessed changes in heart function at the early phase of reperfusion. At the end of 30 minutes of ischemia, the peak LV systolic pressure (LVSP) and maximum and minimum dP/dt (dP/dtmax and dP/dtmin, respectively) were not significantly different between groups. However, at 30 and 45 minutes after reperfusion, LVSP, dP/dtmax, and the absolute value of dP/dtmin were significantly lower in the tTA single-Tg I/R group than in the single-Tg sham group. Importantly, these decreases did not occur in the I/R compared with the sham group in PA28α/tTA double-Tg mice (Figure 8, A–C). A comparable I/R procedure caused significantly less myocardial infarct in double-Tg mice with PA28αOE than in single-Tg controls (P < 0.01; Figure 8, E and F). These findings indicate that CR-PA28αOE protects against myocardial I/R injury.
Figure 8. Enhancing cardiac proteasome function protects against myocardial I/R injury.
I/R injuries were created on tTA single-Tg control or PA28α/tTA double-Tg mice by left anterior descending artery ligation (30 minutes) and release. (A–D) A pressure transducer catheter was inserted into LV via the carotid artery, and LV pressure and dP/dt were monitored. Shown are (A) LVSP, (B) dP/dtmax, (C) dP/dtmin, and (D) HR at baseline, 30 minutes after left anterior descending artery ligation (Isc), and 30 and 45 minutes after reperfusion (Rep). n = 6 or 7 mice/group. *P < 0.05. (E and F) Myocardial ischemia and infarct size were assessed at 24 hours of reperfusion. Phthalocyanine blue perfusion after left anterior descending artery religation at the terminal experiment defined the AAR as the area not perfused. Within the AAR, triphenyltetrazolium chloride staining demarcated the infarcted area (IA; white) and viable (red) myocardium. Shown are representative section series images (E) and quantitative data (F). Scale bar: 10 mm. **P < 0.01.
Discussion
The present study demonstrated, for the first time to our knowledge in intact animals, that forced PA28αOE is sufficient to upregulate 11S proteasomes in cardiomyocytes in which overexpressed PA28α binds and stabilizes its partner, PA28β. By monitoring degradation of a surrogate misfolded protein substrate of the UPS, we found that PA28αOE enhanced proteasome proteolytic function in cardiomyocytes in intact mice without causing a concomitant pathology. We thus established what we believe to be a novel inducible Tg mouse model of benign cardiac proteasome functional enhancement. Given the prevalence of proteasome dysfunction in heart disease (3), this mouse model should provide a valuable tool for exploring the therapeutic potential of manipulating cardiac proteasomal activity and for determining the contribution of proteasome dysfunction in cardiac pathogenesis. Using this mouse, we demonstrated that improving proteasome function inhibited aberrant protein aggregation, attenuated cardiac hypertrophy, and delayed premature death in a well-established DRC mouse model, demonstrating that PFI plays a critical role in the genesis of DRC, a bona fide cardiac proteinopathy. We were also able to demonstrate, for the first time to our knowledge, that PA28αOE protected against myocardial I/R injury in intact animals.
An 11S subcomplex can be either a homoheptamer of PA28γ or a heteroheptamer of PA28α and PA28β (26). In the present study, we found that upregulation of the 11S could be achieved by forced CR-PA28αOE in intact mice. We hypothesize that concurrent upregulation between PA28α and PA28β is mediated by mutual stabilization at the protein level. Transcript levels of PA28β were not significantly changed during forced PA28αOE. As shown previously and in the present study, PA28α and PA28β proteins interact with each other. The degradation rate of PA28β was also significantly decreased by PA28αOE in cultured cardiomyocytes (23), which is consistent with the proposed hypothesis. PA28α and PA28β interaction may prevent both from being recognized by the degradation machinery and thus result in mutual stabilization. This is also consistent with a previous report that PA28α protein cannot be detected in PA28β knockout mice (29).
Damaged or misfolded proteins usually undergo conformational changes with exposure of their hydrophobic sequences and subsequent ubiquitination (12). This process is mimicked by the degron CL1 fused GFP (GFPdgn or GFPu). By manipulating PA28αOE in mouse hearts, we demonstrated here that GFPdgn levels were inversely correlated to protein levels of PA28α. The decreases in GFPdgn protein levels were caused by a posttranslational mechanism, because neither the steady-state level nor the polysomal association of GFPdgn mRNA in the heart was significantly altered by CR-PA28aOE. Indeed, in an ancillary study using cultured cardiomyocytes, we found that PA28αOE destabilizes a similarly modified GFP and reduces the steady level of oxidized proteins during oxidative stress (23). These findings suggest that upregulation of the 11S, at least those formed by PA28α and PA28β, can enhance degradation of misfolded proteins in cardiomyocytes. This is further supported by our findings from cultured cardiomyocytes that PA28αOE enhanced the protein degradation of CryABR120G, a bona fide misfolded protein (36).
Consistent with our in vitro findings that the abundance of the bona fide substrate normal proteins of the UPS was not affected by PA28αOE-induced proteasome functional enhancement (23), no significant changes in cardiac gene expression, cardiac growth, cardiac histology, or heart function were observed in the hearts of mice with PA28αOE, which indicates that the intracellular homeostasis of normal proteins is not markedly perturbed by PA28αOE. We conclude that the increase of proteasome proteolytic function by upregulation of the 11S enhances the removal of abnormal proteins, but has little effect on the turnover of normal proteins.
Upregulation of 11S proteasomes via CR-PA28αOE protected against the pathogenesis, and thereby improved the outcome, of a well-documented mouse model of proteinopathy. Expression of CryABR120G, a bona fide misfolded protein, triggers aberrant aggregation and thereby damages the cell via a number of potential mechanisms (14, 37, 38). One of the suspects is PFI, which leads to accumulation of ubiquitinated proteins and further facilitates aberrant protein aggregation, forming a vicious cycle (1). Conceivably, severe PFI can have adverse effects on cell function. It was recently shown that PFI can activate the calcineurin/NFAT pathway and facilitate adverse remodeling of a pressure-overloaded heart in mice (39). We have also recently demonstrated that disruption of the COP9 signalosome induced cardiomyocyte-restricted UPS impairment, which subsequently led to cardiomyocyte death, dilated cardiomyopathy, and premature death in mice (40). Here, PFI attenuation in DRC hearts via CR-PA28αOE broke the vicious cycle between PFI and aberrant protein aggregation; therefore, the steady levels of the ubiquitinated proteins and detergent-resistant oligomers (Figure 5) as well as the abundance of microscopic protein aggregates (Figure 6) were all substantially reduced. Remarkably, enhancement of proteasome function via CR-PA28αOE substantially slowed the progression of CryABR120G-based DRC, as evidenced by decreased cardiac hypertrophy and prolonged survival (Figure 4). These findings revealed a role for PFI in DRC development and the compelling potential of modulating the 11S proteasome as a therapeutic strategy to treat proteinopathies.
Previous studies showed PFI in I/R hearts (20, 21). Here, we found that CR-PA28αOE prevented and/or attenuated I/R injury–induced cardiac malfunction and significantly reduced infarct size, demonstrating that enhancing proteasome proteolytic function in cardiomyocytes protects against acute I/R injury. Increased oxidative stress is a major pathogenic factor in I/R injury (35). We have recently shown that PA28αOE protects against oxidative stress in cultured cardiomyocytes, likely through enhancing the removal of oxidized proteins (23). On one hand, both oxidative stress and oxidized proteins impair proteasome function (21). On the other hand, proteasome impairment slows down the removal of the toxic oxidized proteins, thereby forming a vicious cycle in I/R hearts. Enhancing proteasome function and subsequently protecting against I/R injury interrupted and attenuated the pathogenic cycle. Alternatively, the observed protection of CR-PA28αOE against acute I/R injury may be interpreted as a preconditioning-like effect. This interpretation implies 2 possibilities: first, CR-PA28αOE represents a mild insult to the heart and triggers mechanisms that protect the heart from I/R injury; and second, CR-PA28αOE–induced proteasome functional enhancement acts like a mediator and/or executor of the preconditioning process to counter the pathogenic factors of I/R injury. The data in the present study and reported by others overwhelmingly favor the latter possibility, because (a) preserving proteasome function was recently shown as an important mechanism underlying ischemic preconditioning (20); (b) our comprehensive baseline characterization of mice with CR-PA28αOE during their first year did not detect any adverse effects; and (c) we demonstrated here that the same enhancement of cardiac proteasome function remarkably rescued CryABR120G-based DRC. Hence, this study provides compelling evidence that PFI plays an important role in acute myocardial I/R injury.
The mechanism by which PA28αOE enhances UPS-mediated degradation of abnormal proteins is not clear at this time, but our data support the notion that the 11S particle formed by PA28α and PA28β functions as an alternative activator for 20S and increases proteasome proteolytic activity directed at the abnormal or denatured/misfolded proteins. The 20S core can be capped by 11S at one end and 19S at the other to form a hybrid proteasome, or by 11S at both ends (25, 41). Association of the 11S with the 20S subcomplex stimulates the 20S peptidase activity (25, 41). Upregulation of 11S proteasomes in cardiomyocytes did not cause detectable changes in the protein levels of representative 19S or 20S subunits. Our gel filtration experiments using native myocardial protein extracts revealed that CR-PA28αOE increased the hybrid proteasome and the subpopulation of 20S proteasomes with both ends capped with the 11S. This was further demonstrated by our 20S IP experiments. More PA28α, but less Rpt6 (a bona fide 19S subunit), was co-immunoprecipitated with the α4 subunit of 20S from mouse hearts with CR-PA28αOE compared with that from littermate control hearts without PA28αOE. Our previous in vitro study showed that this increased association of the 11S with the 20S increases ATP-dependent and -independent proteasome peptidase activities in cultured cardiomyocytes (23).
At the molecular level, an important shared pathological change between CryABR120G-based DRC and I/R injury was increased production of misfolded/damaged proteins, which places increased demands on UPS-mediated protein degradation. Like normal proteins, misfolded proteins (with some exceptions) are generally ubiquitinated first and then degraded by the proteasome, in which the 19S is required for uptake of the ubiquitinated proteins. We hypothesize that PA28αOE increases the subpopulation of hybrid proteasomes (i.e., 19S-20S-11S), thereby allowing the UPS to respond to the increased demand. However, the functional significance of the hybrid proteasome has not been formally determined. We speculate that the hybrid proteasome is better equipped to degrade misfolded proteins than is the conventional 26S proteasome (19S-20S-19S or 19S-20S). It was previously shown that the association of 11S increases the peptidase activities of the 20S (42); meanwhile, the associated 19S allows the hybrid proteasome to uptake ubiquitinated protein substrates. The rate-limiting step for UPS-mediated degradation of a native protein is the ubiquitination step, which often requires exposure or posttranslational maturation of its ubiquitination signal. For a misfolded protein, its ubiquitination signals, such as surface exposure of a patch of hydrophobic residues or cryptic ubiquitination signals that are normally buried in properly folded proteins, are born with the misfolding; hence, proteasome is conceivably the rate-limiting step (4). This may explain why the homeostasis of normal proteins is not perturbed in PA28αOE cells and hearts, but remains a hypothesis to be further tested.
Notably, 11S proteasomes can be upregulated by IFN-γ (41, 43), a cytokine that has been clinically used to treat disease (44). IFN-γ also increases expression of the inducible proteasome peptidase subunits (β1i, β2i, and β5i), which leads to the replacement of corresponding conventional peptidase subunits (β1, β2, and β5) and thereby the formation of immunoproteasomes (43). Interestingly, a recent report compellingly demonstrated that upregulation of the immunoproteasome by IFNs not only plays a previously recognized role in helping antigen presentation, but also facilitates the removal of damaged proteins generated by IFN-induced oxidative stress (45). Hence, it will be interesting to test whether upregulation of 11S proteasomes by pharmacological means mitigates the progression of heart disease in a model that displays PFI.
Methods
Tg mouse models.
The cardiac-specific inducible Tg system reengineered by Sanbe et al. (31) was used to create FVB/N Tg mouse lines that allow Dox-regulated CR-PA28αOE. All mouse data were collected at 2 months of age unless indicated otherwise. Where applicable, Dox (0.5 g/l) was administered via drinking water containing 1% sucrose to suppress PA28α Tg expression.
The creation and initial characterization of stable FVB/N Tg mouse lines with cardiomyocyte-restricted overexpression of CryABR120G or WT CryAB (15), and the creation and validation of the GFPdgn Tg mice (10), were described previously.
SDS-PAGE and Western blot analysis.
SDS-PAGE and Western blot were performed as we previously reported (14). IBs used the primary Ab against PA28α, PA28β (Affiniti Research Products), α-actinin (Sigma-Aldrich), RPN2, RPT-6 (BIOMOL), GATA4, GFP (Santa Cruz), Akt, PTEN (Cell Signaling), mouse Psmb5 (customized Ab; ref. 14), or 20S proteasome core subunits (this Ab recognizes 20S α5/α7, β1, β5, β5i, and β7 subunits; BIOMOL).
Reciprocal IP.
Mouse ventricular tissue samples were homogenized in the radioimmunoprecipitation assay buffer as recently described (40). The supernatant was collected, precleared with rabbit serum, and then incubated with an Ab against 20S proteasome α3-subunit (gift of P. Ping, UCLA, Los Angeles, California, USA), anti-PA28α, or anti-PA28β Ab and protein A/G-conjugated agarose beads (protein A/G PLUS-Agarose IP Reagent; Santa Cruz) overnight at 4°C. IP products were then fractionated by SDS-PAGE and detected by IB.
Transcript analysis.
Total RNA was extracted from mouse ventricles using TRI Reagent (Molecular Research Center Inc.). To examine the transcript levels of PA28α or PA28β in the total RNA, Northern blot analyses were employed using p32-labeled PA28α or PA28β probes generated with the nick translation kit (Roche). The mRNA levels of the fetal gene program and GFPdgn transcript levels were measured by RNA dot blot analyses (10). Semiquantitative RT-PCR was carried out using specific primers toward GFPdgn, PA28α, and PA28β for their transcript levels. Relative transcript levels were obtained with normalization to 18S ribosome RNA or GAPDH transcript levels.
Assessment of GFPdgn mRNA polysomal distribution.
Polysomes were isolated from mouse myocardium as previously described (46), with minor modifications. Briefly, ventricular myocardium was washed with cold PBS supplemented with 100 μg/ml cycloheximide and then homogenized in buffer consisting of 10 mM Tris–HCl (pH 7.5), 250 mM KCl, 10 mM MgCl2, 0.5%Triton X-100, 2 mM dithiothreitol, 100 μg/ml cycloheximide, and 2 U RNasin (Promega). After homogenization, Tween-80 (10% v/v) and deoxycholate (5% w/v) were added into the homogenates to further break down membrane structures. The supernatants were collected by centrifugation at 12,000 g for 5 minutes. A total of 15 OD260 units of the lysates were loaded on the top of the 15%–50% (w/v) linear sucrose gradients. A total of 24 fractions (0.5 ml/fraction) were sequentially recovered from the bottom of the sucrose gradients, and for RNA extraction, every 2 adjacent fractions were combined. The RNAs were then extracted with TRIzol reagent and reverse transcribed to cDNA using the SuperScript III First-Strand RT-PCR kit (Invitrogen). The amount of reverse-transcribed cDNA was quantified by semiquantitative PCR analysis.
Gel filtration.
Ventricular tissue homogenates were fractioned by gel filtration following the method previously described (40).
Echocardiography.
Trans-thoracic echocardiography was performed on mice using the VisualSonics Vevo 770 system and a 30-MHz probe as previously described (40).
Filter trapping assay.
Ventricular myocardium was lysed on ice for 30 minutes in 500 μl lysis buffer (50 mM Tris-HCl [pH 8.8], 100 mM NaCl, 5 mM MgCl2, 0.5% [v/v] NP-40, 1 mM EDTA) containing the protease inhibitor cocktail (Roche). After centrifugation for 5 minutes at 10,000 g, pellets containing insoluble materials were resuspended and incubated in 100 μl DNase buffer (20 mM Tris-HCl [pH 8.0], 15 mM MgCl2, 0.5 mg/ml DNase I) at 37°C for 1 hour. Incubations were terminated by adjusting the mixtures to 20 mM EDTA, 2% SDS, followed by boiling for 5 minutes. Using a dot blot apparatus (Bio-Rad), the protein extracts (5 μg) were filtered through nitrocellulose membrane with a pore diameter of 0.22 μm (Millipore). The membrane was then washed twice in PBS supplemented with 0.1% SDS before being immunoprobed for CryAB or ubiquitin (33).
Immunofluorescence confocal microscopy and quantification of CryAB aggregates in mouse hearts.
3 mouse hearts per group were perfusion-fixed with 3.8% paraformaldehyde. Cryosections obtained from 3 representative areas of each heart were immunofluorescence-labeled for CryAB (green) and sarcomeric α-actinin (red) as we previously described (33). 3 fields per representative longitudinal or cross section were imaged. Because mutant CryAB is highly concentrated in the protein aggregates, the imaging gain setting had to be set at such a low level that the images of CryAB-positive protein aggregates were not saturated/overexposed, while the endogenous CryAB staining became invisible. The aggregate areas and the myofibril areas in each acquired image were quantified by MetaMorph software (Molecular Devices). The aggregate areas were presented as the percentage of the myofibril area in which they were located. The primary antibodies used were anti-CryAB (Enzo Life Sciences) and anti–α-actinin (Sigma-Aldrich). Fluorescence-conjugated secondary antibodies were purchased from Molecular Probes.
NRCM culture and recombinant Ad infection.
NRCM culture and recombinant Ad infection were performed as we previously described (14). We previously described the creation and application of the recombinant Ads harboring expressing PA28α (Ad-PA28α) (23), HA-tagged CryABR120G (Ad–HA-CryABR120G) (14), or β-gal (Ad–β-gal) (14).
siRNA transfection in NRCMs.
To knock down PA28α in NRCMs, 2 siRNAs with different target sequences against rat PA28α (siPA28α-1 and siPA28α-2, 50 pmol each) were transfected into NRCMs, using lipofectamine 2000 (Invitrogen) as the transfection reagent. The knockdown effect was achieved at 48 hours and remained up to 120 hours after transfection. The sense strand sequence of siPA28α-1 was gctctcaacgaagccaacc, and that of siPA28α-2 was ggatgggaataattttggc; both were synthesized by Qiagen. Double-stranded siRNAs specific for luciferase (Luc; Qiagen) were used as control siRNA (47).
Myocardial I/R mouse model and infarct size determination.
Myocardial I/R mouse model and infarct size determination were performed as previously reported (48). PA28α/tTA double-Tg mice and littermate tTA single-Tg mice at 8 weeks old were anesthetized with 1% isoflurane and ventilated through intubation via mouth. A slipknot was made around the left anterior descending coronary artery against a PE10 tubing with an 8-0 nylon suture. Sham-operated animals were subjected to the same procedure, except that the suture was not tied. After 30 minutes of ligation, the ligature was released to allow reperfusion. At the end of 24 hours of reperfusion, the suture was retied, and 250 μl of 5% phthalocyanine blue was injected into the LV chamber. The heart was quickly excised, immediately frozen in a –20°C freezer for 30 minutes, and sliced into 5 short-axis sections, which were incubated in 1% triphenyltetrazolium chloride (Sigma-Aldrich) solution for 15 minutes. Each section was weighed and digitally photographed. The area not at risk (phthalocyanine blue stain), the area at risk (AAR; including viable myocardium and infarcted area), and the total LV areas from both sides of each section were measured using Image-Pro plus software. The AAR was expressed as percent of total LV, and infarct size was expressed as percent of the AAR and LV.
LV catheterization and hemodynamic measurements.
Mice were anesthetized and ventilated as described above. The right carotid artery was then cannulated with a high-fidelity 1.4F Millar Mikro-Tip catheter transducer (model SPR-835; Millar Instruments) and advanced to the LV chamber. LV pressure and its first derivatives (dP/dt) were recorded before and during I/R surgery using a Powerlab data acquisition system (ADInstruments). The animal rectal temperature was maintained throughout. From the LV pressure waveforms, heart rate (HR), LVSP, dP/dtmax, and dP/dtmin were all measured with Powerlab software. Hemodynamic parameters were assessed before induction of ischemia and every 15 minutes during the ischemia and reperfusion stages and terminated at 45 minutes after reperfusion.
Statistics.
Unless indicated otherwise, all quantitative data represent mean ± SEM. Where applicable, 2-tailed Student’s t test or 1- or multiple-factor ANOVA was performed for statistical significance tests using Statmost3.6 software. The Holm-Sidak test (1-way ANOVA) or Duncan post-hoc tests (2-way ANOVA) were used for post-hoc comparisons. A P value less than 0.05 was considered statistically significant.
Study approval.
The protocol using animals was approved by the Institutional Committee of Animal Care and Use of University of South Dakota.
Supplementary Material
Acknowledgments
We thank Andrea Jahn for technical assistance in managing colonies and Mark Ranek for assistance in preparation of this manuscript. X. Wang is an Established Investigator of the American Heart Association. This work was supported in part by NIH grants R01HL072166, R01HL085629, and R01HL068936 and by American Heart Association grants 0740025N (to X. Wang) and 0625738Z and 11SDG6960011 (to H. Su).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(9):3689–3700. doi:10.1172/JCI45709.
References
- 1.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99(12):1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 2.Scruggs SB, Ping P, Zong C. Heterogeneous cardiac proteasomes: mandated by diverse substrates? Physiology (Bethesda). 2011;26(2):106–114. doi: 10.1152/physiol.00039.2010. [DOI] [PubMed] [Google Scholar]
- 3.Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C. Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res. 2010;106(3):463–478. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: a quality control perspective. Cardiovasc Res. 2010;85(2):253–262. doi: 10.1093/cvr/cvp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldfarb LG, Dalakas MC. Tragedy in a heartbeat: malfunctioning desmin causes skeletal and cardiac muscle disease. J Clin Invest. 2009;119(7):1806–1813. doi: 10.1172/JCI38027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tannous P, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117(24):3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 8.Sanbe A, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101(27):10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gianni D, et al. Protein aggregates and novel presenilin gene variants in idiopathic dilated cardiomyopathy. Circulation. 2010;121(10):1216–1226. doi: 10.1161/CIRCULATIONAHA.109.879510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumarapeli AR, et al. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. Faseb J. 2005;19(14):2051–2053. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 11.Gilon T, Chomsky O, Kulka RG. Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol. 2000;20(19):7214–7219. doi: 10.1128/MCB.20.19.7214-7219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9(9):679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. Faseb J. 2006;20(2):362–364. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97(10):1018–1028. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, et al. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89(1):84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, et al. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103(19):2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40(4):451–454. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Bahrudin U, et al. Ubiquitin-proteasome system impairment caused by a missense cardiac myosin-binding protein C mutation and associated with cardiac dysfunction in hypertrophic cardiomyopathy. J Mol Biol. 2008;384(4):896–907. doi: 10.1016/j.jmb.2008.09.070. [DOI] [PubMed] [Google Scholar]
- 19.Mearini G, Schlossarek S, Willis MS, Carrier L. The ubiquitin-proteasome system in cardiac dysfunction. Biochim Biophys Acta. 2008;1782(12):749–763. doi: 10.1016/j.bbadis.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Divald A, et al. Myocardial ischemic preconditioning preserves postischemic function of the 26S proteasome through diminished oxidative damage to 19S regulatory particle subunits. Circ Res. 2010;106(12):1829–1838. doi: 10.1161/CIRCRESAHA.110.219485. [DOI] [PubMed] [Google Scholar]
- 21.Bulteau AL, et al. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276(32):30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 22.Predmore JM, et al. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121(8):997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Powell SR, Wang X. Enhancement of proteasome function by PA28a overexpression protects against oxidative stress. FASEB J. 2011;25(3):883–893. doi: 10.1096/fj.10-160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cascio P, Call M, Petre BM, Walz T, Goldberg AL. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. Embo J. 2002;21(11):2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Realini C, et al. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J Biol Chem. 1997;272(41):25483–25492. doi: 10.1074/jbc.272.41.25483. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Krutchinsky A, Endicott S, Realini C, Rechsteiner M, Standing KG. Proteasome activator 11S REG or PA28: recombinant REG alpha/REG beta hetero-oligomers are heptamers. Biochemistry. 1999;38(17):5651–5658. doi: 10.1021/bi990056+. [DOI] [PubMed] [Google Scholar]
- 28.Kloetzel PM. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol. 2004;5(7):661–669. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- 29.Preckel T, et al. Impaired immunoproteasome assembly and immune responses in PA28–/– mice. . Science. 1999;286(5447):2162–2165. doi: 10.1126/science.286.5447.2162. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26(6):843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92(6):609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 32.Drews O, Tsukamoto O, Liem D, Streicher J, Wang Y, Ping P. Differential regulation of proteasome function in isoproterenol-induced cardiac hypertrophy. Circ Res. 2010;107(9):1094–1101. doi: 10.1161/CIRCRESAHA.110.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H, et al. Doxycycline attenuates protein aggregation in cardiomyocytes and improves survival of a mouse model of cardiac proteinopathy. J Am Coll Cardiol. 2010;56(17):1418–1426. doi: 10.1016/j.jacc.2010.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takagi H, et al. Activation of PKN mediates survival of cardiac myocytes in the heart during ischemia/reperfusion. Circ Res. 2010;107(5):642–649. doi: 10.1161/CIRCRESAHA.110.217554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turer AT, Hill JA. Pathogenesis of myocardial ischemia-reperfusion injury and rationale for therapy. Am J Cardiol. 2010;106(3):360–368. doi: 10.1016/j.amjcard.2010.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bova MP, et al. Mutation R120G in alphaB-crystallin, which is linked to a desmin-related myopathy, results in an irregular structure and defective chaperone-like function. Proc Natl Acad Sci U S A. 1999;96(11):6137–6142. doi: 10.1073/pnas.96.11.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maloyan A, et al. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha-B-crystallin desmin-related cardiomyopathy. Circulation. 2005;112(22):3451–3461. doi: 10.1161/CIRCULATIONAHA.105.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajasekaran NS, et al. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130(3):427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang M, et al. Proteasome functional insufficiency activates the calcineurin–NFAT pathway in cardiomyocytes and promotes maladaptive remodelling of stressed mouse hearts. Cardiovasc Res. 2010;88(3):424–433. doi: 10.1093/cvr/cvq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su H, et al. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108(1):40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J Biol Chem. 2000;275(19):14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 42.Stohwasser R, Salzmann U, Giesebrecht J, Kloetzel PM, Holzhutter HG. Kinetic evidences for facilitation of peptide channelling by the proteasome activator PA28. Eur J Biochem. 2000;267(20):6221–6230. doi: 10.1046/j.1432-1327.2000.01706.x. [DOI] [PubMed] [Google Scholar]
- 43.Murata S, et al. Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. Embo J. 2001;20(21):5898–5907. doi: 10.1093/emboj/20.21.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Liu X. The role of interferon gamma in regulation of CD4+ T-cells and its clinical implications. Cell Immunol. 2009;254(2):85–90. doi: 10.1016/j.cellimm.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Seifert U, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142(4):613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi S, et al. Diminished GATA4 protein levels contribute to hyperglycemia-induced cardiomyocyte injury. J Biol Chem. 2007;282(30):21945–21952. doi: 10.1074/jbc.M703048200. [DOI] [PubMed] [Google Scholar]
- 47.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109(3):296–308. doi: 10.1161/CIRCRESAHA.111.244707. [published online ahead of print June 9, 2011]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohl S, Medway DJ, Schulz-Menger J, Schneider JE, Neubauer S, Lygate CA. Refined approach for quantification of in vivo ischemia-reperfusion injury in the mouse heart. Am J Physiol Heart Circ Physiol. 2009;297(6):H2054–H2058. doi: 10.1152/ajpheart.00836.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.