Abstract
Two functionally related genes, FOXP2 and CNTNAP2, influence language abilities in families with rare syndromic and common nonsyndromic forms of impaired language, respectively. We investigated whether these genes are associated with component phenotypes of dyslexia and measures of sequential motor ability. Quantitative transmission disequilibrium testing (QTDT) and linear association modeling were used to evaluate associations with measures of phonological memory (nonword repetition, NWR), expressive language (sentence repetition), reading (real word reading efficiency, RWRE; word attack, WATT), and timed sequential motor activities (rapid alternating place of articulation, RAPA; finger succession in the dominant hand, FS-D) in 188 family trios with a child with dyslexia. Consistent with a prior study of language impairment, QTDT in dyslexia showed evidence of CNTNAP2 single nucleotide polymorphism (SNP) association with NWR. For FOXP2, we provide the first evidence for SNP association with component phenotypes of dyslexia, specifically NWR and RWRE but not WATT. In addition, FOXP2 SNP associations with both RAPA and FS-D were observed. Our results confirm the role of CNTNAP2 in NWR in a dyslexia sample and motivate new questions about the effects of FOXP2 in neurodevelopmental disorders.
Keywords: Written language, Spoken language, Modality-specific motor sequencing, Quantitative transmission disequilibrium, Linear modeling
Introduction
CNTNAP2 is a gene on chromosome (chr) 7 that encodes a protein belonging to the neurexin family. Mutations in CNTNAP2 cause an autosomal recessive syndrome with mental retardation and seizures (Strauss et al. 2006), and the gene has also been implicated in autism (Alarcon et al. 2008; Arking et al. 2008; Bakkaloglu et al. 2008) and language impairment (Vernes et al. 2008). Language impairment is a disorder affecting comprehension and formulation of spoken language in the absence of hearing impairment, neurological impairment, or psychiatric disturbance (Tomblin et al. 1997; Rice et al. 2009). Difficulty with nonsense-word repetition (NWR), a task that requires reproducing a pronounceable but meaningless word in response to a spoken model, is a robust endophenotype of language impairment (Newbury et al. 2005). The role of CNTNAP2 in language impairment was recently investigated with quantitative disequilibrium testing (QTDT) in 184 families with common forms of the disorder. Results showed that CNTNAP2 was significantly associated with NWR. CNTNAP2 was also associated with composite measures of expressive and receptive language ability, but to a much smaller extent than with NWR (Vernes et al. 2008). Thus, CNTNAP2 may be associated with processes underlying phonologic memory and other developmental disabilities, which include but are not restricted to language impairment.
The rationale to study the effects of CNTNAP2 in language impairment was based on the discovery that this gene is regulated by another gene, FOXP2, also located on chr 7 (Vernes et al. 2008). Mutations in FOXP2 have been shown to cause severe but rare syndromic deficits in language comprehension and expression, concomitant with apraxic speech and orofacial praxis deficits including simultaneous and sequential movements (Alcock et al. 2000; Lai et al. 2001; MacDermot et al. 2005). FOXP2 sequence or structural changes, however, have not been identified in common nonsyndromic forms of language impairment (Newbury et al. 2002). In a family sample with language impairment (Rice et al. 2009), neither linkage nor association analyses provided evidence for FOXP2 influences on NWR. Similarly, even though FOXP2 mutations cause severe syndromic speech difficulties; they are seen very rarely in common nonsyndromic forms of speech sound disorder (MacDermot et al. 2005), defined as difficulty developing speech that can be readily understood, due to speech sound omissions, distortions, or substitutions, in the absence of known causes (Pennington and Bishop 2009). FOXP2, hence, appears to be necessary but not sufficient for the development of typical language and speech abilities.
In animal studies, FOXP2 has been found to play an important role in a variety of behaviors related to vocalization and motor sequencing. For instance, songbirds require upregulation of FOXP2 in a crucial basal ganglia region during the song-learning phase (Teramitsu et al. 2004). In zebra finches with FOXP2 knocked down, the song imitations are incomplete and inaccurate (Haesler et al. 2007). In mice with an induced FOXP2 mutation analogous to that causing speech and language impairments in humans, homozygotes showed severe cerebellar growth reductions and heterozygotes had deficits in motor learning as measured by rapid motor sequencing on running wheels, along with impaired and abnormal plasticity in striatal and cerebellar circuits (Groszer et al. 2008). These findings suggest that FOXP2 might play a role in sequential motor ability across motor systems. It is unknown whether its transcriptional target CNTNAP2 influences motor processes.
Dyslexia is a specific disability that interferes with the acquisition of written language at the word level, characterized by deficits in accurate and/or fluent word recognition, decoding, and spelling in the presence of grossly normal oral language skills (Lyon et al. 2003; Shaywitz and Shaywitz 2003). Difficulty with phonological processing is one of the hallmarks of dyslexia. Multiple groups, including ours, have observed that NWR performance is frequently impaired in children and adults with this disorder (Kamhi and Catts 1986; Hulslander et al. 2004; Ramus et al. 2003; Szenkovits and Ramus 2005; Brkanac et al. 2008). Because NWR deficit is a central characteristic of language impairment, it could be argued that the NWR deficits observed in dyslexia reflects concurrent language problems. However, NWR probes a number of cognitive component abilities including phonological awareness, which is a core requisite for reading. Furthermore, the dyslexia family subject set from which our sample was drawn specifically excluded comorbid low language ability (Berninger et al. 2001a, 2006). Our previous aggregation study in this sample showed that NWR was among the traits with the strongest evidence for a genetic basis (Raskind et al. 2000). The fact that NWR deficits persist into adulthood facilitates investigation of NWR in family-based dyslexia genetics studies. In our genome-wide linkage scan for NWR in a family sample with dyslexia (Brkanac et al. 2008), a locus on chr 7q at 190.9 cM approached but did not quite reach status as a region of interest. The variance component maximum LOD signal maximized approximately 23 and 57 cM from CNTNAP2 and FOXP2, respectively. It is thus unlikely that either gene contributed to that signal.
Another type of deficit, difficulty with sequential finger movements, characterizes dyslexia and other neurodevelopmental disorders (Denckla 1973; Wolff et al. 1983). Finger motor sequencing was also shown to predict writing problems in an unreferred sample of typically developing children (Berninger and Rutberg 1992). Thus, there appears to be an underlying association between literacy skill and sequential finger movements.
As recently reviewed (Pennington and Bishop 2009; Peterson et al. 2007; Lewis et al. 2006), many children who are diagnosed with language impairment and/or speech sound disorder in early childhood show difficulty learning to read and spell once they enter school (Catts et al. 2002; Tomblin et al. 2000). Problems with oral language, speech, and reading can occur in isolation or co-occur in the same child, although not all observed reading problems are well captured by the dyslexia definition provided by the International Dyslexia Association (Lyon et al. 2003) that is focused on impairments in word-level reading and spelling skills.
The presence of comorbidity and shared impairments on some component traits in dyslexia, language impairment, and speech sound disorder, along with the functional relationship between CNTNAP2 and FOXP2, provided a rationale to investigate the possibility of CNTNAP2 and FOXP2 associations with performance on measures of NWR, written language, and sequential motor abilities in families ascertained through a child with dyslexia. Specifically, we investigated (a) whether CNTNAP2 and/or FOXP2 are associated with NWR, sentence repetition (SR), and measures of reading in a family sample with dyslexia, which would replicate findings from the family study of language impairment (Vernes et al. 2008) and extend them to dyslexia; and (b) whether CNTNAP2 and/or FOXP2 variants are associated with motor sequencing abilities not only in oral but also in finger tasks, which might imply a more systemic involvement in motor systems beyond the oral system. Based on the finding that language deficits are associated with FOXP2 in rare cases and CNTNAP in common cases, our expectation for the dyslexia sample was that CNTNAP2 variants would be more likely than FOXP2 variants to show associations with phenotypes related to spoken and written language as well as sequential motor tasks.
Methods
Participants
Data for this study were collected by the University of Washington Learning Disabilities Center (UWLDC) and with the approval of the University of Washington’s Human Subjects Institutional Review Board. The participant recruitment process and the phenotyping and genotyping procedures have been described in detail elsewhere (Berninger et al. 2006; Raskind et al. 2000). Each participating family was identified through a proband child in grades 1 through 9 who met dual criteria of discrepancy from the Verbal Comprehension Index (prorated verbal IQ, VIQ) and low achievement (below the population mean and typically well below) for dyslexia in accuracy or rate of orally reading pseudowords, real words, or passages, or spelling and/or for dysgraphia in automatic alphabet letter writing or spelling. The VIQ-reading discrepancy criterion is not sufficient for a clinical diagnosis of dyslexia because identification of hallmark phenotype impairments is also required (Berninger et al. 2001b), but IQ-discrepant reading deficits are more likely to provide significant heritability estimates, compared to reading deficits without such a discrepancy (Olson et al. 1999). The probands’ average word reading and decoding skills on nationally normed measures were more than .67 SD below the population mean and ranged from 1.5 to 1.8 SD below their VIQ. On average, probands met inclusionary criteria on 6.0 of 10 measures of reading and spelling and on 4.1 of 6 measures of writing (Berninger et al. 2006). To participate in the study, the proband was required to have a VIQ of at least 90 on the Wechsler Intelligence Scale for Children-3rd Edition (Wechsler 2008), to exclude children with a higher probability of having an undiagnosed neurologic or developmental disorder. Children with neurological or psychiatric disorders or other medical conditions, or preschool history of significant difficulty in learning language or other developmental milestones, which could have impaired their reading or writing for reasons other than dyslexia or dysgraphia were excluded, except for attention deficit/hyperactivity disorder. The protocol for the UWLDC study included measures of verbal reasoning/comprehension, reading and writing and phenotypes known to be associated with dyslexia or dysgraphia organized in a working memory architecture (Berninger et al. 2006). In addition, blood samples were collected from participants for DNA analyses. For the present study, 188 family trios from separate families of predominantly Caucasian ethnic background were selected. Per parent report, the probands were of the following ethnic backgrounds: 92.0% Caucasian, 2.1% Asian, 1.1% African American, 1.1% Native American, .5% Hispanic, and 3.2% other or unknown. These trios were the same as those reported in a previous study on associations of dyslexia to the DYX1 locus and DYX1 candidate genes (Brkanac et al. 2007). Tests were administered by graduate student research assistants. All were from an APA-accredited and NASP-approved school psychology program and had completed assessment courses. Many had school psychology certification and were trained and closely supervised by the family genetics clinical coordinator, a licensed or license-eligible psychologist, and the PI of the clinical core who is a licensed psychologist.
Phenotypes
Six phenotypes (Table 1) were tested for SNP association with CNTNAP2 and/or FOXP2 polymorphisms. To evaluate the role of NWR, we selected the Nonword Repetition subtest from the Comprehensive Test of Phonological Processing (Wagner et al. 1999). In this task, participants hear a pronounceable but meaningless pseudoword and imitate it once. This task requires storage and processing of the phonological information in short-term memory. SR, also referred to as sentence imitation, is considered a sensitive measure of expressive language ability and difficulty with this task is one of the psycholinguistic markers of language impairment (Conti-Ramsden et al. 2001). It is one of the subtests contributing to the composite measure of expressive language reported in the language impairment study by Vernes et al. (2008). The task requires reproduction of sentences verbatim in response to a given model, once for each sentence. For the purposes of this study, the Sentence Memory subtest from the Stanford-Binet Intelligence Scale (Thorndike et al. 1986) was selected as a measure of SR. The score reflects the ability to reproduce sentences of increasing length and complexity. Performance in this task depends on memory factors in the context of lexical meaning and syntactic frameworks and it offers an interesting theoretical contrast with memory for nonwords as captured with NWR, where lexical meaning or syntactic frameworks are absent.
Table 1.
Descriptive statistics for proband and parent groups
| Age and Phenotype | Population mean (SD) | N Probands | Proband mean (SD) | N Parents | Parent mean (SD) |
|---|---|---|---|---|---|
| Age (months) | N/A | 188 | 129 (22) | 376 | 525 (59) |
| NWR | 10 (3) | 103 | 7.97 (1.73) | 202 | 7.09 (1.81) |
| SR | 50 (8) | 102 | 50.88 (8.72) | 198 | 47.78 (7.99) |
| RWRE | 0 (1) | 188 | −0.69 (1.18) | 372 | 2.14 (1.00) |
| WATT | 100 (15) | 188 | 85.31 (12.11) | 371 | 101.45 (10.88) |
| RAPA | 0 (1) | 103 | −0.30 (1.50) | 199 | −0.14 (1.23) |
| FS-D | 0 (1) | 101 | 0.09 (1.06) | 200 | 0.72 (0.75) |
Norms for RWRE are based on a prepublication version of the TOWRE (Thorndike et al. 1986) and norms for RAPA and FS-D are based on UWLDC measures (Catts et al. 2002). Because the assessment battery was modified during the 10 years in which the participants were evaluated, not all participants were tested for all the measures
To evaluate associations with reading ability, we selected two complementary measures of reading. Rate of real word reading, here termed real word reading efficiency (RWRE), was evaluated with a subtest of the prepublication version of the Test of Word Reading Efficiency (TOWRE) (Torgesen et al. 1999), on which participants orally read as many real words as possible within 45 s. The published version of this subtest is titled Sight Word Efficiency. Accuracy of nonword reading was evaluated with the Word Attack subtest (WATT) of the Woodcock Reading Mastery Test–Revised (Woodcock 1987) on which participants orally read pronounceable but meaningless nonwords from a list under untimed conditions. A number of previous studies including ours (Berninger et al. 2006; Raskind et al. 2005) confirmed that both of these measures detect core impairments in individuals with dyslexia and have a strong genetic component.
Motor sequencing during a speech-like activity was evaluated with a motor speech task requiring rapid repetition of the syllable sequence “pataka” ten times (Fletcher 1978). This measure assesses rapid alternating place of articulation (RAPA) of the three stop consonants (bilabial, alveolar, velar) in this sequence. Performance on this measure (total time for all the repetitions) contributed uniquely to oral passage reading in the children (rate) and adults (accuracy) with dyslexia in a phenotyping study (Berninger et al. 2008a). An analogous measure of motor sequencing for a hand task is the finger succession-dominant hand (FS-D) task (Berninger and Rutberg 1992). FS-D involves timed, sequential finger maneuvers, requiring that participants perform five repetitions of sequential finger to thumb touches with index to middle finger to ring finger, and then pinky. The score in the current study is the total time required for the dominant hand. Because the data were collected under two slightly different protocols, measures for NWR, SR, RAPA, and FS-D were not always available for participants who had completed RWRE and WATT (see Ns in Table 1). Norms for RAPA and FS-D were based on UWLDC measures (Berninger et al. 2006) and norms for RWRE, on a prepublication version of the test. Table 1 summarizes descriptive statistics for each phenotype in the two groups. Several differences between the proband and parent cohorts are noted. The probands had low reading scores whereas the parents had average WATT scores and above-average RWRE scores. The probands achieved FS-D scores near the population mean whereas the parents achieved higher scores. These differences may in part result from the fact that the probands but not the parents were selected for low reading ability. Another consideration is the fact that the test instruments were normed on child and young adult samples, and it is possible that performance on timed measures of reading and finger sequencing increases in adults beyond the highest available norm age. Norming samples covered age 5 through 24 years for NWR, age 3 through 23 for SR, age 6 through 24 for RWRE, grades K through 12 for WATT, grades K through 6 for FS-D, and age 6 through 13 for RAPA. Table 2 shows power to detect biased transmission and association for each phenotype and lists the minimum genetic effects under each of three modes of inheritance, given the sample size, 80% power, p values of 0.05, and two-sided tests.
Table 2.
Minimum genetic effects in variable units and z scores (power = 0.8, p = 0.05, two-sided test) to detect biased transmission and association in the given sample
| Phenotype | Minimum genetic effect for QTDT | Minimum genetic effect for LAM | ||||
|---|---|---|---|---|---|---|
| Additive | Dominant | Recessive | Additive | Dominant | Recessive | |
| NWR | 1.6 (0.53) | 2.2 (0.73) | 2.4 (0.80) | 0.8 (0.27) | 1.0 (0.33) | 1.7 (0.57) |
| SR | 4.3 (0.54) | 5.8 (0.73) | 6.5 (0.81) | 2.0 (0.25) | 2.6 (0.33) | 3.0 (0.38) |
| RWRE | 0.4 (0.40) | 0.6 (0.60) | 0.7 (0.70) | 0.2 (0.20) | 0.3 (0.30) | 0.4 (0.40) |
| WATT | 6.1 (0.41) | 8.3 (0.55) | 9.1 (0.61) | 2.9 (0.19) | 4.0 (0.27) | 4.0 (0.27) |
| RAPA | 0.6 (0.60) | 0.8 (0.80) | 0.8 (0.80) | 0.3 (0.30) | 0.4 (0.40) | 0.4 (0.40) |
| FS-D | 0.6 (0.60) | 0.8 (0.80) | 0.9 (0.90) | 0.3 (0.30) | 0.4 (0.40) | 0.4 (0.40) |
Genotyping
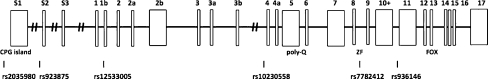
Of the nine CNTNAP2 SNPs with the strongest statistical associations in the language impairment family study (Vernes et al. 2008), five were in complete linkage disequilibrium with neighboring SNPs in this sample. Thus, only rs851715, rs2710102, rs1723623, and rs4431523, all located between exons 13 and 14, were retained for further analysis. In addition, six highly informative SNPs were selected as tags for the following genomic environments of the FOXP2 gene (Fig. 1): rs2035980, rs923875: promoter region and CpG island upstream from exon 1; rs12533005: transcription start site; rs10230558: polyQ region between exons 7 and 8; rs7782412, rs936146: zinc finger region. Two forkhead box SNPs, rs12154391 and rs12705970, were found to be in high linkage disequilibrium with one SNP in the zinc finger region and were excluded from the analysis.
Fig. 1.
Exon architecture of FOXP2 (not drawn to scale) and position of selected SNPs. ZF zinc finger. Exons longer than 200 bp are represented with wide rectangles. Structure adapted from recent studies of FOXP2 elements (Bruce and Margolis 2002; Schroeder and Myers 2008)
In both the proband and parent cohorts, SNP genotypes showed two blocks of correlated SNPs for each of the two selected genes. The following FOXP2 SNPs were mutually significantly correlated: rs12533005, rs10230558, rs7782412, and rs936146. The absolute magnitude of the correlation coefficients in neighboring SNPs ranged from .34 to .63 in the probands and from .35 to .61 in the parents (all p values <.0001). This indicates that the two SNPs near the promoter and CPG island were neither in linkage disequilibrium with each other nor with the remaining SNPs in this gene, whereas the SNPs located near the transcription start site, PolyQ region, zinc finger region, and, by inference, the forkhead box region, were significantly correlated as linear pairs. The four selected CNTNAP2 SNPs were significantly correlated, with absolute correlation coefficients between neighboring SNPs ranging from .64 to .83 in the probands and from .70 to .81 in the parents (all p values <.0001). This pattern is not surprising, given that these SNPs are all located between exons 13 and 14.
All SNPs were in Hardy–Weinberg equilibrium, as calculated in the parent cohort only and using an exact test of the probability of observing the number of heterozygotes conditional on the number of minor allele copies, as implemented in PEDSTATS (Wigginton and Abecasis 2005). Table 3 lists the SNPs, their physical positions (dbSNP130, NCBI Build 36), minor allele frequencies in the parent cohort, and p values from Hardy–Weinberg equilibrium testing in the parent cohort.
Table 3.
Summary of selected FOXP2 and CNTNAP2 SNPs
| Gene | Marker, AB ID | Physical position (dbSNP130) | Genetic environment | SNP alleles (Major/Minor) | Minor allele frequency | Hardy–Weinberg test p value |
|---|---|---|---|---|---|---|
| FOXP2 | rs2035980, hCV1189578 | 113495736 | Promoter, CPG Island | T/C | 0.3630 | 0.2215 |
| rs923875, hCV1115584 | 113522272 | Promoter, CPG Island | A/C | 0.4348 | 0.4624 | |
| rs12533005, hCV220195 | 113843291 | Transcription Start Site | G/C | 0.4535 | 0.0963 | |
| rs10230558, hCV1604741 | 114032985 | PolyQ Region | A/T | 0.4880 | 0.6047 | |
| rs7782412, hCV1604757 | 114077651 | Zinc Finger | T/C | 0.4420 | 0.5996 | |
| rs936146, hCV7598355 | 114081641 | Zinc Finger | G/C | 0.4144 | 0.1657 | |
| CNTNAP2 | rs851715, hCV2669966 | 147157839 | Betw. Exons 13 and 14 | A/G | 0.3413 | 1.0000 |
| rs2710102, hCV11430714 | 147205323 | Betw. Exons 13 and 14 | C/T | 0.4827 | 0.9181 | |
| rs17236239, hCV34470590 | 147213238 | Betw. Exons 13 and 14 | A/G | 0.3493 | 0.8202 | |
| rs4431523, hCV32080083 | 147228099 | Betw. Exons 13 and 14 | T/C | 0.3293 | 0.8150 |
Following the TaqMan Genotyping Master Mix Protocol for a 10-μL reaction, PCR amplification and allele identification were carried out with an ABI 7500 Real-Time PCR system and ABI Sequence Detection Software Version 1.31. The reaction mixture contained 5 μL of TaqMan Genotyping Mix, 2.5 μL of water, 0.5 μL of an AB Validated TaqMan SNP Genotyping Assay probe mixture, and 10 ng of DNA. Enzyme activation occurred at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, and annealing and extension at 60°C for 1 min. PEDSTATS (Wigginton and Abecasis 2005) was used to check for genotyping errors and pedigree inconsistencies.
Statistical analysis
To explore associations among the selected traits, pairwise correlations were calculated. Because of the familial relationships among the participants, these calculations were carried out separately in the parents and in the probands. A Bonferroni correction was used to correct for 15 comparisons in two groups, arriving at testwise significance level of alpha = .0017.
For the association analyses, relevant covariates for each phenotype variable were explicitly determined using linear regression modeling. Age and sex have previously been found to affect many of the variables in the family participant set from which this sample was drawn, despite age adjustments in the normed test instruments and were, therefore, included as covariates in our previous studies (Brkanac et al. 2008; Raskind et al. 2000; Chapman et al. 2004). For this reason, age in months, sex, and an interaction between age and sex were evaluated for significant contributions to the score distributions of each of the selected phenotype variables, separately in the probands and parents. For the proband cohort, age was identified as a minor but statistically significant covariate for RWRE and RAPA. For the parent cohort, sex was identified as a covariate for NWR, SR, RWRE, and WATT, and age was also a minor but statistically significant covariate for NWR and SR. All measures had been preadjusted for age but not sex. One plausible reason for the need to add age as a covariate is that test norms, as mentioned, were based on child or young adult data and standard scores for adults were typically based on the highest normed age.
Because the data were collected at different time points under slightly different phenotyping protocols, not all tasks were completed by all families. In general, presence or absence of data in a given variable affected all members of a trio equally, because the phenotype was not assessed due to protocol differences. Trios with missing data for all three members were dropped from the statistical analysis. In rare cases, data were not available for a parent. For instance, in four trios, NWR data were not available for one of the parents. Missing parental phenotype data have no effect on QTDT, and in the linear association models, these missing datapoints were dropped.
To replicate the design in the language impairment study of CNTNAP2 (Vernes et al. 2008), preferential SNP allele transmission for the selected phenotypes was evaluated with a family-based QTDT as implemented in MERLIN (Abecasis et al. 2002), selecting the Abecasis orthogonal model. This model depends only on within-family variance and a linear term describing parental mating type. The following alternative hypotheses are specified in terms of mating type (Ewens et al. 2008):
 |
where
- Y
is the quantitative phenotype
- μ
is the population mean
- α
is the linear term describing the expected mean offspring phenotype based on the parental genotypes and an additive model for the number of risk alleles
- W
denotes the number of excess risk alleles in a child
- E
is a random residual term with mean zero and variance σ2 and the null hypothesis being tested is β = 0.
Because this model is based on the genotypes of parents and offspring but the phenotypes of only the offspring, covariates were selected based on their relevance to the probands only. Consequently, RWRE and RAPA were adjusted for participant age. Parental phenotypes were explicitly excluded from the analysis. We generated empirical p values for the within-family component w of the association, defined as excess transmission of the allele arbitrarily coded as “1” (Abecasis et al. 2000), as implemented in MERLIN (Abecasis et al. 2002). In this permutation framework, each within-family component w is randomly replaced with itself or with its mirror image, −w, with equal probability (Abecasis et al. 2000).
Because our sample consisted predominantly of families of Caucasian descent as per parent report, we were able to add more powerful analysis tools based on linear association modeling (LAM) that were not restricted by the presence of population stratification. LAM is based on simple linear regression of phenotypes on genotypes and has the advantage of greater statistical power to detect association, compared to QTDT, because genotypes and phenotypes of all participants can be considered, whereas QTDT disregards all double homozygous parental mating types and all parental phenotypes. Associations were evaluated using PLINK version 1.06 (Purcell et al. 2007). In these procedures, covariates with relevance for each of the two cohorts and for each phenotype were incorporated. When LAM was run on the genotypes and phenotypes from the entire sample, the procedure was corrected for family structure by permuting parental phenotypes (Purcell et al. 2005), as implemented in PLINK. To further investigate differences between the two cohorts, LAM was conducted in the proband and parent cohort separately. All empirical p values in the QTDT and association testing were based on 100,000 permutations.
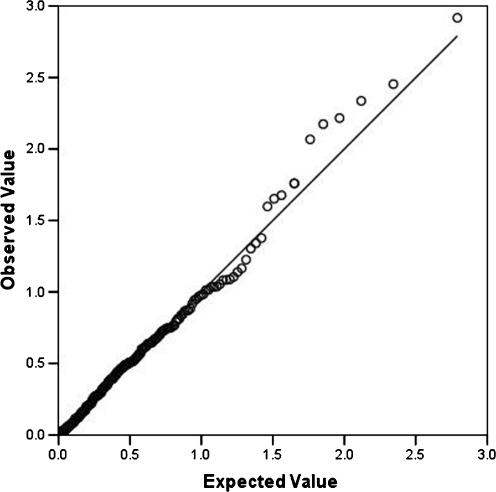
Given the lack of independence among the selected phenotypic traits and also among the SNPs, as documented with significant correlations, Bonferroni adjustments are unnecessarily stringent. Therefore, empirical significance was determined using Monte-Carlo global assessments of significance (McIntyre et al. 2000), as implemented in MERLIN (Abecasis et al. 2002), for an experimentwise alpha of.05. To evaluate the role of CNTNAP2 in the six phenotypes, a single test p value of.0027 was required (.0014 for each cohort separately). To evaluate the role of FOXP2 in these traits, a single test p value of .0015 was required (.0008 for each cohort separately). To further evaluate the distribution of the p values and their conformity to the expected distribution under the null hypothesis, they were compared to a distribution of –log10 p values expected under the null hypothesis of no association, using a QQ plot. The Monte-Carlo assessment of significance takes the lack of independence among the genotypes into account but not the correlations among the phenotypes, justifying the use of a QQ plot as an additional indication of significance.
Results
Pairwise correlation calculations among the six selected traits (Table 4) showed several statistically significant correlations of moderate magnitude. In the probands as a group, four of the 15 pairwise correlations were statistically significant. In the parent group, which consisted of twice as many participants as the proband group, nine of the 15 pairwise correlations were statistically significant. All correlations except one were of the same sign in the parents and children, even if they were not statistically significant. Overall, ten of the 13 significant correlations involved measures of reading ability. In the probands, neither RAPA nor FS-D were correlated with each other or any of the reading, nonword, or sentence repetition tasks, but in the parents, RAPA was significantly correlated with FS-D. It was also correlated with RWRE, which is consistent with previous findings (Berninger et al. 2008a). Also in the parents, FS-D was correlated with WATT, a different reading skill shown to be related to poor spelling (Garcia et al. in press), the persistent impairment in adults with a history of dyslexia (Berninger et al. 2008a).
Table 4.
Pairwise correlation coefficients (and corresponding nominal asymptotic p values) for the selected traits
| NWR | SR | RWRE | WATT | RAPA | FS-D | |
|---|---|---|---|---|---|---|
| NWR | 0.3626 | 0.2516 | 0.3662 | 0.1559 | 0.1336 | |
| (<0.0001) | (0.0003) | (<0.0001) | (0.0283) | (0.0607) | ||
| SR | 0.2058 | 0.2428 | 0.3830 | 0.1165 | −0.0282 | |
| (0.0380) | (0.0006) | (<0.0001) | (0.1040) | (0.6946) | ||
| RWRE | 0.2723 | 0.3110 | 0.5079 | 0.2305 | 0.1922 | |
| (0.0054) | (0.0015) | (<0.0001) | (0.0011) | (0.0064) | ||
| WATT | 0.3322 | 0.5374 | 0.5022 | 0.2171 | 0.2344 | |
| (0.0006) | (<0.0001) | (<0.0001) | (0.0021) | (0.0008) | ||
| RAPA | 0.1001 | 0.1770 | 0.1710 | 0.2858 | 0.3407 | |
| (0.3142) | (0.0751) | (0.0841) | (0.0034) | (<0.0001) | ||
| FS-D | 0.1781 | 0.1283 | 0.2396 | 0.1375 | 0.1433 | |
| (0.0748) | (0.2033) | (0.0158) | (0.1704) | (0.1529) |
Data from the parents are listed in the upper right triangle and data from the probands, in the left lower triangle. Statistical significance is indicated by bold font
Empirical p values from all association tests with values <.05 are listed in Table 5. One CNTNAP2 SNP, rs2710102, was associated with NWR at p = .0174, as calculated with QTDT. This association was nominally significant. Although it did not reach the adjusted alpha threshold of.0027, it exceeded expectation under the null hypothesis as seen in the QQ plot (Fig. 2). No other CNTNAP2 association with a p value <.05 was found in any of the selected phenotypes.
Table 5.
Association results
| Gene | Task | Marker | Risk Allele | QTDT | LAM (Parental Phenotypes Permuted) | LAM (Probands Only) | LAM (Parents Only) |
|---|---|---|---|---|---|---|---|
| CNTNAP2 | NWR | rs2710102 | Minor | 0.0174 | |||
| FOXP2 | NWR | rs7782412 | Major | 0.0174 | |||
| rs936146 | Minor | 0.0067 | |||||
| RWRE | rs923875 | Major | 0.0046 | ||||
| rs7782412 | Major | 0.042 | 0.0086 | 0.0012 | |||
| RAPA | rs12533005 | Major | 0.0061 | ||||
| rs10230558 | Major | 0.0498 | |||||
| FS-D | rs923875 | Major | 0.0211 | 0.0035 | |||
| rs12533005 | Major | 0.0223 | |||||
| rs7782412 | Major | 0.0456 | |||||
| rs936146 | Major | 0.0253 |
Nominal empirical p values <.05 are reported; bold italicized type indicates significance based on QQ evidence
Fig. 2.
QQ plot of quantiles from observed -10log values of the empirical p values compared to expected values under the null hypothesis
Regarding the role of FOXP2 in NWR and SR, QTDT and LAM resulted in several NWR associations. NWR was associated with rs7758412 at p = .0174 as tested with QTDT. LAM showed an NWR association with rs936146 in the proband cohort at p = .0067. These results met nominal and QQ criteria for significance. No FOXP2 SNP showed an association with SR that met nominal, QQ, or adjusted alpha criteria for significance.
One measure of reading, RWRE, was associated with FOXP2 SNPs. QTDT showed a nominally significant association with rs7782412. LAM within the entire sample confirmed this association with p = .0086. LAM within the cohorts separately found strong evidence of this association in the proband cohort at p = .0012. Within the parent cohort, rs923875 was associated with RWRE at 0 = .0046. All RWRE SNP associations calculated with LAM met nominal and QQ criteria for significance. No associations with p values <.05 were found for WATT.
Both measures of timed sequential movement were associated with FOXP2 SNPs. Within the parent cohort, LAM showed that rs12533005 was associated with RAPA at p = .0061, which met nominal and QQ criteria for significance, and a neighboring SNP, rs10230558, was nominally significantly associated with RAPA (p = .0498). QTDT showed a nominally significant association with FS-D for rs778412 and LAM within the parent cohort showed a nominally significant association with FS-D for a neighboring SNP, rs936146. LAM showed associations with FS-D for rs923875 in the entire sample at p = .0211 and within the parent cohort at p = .0035. A neighboring SNP, rs12533005, was also associated with RS-D in the parent cohort at p = .0223. All three of these associations met nominal and QQ criteria for significance.
Discussion
The purpose of this study was to evaluate associations of polymorphisms in CNTNAP2 and FOXP2 with multiple traits in a sample of 188 family trios with dyslexia. We investigated whether CNTNAP2 and/or FOXP2 variants are associated with NWR, SR, reading, and sequential oral and finger movements in dyslexia, using QTDT and LAM.
Regarding CNTNAP2 associations with the selected phenotypes, the QTDT analysis identified a NWR association with one SNP, rs2710102, which was also shown to be associated with variations in NWR in the language impairment sample reported by Vernes et al. (2008). This result confirms the findings in the language impairment sample and extends them to a sample with dyslexia, implying that NWR in both disorder types may be influenced by the same genetic mechanism. In the present study, only one SNP showed this association and with lower statistical significance, compared to the language impairment study. There are several possible explanations for this discrepancy. It is possible that the strength of association increases with increasing severity of disability; the probands in the language impairment sample (Vernes et al. 2008) were more severely impaired (average—1 SD) than the probands in our study (average—.75 SD). In addition, NWR data were not available from all participants in the present study, which lowered the power to detect association. There were also methodological differences. Our study included one trio per family whereas the language impairment study included siblings, whether affected or not. This larger and more broadly based sample may have provided greater variability in the data, conceivably increasing the power. Furthermore, the studies used different sets of nonwords for stimuli. Future studies should replicate this experiment in a larger dyslexia sample.
The hypothesis regarding an association between SR and CNTNAP2 polymorphisms was not supported by the results, despite adequate power to detect association in this sample. The expectation to find this association was based on the Vernes et al. (2008) finding that a composite measure of expressive language was associated with one of the CNTNAP2 SNPs. One possible reason for the absence of this association in our dyslexia sample is our use of a narrower phenotype, sentence memory, that is analogous to one of the subtests incorporated in the composite measure in the language impairment study. The association reported by Vernes et al. (2008) for the composite language measure was seen in only one of the SNPs and of much smaller magnitude than that for NWR. The fact that the present study showed one nominally significant CNTNAP2 association with NWR but none with SR is consistent with this pattern.
The expectation to find FOXP2 SNP associations with measures of spoken and written language and motor sequencing was based on the functional relationships between this gene and CNTNAP2, which is known to influence component traits in nonsydromic forms of language impairment. Because the dyslexia sample contained nonsyndromic forms of that disorder, we expected to find primarily CNTNAP2 SNP associations but unexpectedly found FOXP2 SNP associations for several of the phenotypes. For instance, we found a nominal FOXP2 SNP association with NWR, whereas in a previous study of language impairment, no evidence for FOXP2 SNP associations with NWR or composite measures of expressive and receptive language was found (Newbury et al. 2002).
This study provides supportive evidence for the hypothesis that FOXP2 influences performance on RWRE, a measure of reading that incorporates an element of rapid motor ability in the speech production domain. The task is to orally read words on a list under timed conditions and higher scores indicate a more rapid rate of reading the words. This task, however, also requires coordination of phonological, orthographic, and morphological codes and semantic representations (Berninger et al. 2008b). In contrast, no association was found for WATT, a reading task that also requires coordination of phonological, orthographic, and morphological codes but without access to semantic representation (Berninger et al. 2008b). This task is not timed and thus is less likely to tax the rate of motor output in the speech production system.
Our results provide suggestive evidence for the hypothesis that FOXP2 variants are associated with timed sequential finger-to-thumb tapping. Linear association testing in the parent group showed three SNPs that were associated with FS-D. One of them, rs12533005, was also associated with RAPA. Given what is known about impaired oral praxis in individuals with FOXP2 mutations, we expected to find an association with the RAPA measure. A FOXP2 SNP association with motor sequencing in a finger task is consistent with a more general influence on motor systems beyond the oral and verbal systems. FOXP2 associations with measures of movement sequencing across motor systems are consistent with the finding that mice with induced FOXP2 mutations showed cerebellar growth reductions and gross motor impairments (Groszer et al. 2008). These associations are also consistent with the finding that, in individuals with FOXP2 mutations, gray matter density was reduced in the cerebellum and the caudate nucleus, both of which are crucial for motor control (Belton et al. 2003).
To integrate the findings from FOXP2 into a coherent picture, gene regions and cohort characteristics should be considered. Regarding gene regions, it is possible that the region surrounding the promoter and transcription start site harbors a variant that influences motor sequencing ability shared by RWRE, RAPA, and FS-D, all of which were collected under timed conditions and high scores require not only accurate but also rapid performance. RAPA and FS-D sample similar motor skills in that they require sequential movements in repeated sets but they differ in that RAPA taxes the motor speech system whereas FS-D involves finger movements in the grapho-motor system. Reading real words from a list as fast as possible involves rapid sequential articulation of single words in addition to components of reading such as written word recognition. In this context, it is noteworthy that associations between FS-D and measures of writing have been documented, as FS-D brain activation was found to be significantly predicted by behavioral measures of word spelling in a sample that includes children with and without dysgraphia (Richards et al. 2009). By contrast, the zinc finger region and, because of its strong linkage disequilibrium with the probed SNPs downstream, perhaps also the forkhead box region may harbor a variant that influences the more linguistic aspects of reading ability. In the zinc finger region, association signals were seen in rs7782412 for NWR and RWRE and in rs936146 for NWR.
Regarding the associations in the two cohorts separately, one obvious difference, as mentioned, is the fact that the parent cohort consisted of twice as many participants as the proband cohort, which implies greater power to detect association. Furthermore, it is possible that the adults expressed variability in the more motoric aspects of word reading efficiency whereas the probands expressed variability in the more linguistic aspects of word reading efficiency. Adults have often reached their biological limits of motor speeds and their scores are more likely to represent a biological substrate of genes that are not exhibiting developmentally related changes in expression. In children, motor and other performance is still undergoing maturation and may vary due to other developmental changes that reflect the involvement of additional genes, for example those that affect myelination rates. Consistent with this hypothesis is our observation that FS-D and RAPA were significantly correlated in the parents but not in the probands. An increasing trajectory for motor speed in children, leveling off in adults and decreasing in older adults in direct parallel with myelin disintegration, has been described in the finger tapping literature (Gualtieri and Johnson 2006; Bartzokis et al. 2010; Prigatano 2007). Children may be more likely than adults to express the linguistic aspects of reading as a true representation of their biological limits. The children in this study all had dyslexia and the variability in their scores may be more tightly linked to the biological cause of their reading disability. Not all adults had dyslexia and the variability in their scores may represent other more randomly varying factors. We reiterate that the tests used in this study were normed largely on children and standard scores for adults are less reliable.
Together, these results provide support for a CNTNAP2 SNP association with NWR in family trios with dyslexia, a sample not previously evaluated for this association. This study also provides suggestive evidence that FOXP2 influences NWR, rapid oral reading, and measures of rapid motor sequencing ability beyond the oral motor and motor speech domain to also include finger movements. The findings motivate new research regarding the role of FOXP2 in motor systems besides speech-related and hand movements, not only in samples with reading impairments but also with speech and language impairments, and also regarding differential CNTNAP2 and FOXP2 effects on various aspects of reading ability, including phonemic awareness, working memory, spelling, and handwriting.
Acknowledgments
We thank the families whose participation made this study possible, and Drs. Jennifer Thompson and Kathleen Nielsen and the many graduate students in Educational Psychology who administered the tests. We gratefully acknowledge the following funding sources: NIHDCD T32DC00033 (B. Peter), K08HD049342 (Z. Brkanac), R01HD054562 (V.W. Berninger, M. Lisowski, W.H. Raskind, T. Vu, E.M. Wijsman).
References
- Abecasis GR, Cookson WO, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet. 2000;8(7):545–51. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, et al. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Alarcon M, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–9. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock KJ, et al. Oral dyspraxia in inherited speech and language impairment and acquired dysphasia. Brain Lang. 2000;75(1):17–33. doi: 10.1006/brln.2000.2322. [DOI] [PubMed] [Google Scholar]
- Arking DE, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–4. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkaloglu B, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82(1):165–73. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartzokis G, et al. Lifespan trajectory of myelin integrity and maximum motor speed. Neurobiol Aging. 2010;31(9):1554–62. doi: 10.1016/j.neurobiolaging.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton E, et al. Bilateral brain abnormalities associated with dominantly inherited verbal and orofacial dyspraxia. Hum Brain Mapp. 2003;18(3):194–200. doi: 10.1002/hbm.10093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger VW, Rutberg J. Relationship of finger function to beginning writing: application to diagnosis of writing disabilities. Dev Med Child Neurol. 1992;34(3):198–215. doi: 10.1111/j.1469-8749.1992.tb14993.x. [DOI] [PubMed] [Google Scholar]
- Berninger VW, et al. Language phenotype for reading and writing disability: a family approach. Scientific Studies of Reading. 2001;5(1):59–106. doi: 10.1207/S1532799XSSR0501_3. [DOI] [Google Scholar]
- Berninger V, et al. Assessment for reading and writing intervention: a 3-tier model for prevention and intervention. In: Andrews JS, Saklofske HD, Janzen H, et al., editors. Handbook of psychoeducational assessmen. New York: Academic; 2001. pp. 195–223. [Google Scholar]
- Berninger VW, et al. Modeling developmental phonological core deficits within a working-memory architecture in children and adults with developmental dyslexia. Scientific Studies in Reading. 2006;10:165–98. doi: 10.1207/s1532799xssr1002_3. [DOI] [Google Scholar]
- Berninger VW, et al. Writing problems in developmental dyslexia: under-recognized and under-treated. J Sch Psychol. 2008;46(1):1–21. doi: 10.1016/j.jsp.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger VW, et al. A multidisciplinary approach to understanding developmental dyslexia within working-memory architecture: genotypes, phenotypes, brain, and instruction. Dev Neuropsychol. 2008;33(6):707–44. doi: 10.1080/87565640802418662. [DOI] [PubMed] [Google Scholar]
- Brkanac Z, et al. Evaluation of candidate genes for DYX1 and DYX2 in families with dyslexia. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(4):556–60. doi: 10.1002/ajmg.b.30471. [DOI] [PubMed] [Google Scholar]
- Brkanac Z, et al. Genome scan of a nonword repetition phenotype in families with dyslexia: evidence for multiple loci. Behav Genet. 2008;38(5):462–75. doi: 10.1007/s10519-008-9215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce HA, Margolis RL. FOXP2: novel exons, splice variants, and CAG repeat length stability. Hum Genet. 2002;111(2):136–44. doi: 10.1007/s00439-002-0768-5. [DOI] [PubMed] [Google Scholar]
- Catts HW, et al. A longitudinal investigation of reading outcomes in children with language impairments. J Speech Lang Hear Res. 2002;45(6):1142–57. doi: 10.1044/1092-4388(2002/093). [DOI] [PubMed] [Google Scholar]
- Chapman NH, et al. Linkage analyses of four regions previously implicated in dyslexia: confirmation of a locus on chromosome 15q. Am J Med Genet B Neuropsychiatr Genet. 2004;131B(1):67–75. doi: 10.1002/ajmg.b.30018. [DOI] [PubMed] [Google Scholar]
- Conti-Ramsden G, Botting N, Faragher B. Psycholinguistic markers for specific language impairment (SLI) J Child Psychol Psychiatry. 2001;42(6):741–8. doi: 10.1111/1469-7610.00770. [DOI] [PubMed] [Google Scholar]
- Denckla MB. Development of speed in repetitive and successive finger-movements in normal children. Dev Med Child Neurol. 1973;15(5):635–45. doi: 10.1111/j.1469-8749.1973.tb05174.x. [DOI] [PubMed] [Google Scholar]
- Ewens WJ, Li M, Spielman RS. A review of family-based tests for linkage disequilibrium between a quantitative trait and a genetic marker. PLoS Genet. 2008;4(9):e1000180. doi: 10.1371/journal.pgen.1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher SG. The fletcher time-by-count test of diadochokinetic syllable rate. Austin: Pro-Ed; 1978. [DOI] [PubMed] [Google Scholar]
- Garcia N, Abbott R, Berninger VW. Children’s longitudinal growth of orthographic coding and finger sequencing, in press.
- Groszer M, et al. Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr Biol. 2008;18(5):354–62. doi: 10.1016/j.cub.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualtieri CT, Johnson LG. Reliability and validity of a computerized neurocognitive test battery, CNS Vital Signs. Arch Clin Neuropsychol. 2006;21(7):623–43. doi: 10.1016/j.acn.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Haesler S, et al. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol. 2007;5(12):e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulslander J, et al. Sensory processing, reading, IQ, and attention. J Exp Child Psychol. 2004;88(3):274–95. doi: 10.1016/j.jecp.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Kamhi AG, Catts HW. Toward an understanding of developmental language and reading disorders. J Speech Hear Disord. 1986;51(4):337–47. doi: 10.1044/jshd.5104.337. [DOI] [PubMed] [Google Scholar]
- Lai CS, et al. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413(6855):519–23. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Lewis BA, et al. The genetic bases of speech sound disorders: evidence from spoken and written language. J Speech Lang Hear Res. 2006;49(6):1294–312. doi: 10.1044/1092-4388(2006/093). [DOI] [PubMed] [Google Scholar]
- Lyon GR, Shaywitz S, Shaywitz B. A definition of dyslexia. Ann Dyslexia. 2003;53:1–14. doi: 10.1007/s11881-003-0001-9. [DOI] [Google Scholar]
- MacDermot KD, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76(6):1074–80. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre LM, et al. Circumventing multiple testing: a multilocus Monte Carlo approach to testing for association. Genet Epidemiol. 2000;19(1):18–29. doi: 10.1002/1098-2272(200007)19:1<18::AID-GEPI2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Newbury DF, et al. FOXP2 is not a major susceptibility gene for autism or specific language impairment. Am J Hum Genet. 2002;70(5):1318–27. doi: 10.1086/339931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury DF, Bishop DV, Monaco AP. Genetic influences on language impairment and phonological short-term memory. Trends Cogn Sci. 2005;9(11):528–34. doi: 10.1016/j.tics.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Olson RK, Datta H, Gayán J, DeFries J. A behavioral genetic analysis of reading disabilities and component processes. In: Klein RM, McMullen PA, editors. Converging methods for understanding reading and dyslexia. Cambridge: MIT; 1999. pp. 133–51. [Google Scholar]
- Pennington BF, Bishop DV. Relations among speech, language, and reading disorders. Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- Peterson RL, et al. Neuropsychology and genetics of speech, language, and literacy disorders. Pediatr Clin North Am. 2007;54(3):543–61. doi: 10.1016/j.pcl.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Prigatano GP. Cognitive and behavioral dysfunction in children with hypothalamic hamartoma and epilepsy. Semin Pediatr Neurol. 2007;14(2):65–72. doi: 10.1016/j.spen.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Purcell S, Sham P, Daly MJ. Parental phenotypes in family-based association analysis. Am J Hum Genet. 2005;76(2):249–59. doi: 10.1086/427886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramus F, et al. Theories of developmental dyslexia: insights from a multiple case study of dyslexic adults. Brain. 2003;126(Pt 4):841–65. doi: 10.1093/brain/awg076. [DOI] [PubMed] [Google Scholar]
- Raskind WH, et al. Familial aggregation of dyslexia phenotypes. Behav Genet. 2000;30(5):385–96. doi: 10.1023/A:1002700605187. [DOI] [PubMed] [Google Scholar]
- Raskind WH, et al. A genome scan in multigenerational families with dyslexia: Identification of a novel locus on chromosome 2q that contributes to phonological decoding efficiency. Mol Psychiatry. 2005;10(7):699–711. doi: 10.1038/sj.mp.4001657. [DOI] [PubMed] [Google Scholar]
- Rice ML, Smith SD, Gayán J. Convergent genetic linkage and associations to language, speech and reading measures in families of probands with Specific Language Impairment. J Neurodev Disord. 2009;1(4):264–282. doi: 10.1007/s11689-009-9031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TL, et al. Functional magnetic resonance imaging sequential-finger movement activation differentiating good and poor writers. J Clin Exp Neuropsychol. 2009;31(8):967–83. doi: 10.1080/13803390902780201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Myers RM. Multiple transcription start sites for FOXP2 with varying cellular specificities. Gene. 2008;413(1–2):42–8. doi: 10.1016/j.gene.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Shaywitz SE, Shaywitz BA. The science of reading and dyslexia. J AAPOS. 2003;7(3):158–66. doi: 10.1016/S1091-8531(03)00002-8. [DOI] [PubMed] [Google Scholar]
- Strauss KA, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354(13):1370–7. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- Szenkovits G, Ramus F. Exploring dyslexics’ phonological deficit I: lexical vs sub-lexical and input vs output processes. Dyslexia. 2005;11(4):253–68. doi: 10.1002/dys.308. [DOI] [PubMed] [Google Scholar]
- Teramitsu I, et al. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24(13):3152–63. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorndike RL, Hagen EP, Sattler JM. Stanford-binet intelligence scale: fourth edition. Itasca: Riverside; 1986. [Google Scholar]
- Tomblin JB, et al. Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res. 1997;40(6):1245–60. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomblin JB, et al. The association of reading disability, behavioral disorders, and language impairment among second-grade children. J Child Psychol Psychiatry. 2000;41(4):473–82. doi: 10.1111/1469-7610.00632. [DOI] [PubMed] [Google Scholar]
- Torgesen JK, Wagner RK, Rashotte CA. Test of word reading efficiency. Austin: Pro-Ed; 1999. [Google Scholar]
- Vernes SC, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359(22):2337–45. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RK, Torgesen JK, Rashotte CA, CTOPP . Comprehensive test of phonological processing. Austin: PRO-ED; 1999. [Google Scholar]
- Wechsler D. Wechsler adult intelligence scale - fourth edition. San Antonio: Psychological Corporation; 2008. [Google Scholar]
- Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21(16):3445–7. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- Wolff PH, Gunnoe CE, Cohen C. Associated movements as a measure of developmental age. Dev Med Child Neurol. 1983;25(4):417–29. doi: 10.1111/j.1469-8749.1983.tb13786.x. [DOI] [PubMed] [Google Scholar]
- Woodcock R. Woodcock reading mastery tests—revised. Circle Pines: American Guidance Service; 1987. [Google Scholar]