Abstract
Initiation of translation of hepatitis C virus and classical swine fever virus mRNAs results from internal ribosomal entry. We reconstituted internal ribosomal entry in vitro from purified translation components and monitored assembly of 48S ribosomal preinitiation complexes by toe-printing. Ribosomal subunits (40S) formed stable binary complexes on both mRNAs. The complex structure of these RNAs determined the correct positioning of the initiation codon in the ribosomal “P” site in binary complexes. Ribosomal binding and positioning on these mRNAs did not require the initiation factors eIF3, eIF4A, eIF4B, and eIF4F and translation of these mRNAs was not inhibited by a trans-dominant eIF4A mutant. Addition of Met–tRNAiMet, eIF2, and GTP to these binary ribosomal complexes resulted in formation of 48S preinitiation complexes. The striking similarities between this eukaryotic initiation mechanism and the mechanism of translation initiation in prokaryotes are discussed.
Keywords: Hepatitis C virus, mRNA, translation initiation, protein synthesis, ribosome
Protein synthesis begins following assembly of an initiation complex in which the initiation codon of an mRNA and the anticodon of initiator tRNA are base-paired in the ribosomal “P” site. There are similarities and significant differences in the mechanisms of initiation complex formation in prokaryotes and eukaryotes. A universal characteristic is that initiation starts with separated ribosomal subunits.
In prokaryotes, the small (30S) ribosomal subunit binds mRNA and initiator tRNA in random order to form a complex that then undergoes conformational rearrangement, promoting codon–anticodon base-pairing at the P site and joining of the large (50S) subunit (Gualerzi and Pon 1990). Ribosome binding results from interactions between the 30S subunit and multiple recognition elements in the mRNA, such as the Shine-Dalgarno sequence (McCarthy and Brimacombe 1994). It does not depend on initiation factors. Ribosome-binding sites can occur at any position within an mRNA and as a result many prokaryotic mRNAs are polycistronic.
In contrast, the small (40S) ribosomal subunit in eukaryotes requires several eukaryotic initiation factors (eIFs), first to bind initiator tRNA (as a ternary complex with eIF2 and GTP) to form a 43S complex and then to bind mRNA to form a 48S complex (Merrick 1992). The most common mechanism for recruitment of an mRNA is mediated by its capped 5′ end, which is bound by the eIF4E subunit of eIF4F and then by the 43S complex. Ribosomal binding to mRNA and scanning to the initiation codon require ATP hydrolysis and involve eIF4A, eIF4B, and eIF4F. Most mRNAs that use this mechanism of ribosomal binding are monocistronic because initiation is usually limited to the 5′-most AUG codon.
A second, cap-independent mechanism of ribosome binding is used by mRNAs whose 5′-untranslated region (5′UTR) contains an internal ribosomal entry segment (IRES). IRES-mediated initiation is used by several cellular mRNAs that encode regulatory proteins, and has been usurped by numerous viruses (e.g., Pestova et al. 1996a). Two major groups of viral IRESs have been identified; there is no obvious similarity between them or other viral or cellular IRESs. One of these groups is exemplified by the IRES of encephalomyocarditis virus (EMCV), which is ∼450 nucleotides long, highly structured, and lies immediately upstream of the initiation codon. The secondary and/or tertiary structure of the IRES is critical for its ability to promote initiation. We reconstituted EMCV IRES-mediated initiation in vitro from purified translation components and found that this process does not require noncanonical initiation factors. It is ATP-dependent and uses the same set of canonical eIFs as cap-mediated initiation (Pestova et al. 1996a,b). It does not involve interaction between eIF4E and an m7G cap. Instead, binding of the 43S complex to the EMCV IRES involves a specific cap-independent interaction between the 4G subunit of eIF4F and the IRES.
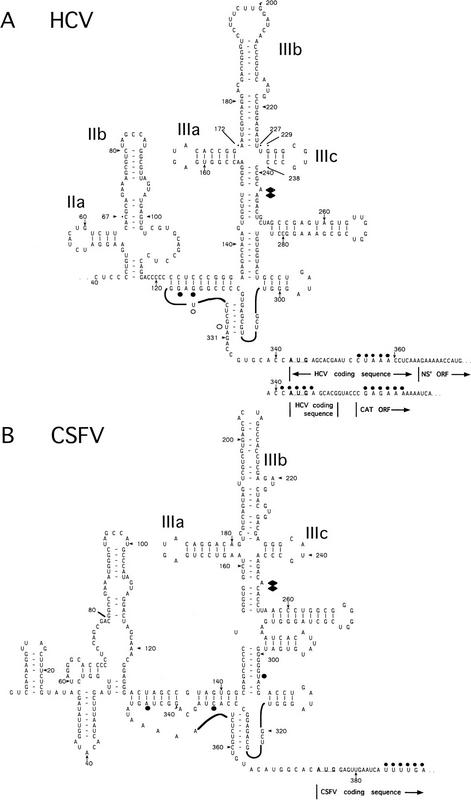
Hepatitis C virus (HCV) and pestiviruses such as classical swine fever virus (CSFV) and bovine viral diarrhea virus constitute another IRES group (Brown et al. 1992; Tsukiyama-Kohara et al. 1992; Poole et al. 1995; Wang et al. 1995; Rijnbrand et al. 1997). Ribosomal entry also occurs at or immediately upstream of the initiation codon on HCV-like IRESs and initiation probably also does not involve scanning (Reynolds et al. 1995, 1996; Rijnbrand et al. 1995, 1996, 1997; Wang et al. 1995; Honda et al. 1996; Lu and Wimmer 1996). Several properties, however, distinguish HCV-like IRESs from EMCV-like IRESs. They are ∼100 nucleotides shorter than EMCV-like IRESs and contain up to 30 nucleotides of coding sequence as well as functionally important structures, such as a pseudoknot upstream of the initiation codon (Fig. 1) that are unrelated to structural elements in the EMCV IRES.
Figure 1.
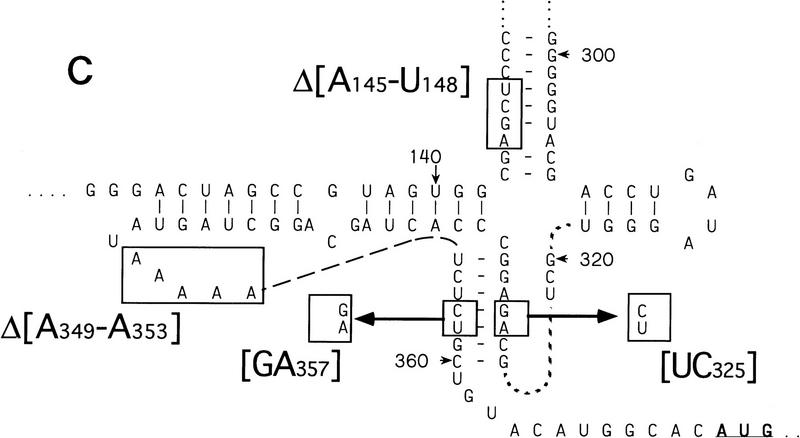
Model secondary and tertiary RNA structures of the 5′NTRs of hepatitis C virus (A) and classical swine fever virus (B), based on proposals by Brown et al. (1992), Honda et al. (1996), and Wang et al. (1995). The nomenclature of domains is as described by Honda et al. (1996). HCV constructs were linked either to NS′ or CAT reporter cistrons–both sequences are shown. An NS′ reporter cistron (not shown) was linked 74 nucleotides downstream of the CSFV initiation codon in all CSFV constructs. HCV and CSFV initiation codons are underlined. Stop-sites where primer extension was arrested on naked RNA only are indicated by open circles; sites of RT arrest caused or enhanced by binding of eIF3 and of 40S subunits are indicated by solid diamonds and circles, respectively. The 3′ border of the HCV nucleotides 26–67 deletion and both borders of the HCV nucleotides 172–227 and HCV nucleotides 229–238 deletions are labeled and indicated by asterisks.
We report here that we have reconstituted HCV and CSFV IRES-mediated initation in vitro up to the stage of 48S complex formation using purified components (Met–tRNAiMet, 40S subunits, initiation factors, and RNA) to identify which factors are requred for this process and begin to characterize their functions during it. Unexpectedly, the mechanism of initiation mediated by HCV and CSFV IRESs differed significantly from both cap- and EMCV IRES-mediated modes of initiation. Multiple interactions between 40S subunits and HCV and CSFV IRESs resulted in formation of stable binary complexes. The complex structure of these IRESs determined the correct positioning of the initiation codon in the ribosomal P site of binary complexes. Ribosomal binding to these mRNAs and positioning on them did not require, and were not affected by, the initiation factors eIF4A, eIF4B, and eIF4F. Complexes (48S) assembled in the absence of eIF4A, eIF4B, and eIF4F were competent to form 80S complexes active in methionylpuromycin synthesis. Moreover, translation of HCV and CSFV mRNAs differed from cap- and EMCV IRES-dependent mRNAs in that it was not inhibited by a trans-dominant eIF4A mutant. Binary ribosomal complexes required addition of only Met–tRNAiMet, eIF2, and GTP to form 48S complexes. These striking similarities between the mechanism of initiation on these viral IRESs and initiation on prokaryotic mRNAs are discussed.
Results
HCV and CSFV RNAs bind 40S ribosomal subunits in the absence of initiation factors
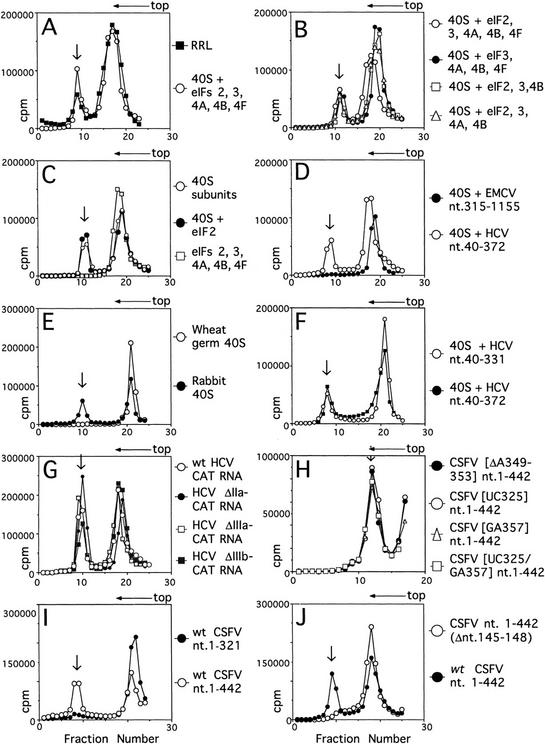
HCV RNA is translated efficiently in rabbit reticulocyte lysate (RRL) (Tsukiyama-Kohara et al. 1992). Assembly of 48S ribosomal preinitiation complexes on the HCV IRES was analyzed initially by incubating HCV (nucleotides 40–372) RNA in RRL under normal translation conditions in the presence of GMP–PNP and then resolving ribosomal and ribonucleoprotein (RNP) complexes by sucrose density gradient centrifugation. GMP–PNP is a nonhydrolyzable GTP analog that causes 48S complexes to accumulate. Complexes (48S) formed under these conditions on up to 30% of the input [32P]UTP-labeled HCV nucleotides 40–372 RNA (Fig. 2A).
Figure 2.
Ribosomal complex formation on HCV and CSFV IRESs. Assays were done using HCV nucleotides 40–372 RNA (A–F) and HCV, CSFV, and EMCV RNAs (D,F–J) as indicated and with RRL or rabbit 40S subunits and eIF2, eIF3, eIF4A, eIF4B, and eIF4F (A) with rabbit 40S subunits and combinations of eIF2, eIF3, eIF4A, eIF4B, and eIF4F (B,C) and with rabbit or (where indicated) wheat germ 40S subunits (D–J). ATP was included only in reactions that contained eIF4A, eIF4B, or eIF4F; Met–tRNAiMet and GMP–PNP were included only in reactions that contained eIF2. Other components described in Materials and Methods were present in all reactions. Sedimentation was from right to left. The positions of ribosomal complexes are indicated by arrows. (G) HCV ΔIIa–CAT, ΔIIIb–CAT, and ΔIIIc–CAT mRNAs are identical to wild-type HCV–CAT mRNA except for deletions of HCV nucleotides 28–67, 172–277, and 229–238, respectively. Sucrose gradients shown in H were centrifuged for a shorter time than others shown here. Fractions from upper parts of the sucrose gradient have been omitted from graphs in H for greater clarity.
The canonical factors eIF2, eIF3, eIF4A, eIF4B, and eIF4F mediate attachment (“internal entry”) of 40S ribosomal subunits to the EMCV IRES (Pestova et al. 1996a). Purified 40S subunits also bound the HCV IRES in the presence of Met–tRNAiMet, ATP, GMP–PNP, and these five factors (Fig. 2A). Each factor and cofactor used to assemble this ribosomal complex was omitted from the reaction to determine whether it was essential. Any one, or even all of them, could be omitted without impairing binding of 40S subunits to the HCV IRES (Fig. 2B,C). In parallel reactions, 40S subunits also bound the CSFV IRES directly without factors or cofactors (Fig. 2H–J) but did not bind the EMCV IRES or to β-globin mRNA under these conditions (Fig. 2D; data not shown). The 40S subunits were therefore not contaminated by initiation factors or by nonspecific RNA-binding proteins. HCV and CSFV IRESs have similar structures that are not related to the EMCV IRES (Fig. 1; Brown et al. 1992; Wang et al. 1995). Deletion of the initiation codon and coding region did not prevent 40S ribosomal subunits from binding to the HCV IRES (Fig. 2F). Neither HCV nor CSFV IRESs bound to active wheat germ 40S subunits (Fig. 1E; data not shown). These results indicate that rabbit 40S ribosomal subunits do not require initiation factors to bind HCV and CSFV IRESs, that their interaction with these RNAs is specific, and that it is stable enough to withstand sucrose density gradient centrifugation.
Binary IRES–40S subunit complexes arrest primer extension within the pseudoknot and downstream of the initiation codon of HCV and of CSFV
Primer extension inhibition (“toeprinting”) has been used to detect binary prokaryotic 30S ribosomal subunit–mRNA complexes (Hartz et al. 1991). We used this method to analyze binding of mammalian 40S ribosomal subunits to HCV and CSFV IRESs. Toeprinting involves cDNA synthesis by reverse transcriptase (RT) on a template RNA to which a ribosome or protein is bound. cDNA synthesis is arrested either directly by the bound complex, yielding a stop or toeprint at its leading edge, or indirectly, by stabilization of adjacent helices (Hartz et al. 1988; Baker and Draper 1995). The resulting toeprints are located on a sequencing gel. Toeprinting is a more stringent assay of RNA–protein interaction than sucrose density gradient centrifugation. For example, cytoplasmic RNA-binding proteins (including initiation factors) form RNP complexes readily on capped eukaryotic mRNAs but do not arrest primer extension (Anthony and Merrick 1992). In all toeprinting experiments described here (except Fig. 8B, below), cDNA products contained a single radioactive moiety derived from the end-labeled primer. The intensity of a toeprint is therefore directly proportional to the frequency of arrest at a specific position. The positions of all the RT stops described below are shown on the structural models of HCV and CSFV IRESs in Figure 1.
Figure 8.
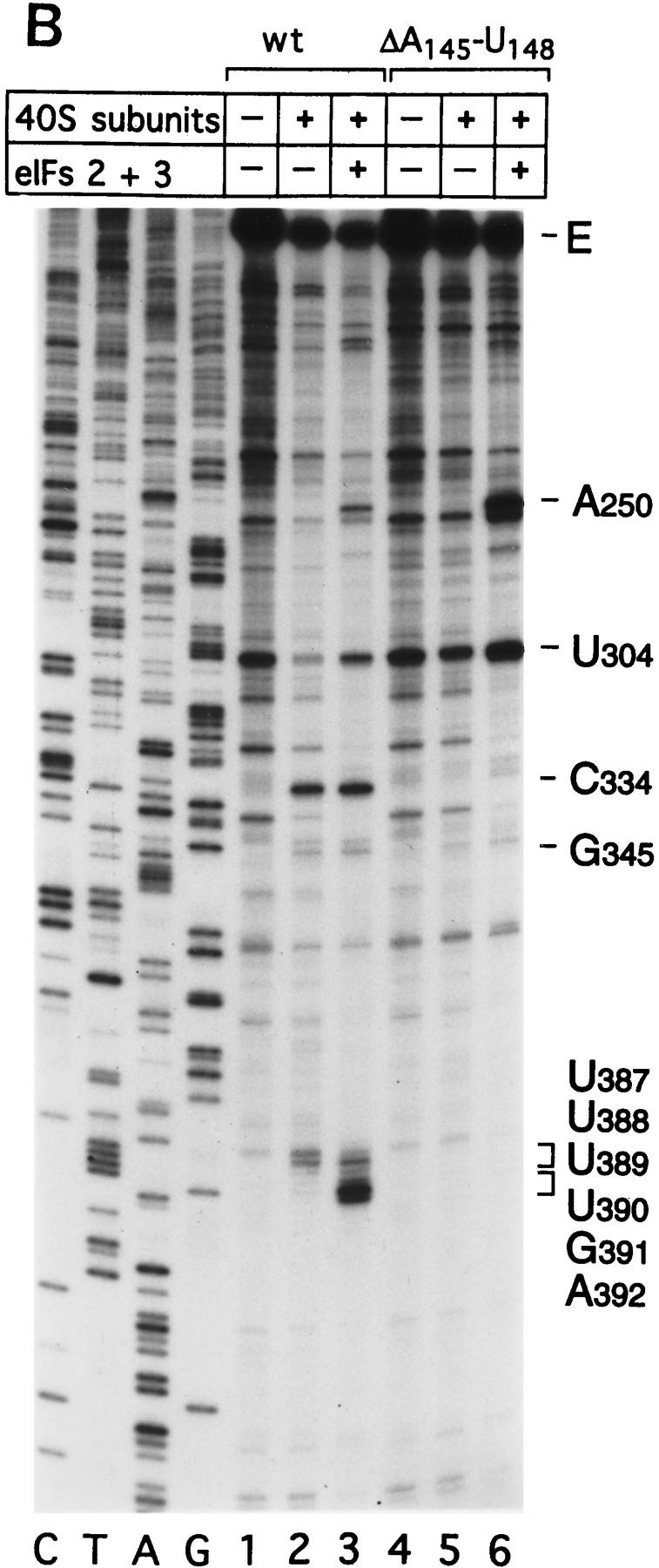
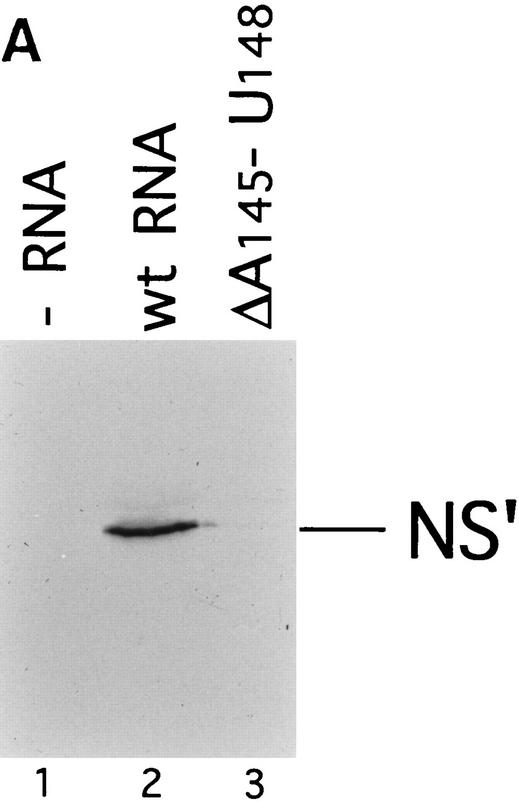
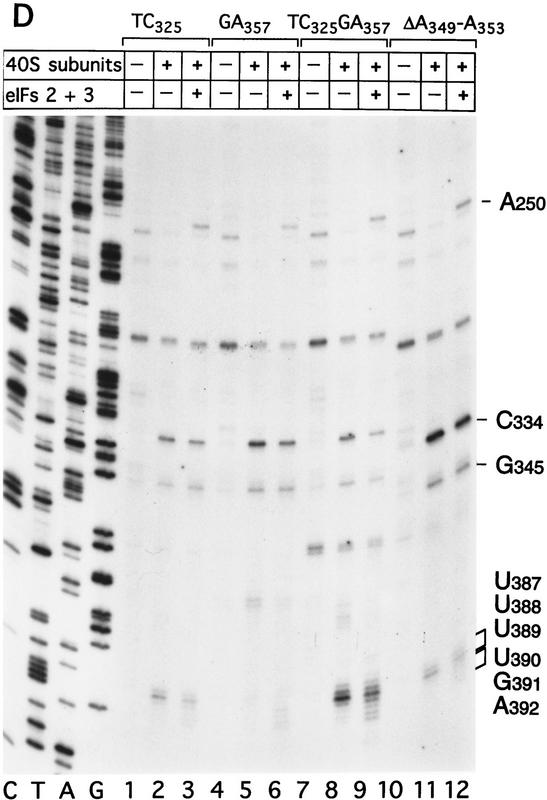
Effect of deleting nucleotides 145–148 on CSFV IRES-mediated translation, binding of 40S subunits and assembly of 48S complexes. (A) RRL (10 μl) was incubated alone (lane 1), with 0.5 μg wild-type CSFV (nucleotides 1–442)–NS′ RNA (lane 2), or with 0.5 μg mutant CSFV (nucleotides 1–442Δ145–148) RNA (lane 3) for 60 min at 30°C as described in Materials and Methods. Translation products were analyzed by autoradiography after electrophoresis on SDS-17% polyacrylamide gel. (B) Wild-type CSFV (nucleotides 1–442)–NS′ (lanes 1–3) and mutant CSFV (nucleotides 1–442Δ145–148) RNAs (lanes 4–6) were incubated with 40S subunits (lanes 2,3,5,6) and with eIF2, eIF3, Met–tRNA iMet and GMP–PNP (lanes 3,6). Reaction conditions are described in Materials and Methods. An unlabeled primer (5′-GGGATTTCTGATCTCGGCG-3′) was annealed to the NS′ reporter cistron and was extended with AMV–RT in the presence of [α-32P]dATP. RT stop sites are indicated on the right. Reference lanes C,T, A, and G depict the CSFV sequence.
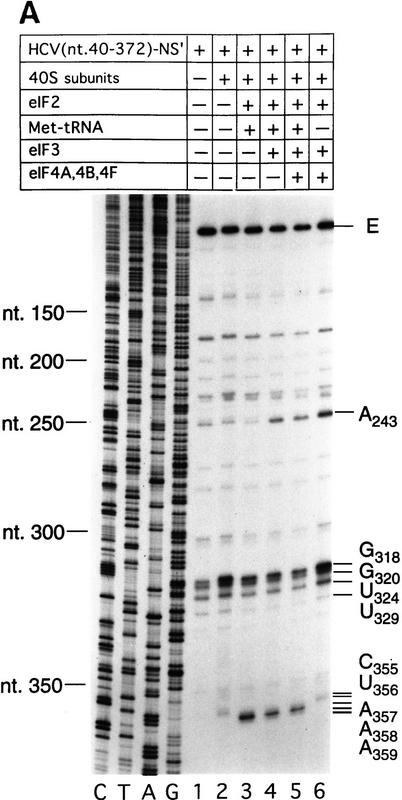
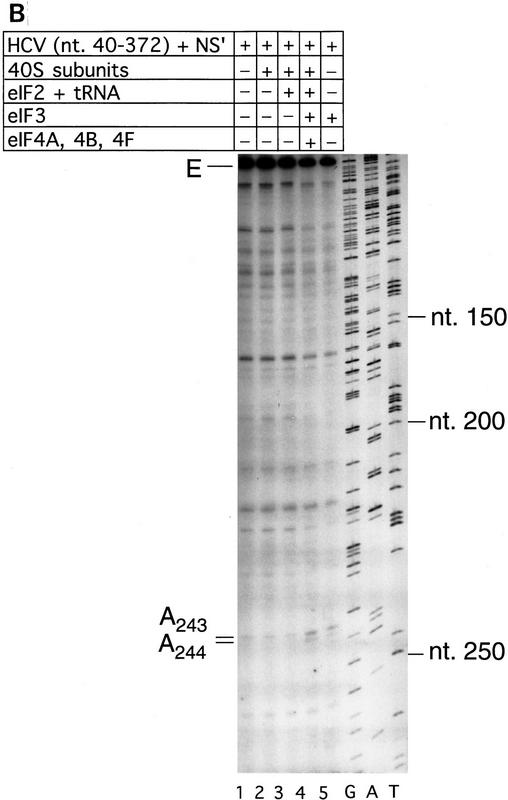
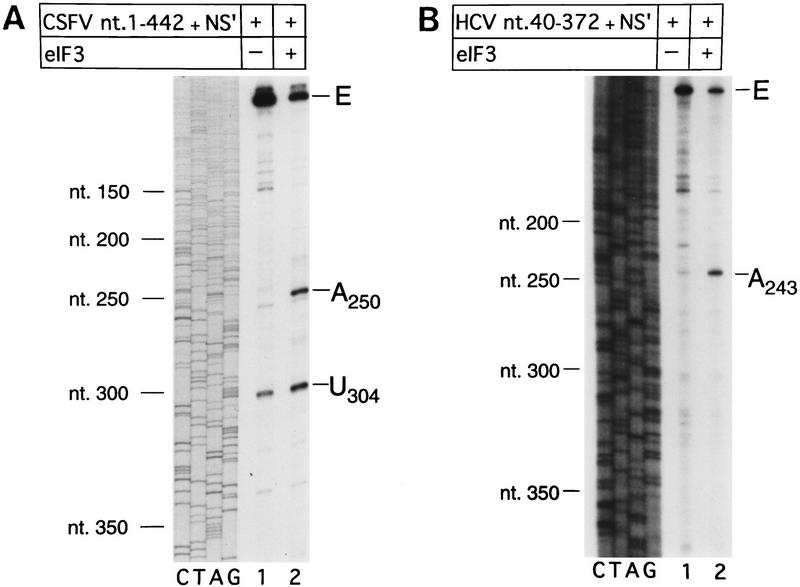
The HCV IRES is highly structured (Fig. 1A) and as a result, RT arrest occurs at a number of sites on naked HCV RNA. In the experiments described here, primer extension on RNA that consisted of HCV nucleotides 40–372 linked to a truncated influenza NS cistron (NS′) was arrested at several stable structures in the IRES, yielding strong stops at positions that included G318, G320, U324, and U329 in the pseudoknot (Fig. 3A, lane 1). Toeprint analysis of binary HCV RNA–40S subunit complexes indicated that 40S subunits enhanced RT stops strongly at G318 and G320 in the pseudoknot and arrested primer extension at C355 and to a lesser extent at U356, downstream of the initiation codon AUG342–344 (Fig. 3A, lane 2).
Figure 3.
Toeprint analysis of 48S complex formation on HCV and CSFV IRESs. (A) Wild-type HCV (nucleotides 40–372)–NS′ RNA was incubated with 40S ribosomal subunits (lanes 2–6), eIF2 (lanes 3–6), Met–tRNAiMet (lanes 3–5), eIF3 (lanes 4–6), and eIF4A, eIF4B, and eIF4F (lanes 5,6); (B) wild-type HCV (nucleotides 40–372)–NS′ RNA was incubated with 40S ribosomal subunits (lanes 2–4), eIF2, Met–tRNAiMet, and GMP–PNP (lanes 3,4), eIF3 (lanes 4,5), and eIF4A, eIF4B, and eIF4F (lane 4); (C) wildtype HCV (nucleotides 40–372)–NS′ (lanes 1–3), (AUG → AAG) mutant HCV (nucleotides 40–372)–NS′ (lanes 4–6), and (AUG → GCG) mutant HCV (nucleotides 40–372)–NS′ RNAs (lanes 7–9) were incubated with 40S ribosomal subunits (lanes 2,3,5,6,8,9) and with eIF2 and eIF3, Met–tRNAiMet and GMP–PNP (lanes 3,6,9); (D) wild-type CSFV (nucleotides 1–442)–NS′ RNA was incubated with 40S ribosomal subunits (lanes 2–6), eIF2 (lanes 3–6), Met–tRNAiMet (lanes 4–6), eIF3 (lanes 5–6), and eIF4A, eIF4B, and eIF4F (lane 6); (E) wild-type CSFV (nucleotides 1–442)–NS′ RNA was incubated with 40S ribosomal subunits (lanes 2–5), eIF2, Met–tRNA iMet and GMP–PNP (lanes 3–5), eIF3 (lanes 4–6), and eIF4A, eIF4B, and eIF4F (lane 5). Reaction conditions are described in Materials and Methods. In A,C, and D, the primers (5′-GGGATTTCTGATCTCGGCG-3′) and (5′-CTCGTTTGCGGACATGCC-3′) were annealed to the NS′ cistron 130 nucleotides downstream of the HCV initiation codon and 110 nucleotides downstream of the CSFV initiation codon, respectively, and were extended with AMV–RT. In B and D, the primers 5′-CGCAAGCACCCTATC-3′ (complementary to HCV nucleotides 295–309) and 5′-CCTGATAGGGTGCTGCAG-3′ (complementary to CSFV nucleotides 309–326) were annealed to HCV and CSFV IRESs, as appropriate, and were extended with AMV–RT. Full-length cDNA is marked E. Other cDNA products terminated at the sites are indicated on the right. Reference lanes C,T,A, and G depict HCV or CSFV sequences, as appropriate.
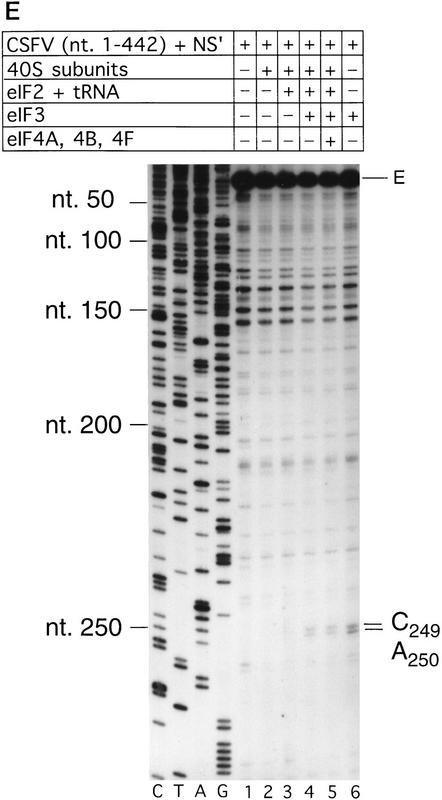
Primer extension on naked CSFV (nucleotides 1–442)–NS′ RNA was arrested strongly at U304 near the base of domain III but not significantly in the pseudoknot (Fig. 3D, lane 1). Analysis of binary CSFV RNA–40S subunit complexes indicated that 40S ribosomal subunits arrested primer extension strongly at C334 and to a lesser extent at G345 in the pseudoknot, as well as strongly at U388 and to a lesser extent at U387 and U389 downstream of the initiation codon AUG373–375 (Fig. 3D, lane 2).
Binding of 40S subunits at these contact points on HCV and CSFV IRESs might prevent detection of additional IRES–40S subunit contacts using the same primers, for example, on the opposite side of the pseudoknot. To address this possibility, and the possibility that additional 40S subunits might bind elsewhere on these IRESs, we carried out additional toeprint analyses using new primers that were chosen to bind upstream of the 5′-most stops described above. The results of these additional experiments are shown in Figure 3, B and E. These panels show that no additional contacts between 40S subunits and these IRESs were identified other than those that were described above. Subunits (40S) do not, therefore, bind anywhere else on these IRES elements other than at the position described above, including the opposite side of the HCV and CSFV pseudoknots and at or near to the 5′ terminii of these RNAs.
These results are summarized in Figure 1. They confirm that 40S ribosomal subunits alone can bind directly to HCV and CSFV IRESs, probably at or close to the pseudoknot. The observation that binary complexes yielded toeprints downstream of HCV and CSFV initiation codons indicate that the coding regions of both RNAs are fixed stably in the mRNA-binding cleft of 40S subunits. This observation may also provide an explanation for the importance of the conserved sequences adjacent to the initiation codon for IRES function (Reynolds et al. 1995; Honda et al. 1996; Lu and Wimmer 1996).
The 43S ribosomal complex binds by itself at the initiation codon of HCV and CSFV IRESs without eIF4A, eIF4B, or eIF4F
Eukaryotic translation initiation is analyzed conventionally using sucrose density gradient centrifugation to resolve RNP, preinitiation, and initiation complexes. This approach is not appropriate for analysis of initiation on HCV and CSFV IRESs because as the results described above have shown, 40S subunits bind directly to these RNAs without aminoacylated initiator tRNA or initiation factors. Primer extension analysis, however, is a sensitive assay of prokaryotic initiation (Hartz et al. 1988) and has been used to assay cap- and IRES-mediated modes of eukaryotic initiation (Anthony and Merrick 1992; Pestova et al. 1996a,b). This method was used to dissect the initiation process on HCV and CSFV IRESs.
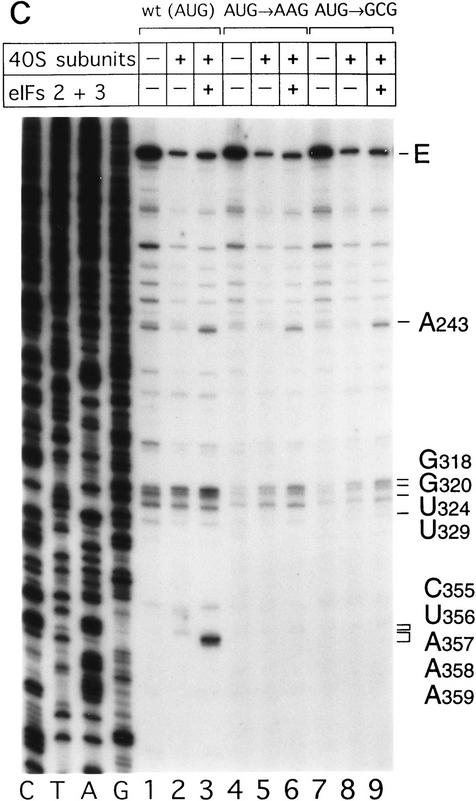
Toeprint analysis of complexes assembled from eIF2, 40S subunits, Met–tRNAiMet and GMP–PNP on HCV (nucleotides 40–372)–NS′ RNA yielded prominent stops at AAA357–359 (Fig. 3A, lane 3). These stops were not observed during toeprint analysis of complexes assembled from the ternary [eIF2/initiator tRNA/GMP–PNP] complex and HCV (nucleotides 40–372)–NS′ mRNA in the absence of 40S subunits (data not shown). This result and the position of these stops relative to the initiation codon indicates that they are caused by a stable ribosomal complex assembled at the initiation codon. The stops that had been detected at C355 and U356 in the binary HCV IRES–40S complex were not apparent and the G318 and G320 stops were weaker (Fig. 3A, lane 2). The same stops at G318, G320, and AAA357–359 were detected on HCV RNA when eIF3 or eIF3, eIF4A, eIF4B, and eIF4F (Fig. 3A, lanes 4,5) were included in assembly reactions in addition to eIF2, Met–tRNAiMet GMP–PNP and 40S subunits. Therefore eIF3, eIF4A, eIF4B, and eIF4F did not alter the interaction of the 40S subunit with the HCV IRES. We have shown previously that these factors are absolutely required for assembly of 48S complexes on the EMCV IRES (Pestova et al. 1996a,b). We therefore used 48S complex formation on the EMCV IRES to confirm that the factors used in reconstituting 48S complex formation on the HCV and CSFV IRESs were all active (data not shown). A band at A243 became more prominent when eIF3 was present in reactions (Fig. 3A, lanes 4–6). Closer inspection revealed that this band consisted of stops at A243 and A244 (Fig. 3B, lanes 4,5). The stops at CU355–356 were apparent, whereas those at AAA357–359 were not when Met–tRNAiMet was not included in a reaction that contained eIF2, eIF3, GMP–PNP, and 40S subunits (Fig. 3A, lane 6). This result suggests that RT arrest at AAA357–359 requires cognate codon–anticodon base-pairing between HCV mRNA and initiator tRNA.
To address this question, 48S complex assembly was analyzed on AUG → AAG and AUG → GCG mutant IRESs that differ from wild type in the initiation codon. These triplets are not active initiation codons in these RNAs (Reynolds et al. 1995). The efficiency of binding of 40S subunits to these wild-type and mutant RNAs was indistinguishable by sucrose density gradient centrifugation (data not shown). The strong stops at AAA357–359 that are characteristic of 48S complex formation at the initiation codon, however, were detected only on wild-type HCV RNA and not on either mutant (Fig. 3C, lanes 3,6,9). These observations strongly support the conclusion that codon–anticodon base-pairing is required for stable IRES-mediated assembly of a 48S complex at the HCV initiation codon.
We next used toeprinting to analyze ribosomal complex formation on the CSFV IRES. Toeprint analysis of complexes assembled from 40S subunits, eIF2, Met–tRNAiMet, and GMP–PNP on CSFV (nucleotides 1–442)–NS′ RNA yielded stops at C334 and G345 in the pseudoknot that were of similar intensity, and stops at UUU387–389 that were weaker than for binary CSFV IRES–40S complexes (Fig. 3D, lane 2,4). In addition, very prominent stops appeared at UGA390–392 (Fig. 3D, lane 4) that were not detected on these binary complexes (Fig. 3D, lane 2). These stops were not detected on analysis of complexes assembled from eIF2, GMP–PNP, and 40S subunits without Met–tRNAiMet (Fig. 3D, lane 3). The position of these stops relative to the initiation codon indicates that 43S complexes alone bind correctly to the CSFV initiation codon AUG373–375, arresting primer extension at UGA390–392. The stops derived from complexes that had assembled on the CSFV IRES from eIF2, Met–tRNAiMet, GMP–PNP, and 40S subunits were similar to those derived from these components and eIF3 (Fig. 3D, lanes 4,5) except that in the latter instance (1) a faint stop was detected at A250 and the stop at U304 was stronger and (2) the stops at UUU387–389 were relatively weaker and those at UGA390–392 were relatively stronger. The first of these results suggest that eIF3 binds specifically to the CSFV IRES. Closer inspection indicated that eIF3 arrested primer extension at A250 and C251 (Fig. 3E, lanes 4–6). The second of these results indicates that formation of 48S complexes is enhanced in the presence of eIF3 vis-á-vis formation of binary IRES–40S subunit complexes, and therefore suggests that a role of eIF3 in CSFV initiation may be to stabilize the 48S complex on the initiation codon. Neither the pattern nor the intensity of stops that were detected on CSFV RNA were altered as a result of inclusion of eIF4A, eIF4B, and eIF4F in reactions that contained eIF2, eIF3, Met–tRNAiMet, GMP–PNP, and 40S subunits (Fig. 3D, cf. lanes 5 and 6). Therefore, these three factors do not affect the interaction of the 43S complex with the CSFV IRES.
In a further series of experiments, toeprinting analysis of complexes assembled on HCV and CSFV IRESs in reactions that contained eIF2, eIF3, Met–tRNAiMet, GMP–PNP, and 40S subunits with or without eIF4A, eIF4B, and eIF4F using a different set of primers revealed that 48S complexes did not assemble either at or near to the 5′ ends of these mRNAs or at or near to any AUG codon other than the authentic initiation codon (Fig. 3B,E).
Taken together, these results indicate that a 43S ribosomal preinitiation complex is able, by itself, to bind directly and stably to the authentic initiation codon on HCV and CSFV IRESs without the involvement of noncanonical factors, or of eIF4A, eIF4B, or eIF4F. Stable binding of this complex at the initiation codon requires initiator tRNA. It may be enhanced by, but does not require, eIF3, which is present in stoichiometric amounts on native 40S subunits (Sundkvist and Staehelin 1975). The 40S subunit itself, however, is responsible for the specificity of binding of the 43S complex to these IRESs.
eIF3 alone binds specifically to HCV and CSFV IRESs
Inclusion of eIF3 in reactions that contained eIF2 and 40S subunits resulted in RT arrest at AC250–251 and U304 on the CSFV IRES and at AA243–244 on the HCV IRES (Fig. 3, A, lanes 4–6; B, lanes 4,5; D, lanes 5,6; E, lanes 4–6). Appropriate RNP complexes were toeprinted to determine whether eIF3 alone binds specifically to these RNAs. eIF3 alone arrested RT at these positions on the CSFV IRES (Fig. 3E, lane 6 and Fig. 4A, lane 2) and on the HCV IRES (Fig. 4B, lane 2). These results indicate that eIF3 binds specifically to domain III of HCV and CSFV IRES elements. The specificity of both of these interactions has been confirmed by chemical and enzymatic footprinting of binary eIF3–IRES complexes (D. Sizova, V. Kolupaeva, T. Pestova, I.N. Shatsky, and C. Hellen, in prep.).
Figure 4.
Primer extension analysis of RNP complexes formed on CSFV and HCV IRES elements as follows. (A) CSFV (nucleotides 1–442)–NS′ and (B) HCV (nucleotides 340–372)–NS′ RNAs were incubated with (lane 2) or without eIF3 (lane 1) under standard conditions. Primers were annealed to the NS′-coding sequences of these RNAs and extended with AMV–RT. The full-length cDNA extension product is marked E. The cDNA products labeled A250, U304, and A243 on the right terminated at these nucleotides. The reference lanes C, T, A, and G depict the CSFV sequence (A) and the HCV sequence (B). Positions of CSFV and HCV nucleotides are indicated on the left of the appropriate panel.
Factor requirements for assembly of active 80S complexes on HCV and CSFV IRESs
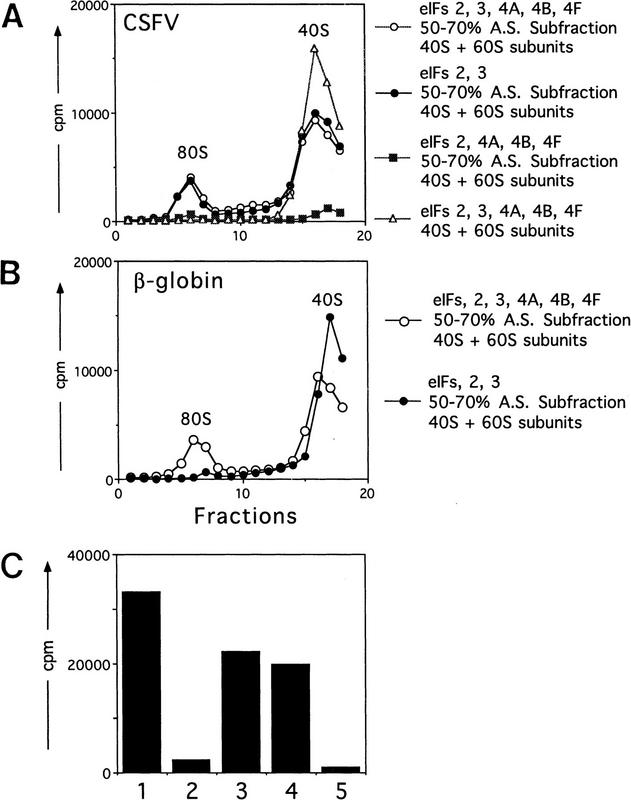
Complexes (48S) assemble accurately at the initiation codon on HCV and CSFV IRESs on inclusion of eIF2, initiator tRNA, and GMP–PNP (or GTP) with 40S subunits. To determine whether these 48S complexes are bona fide intermediates in the initiation process that culminates in formation of the first peptide bond, we assayed the ability of these complexes to join with 60S ribosomal subunits to form 80S complexes and for these 80S complexes to catalyze methionylpuromycin synthesis. This model reaction mimics dipeptide formation. Assembly of active 80S complexes was mediated by a ribosomal salt wash (RSW) fraction (the 0.2–0.8 m KCl phosphocellulose elution fraction derived from the 50%–70% A.S. cut) that contained eIF5 and eIF5A (which are required for subunit joining and peptide bond formation) but that did not contain eIF4A, eIF4B, or eIF4F (Benne et al. 1978). The absence of eIF4A, eIF4B, and eIF4F from this fraction was confirmed by Western blotting (data not shown). Toeprinting assays showed that this RSW fraction had no effect on the amount of 48S complex formed on HCV and CSFV IRESs or on the position of these complexes (data not shown).
There was no requirement for eIF4A, eIF4B, or eIF4F in formation of 80S complexes on the CSFV IRES, but eIF3 was absolutely essential for this process (Fig. 5A). Moreover, omission of eIF3 from the 80S assembly reaction also caused the destabilization of 48S complexes (Fig. 5A). Formation of 80S complexes was accompanied by a decrease in the amount of 48S complexes. The formation of 80S complexes was not observed if the RSW fraction or 60S subunits were omitted from assembly reactions, or if GTP was substituted by GMP–PNP (Fig. 5A; data not shown). Similarly, eIF4A, eIF4B, and eIF4F were not required for assembly of 80S complexes capable of methionylpuromycin synthesis (Fig. 5C). As expected, and consistent with previous reports (Trachsel et al. 1977; Benne and Hershey 1978; Grifo et al. 1982), eIF4A, eIF4B, and eIF4F were essential for assembly of 80S complexes on β-globin mRNA that were competent to catalyze methionylpuromycin synthesis (Fig. 5B,C). This result serves as an additional control that the RSW fraction used to mediate subunit joining was not contaminated by eIF4A, eIF4B, or eIF4F. As described for CSFV, formation of 80S complexes was accompanied by a decrease in the amount of 48S complexes.
Figure 5.
Assembly of 80S ribosomal initiation complexes on the CSFV IRES and on β-globin mRNA and analysis of their activity in stimulating methionylpuromycin synthesis. Assays were done using CSFV (nucleotides 1–442)–NS′ mRNA (A,C) and β-globin mRNA (B,C) and with initiation factors and other translation components as described in Materials and Methods. (A,B) Sedimentation was from right to left, and the formation of complexes was assayed by incorporation of [35S]methionine–tRNA. The positions of ribosomal complexes are indicated. Fractions from upper parts of sucrose gradients have been omitted from graphs in A and B for greater clarity. (C) The methionylpuromycin synthesis assay was done as described in Materials and Methods, using β-globin mRNA (columns 1 and 2); CSFV (nucleotides 1–442)–NS′ mRNA (columns 3–5); eIF2, eIF3, eIF4A, eIF4B, eIF4F (columns 1 and 3), eIF2 and eIF3 (columns 2 and 4); and eIF2, eIF4A, eIF4B, and eIF4F (column 5). All reactions also contained GTP, 40S and 60S ribosomal subunits, and a 50%–70% A.S. RSW subfraction as described in Materials and Methods. A background value determined with charged initiator tRNA alone was subtracted from all panels.
These results confirm that assembly of 48S preinitiation complexes on HCV and CSFV IRESs that are competent to complete all remaining stages in the initiation process does not require eIF4A, eIF4B, or eIF4F. This process therefore differs fundamentally from cap-dependent initiation on β-globin.
Toeprinting assays showed that eIF3 did not influence the efficiency or the fidelity of assembly of 48S complexes at the initiation codon of HCV and CSFV IRESs (Fig. 3A,D), but this factor was required for assembly of 80S complexes (Fig. 5A). Our data are not sufficient to determine whether eIF3 is involved in altering the conformation of the 48S complex so that it can bind to 60S subunits or whether it is involved more directly in joining of subunits.
Translation of HCV and CSFV mRNAs is resistant to inhibition by trans-dominant eIF4A(R362Q)
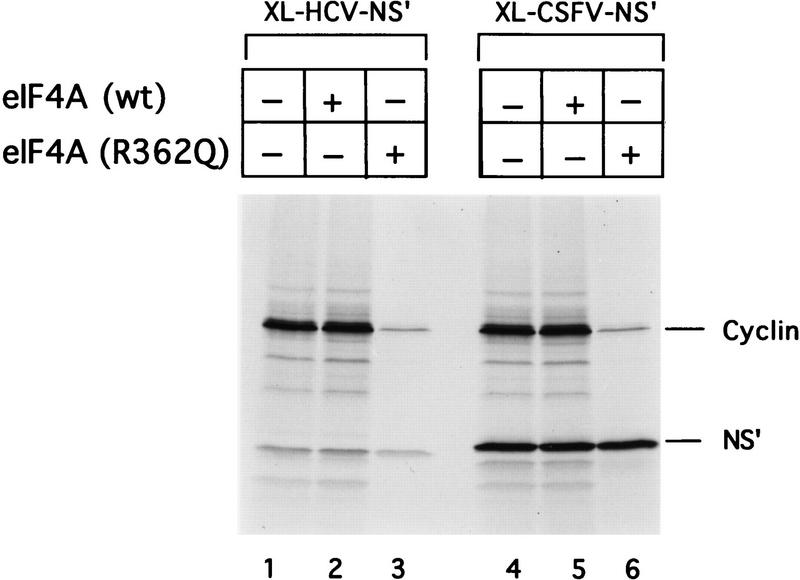
Cap- and EMCV IRES-mediated initiation both require eIF4A, probably as a subunit of eIF4F (Trachsel et al. 1977; Benne and Hershey 1978; Grifo et al. 1983; Pestova et al. 1996b). Both processes are inhibited by the trans-dominant eIF4A (R362Q) mutant; inhibition is relieved by eIF4F, and less effectively, by eIF4A (Pause et al. 1994). The data above indicate that 43S ribosomal complexes bind to HCV and CSFV IRESs without eIF4A, eIF4B, or eIF4F and can then bind 60S ribosomal subunits to form active 80S complexes. Translation of these mRNAs would therefore be expected to be unaffected by eIF4A(R362Q). Addition of eIF4A(R362Q) to RRL programmed with dicistronic cyclin–HCV IRES–NS′ or cyclin–CSFV IRES–NS′ mRNAs strongly impaired translation of the upstream cyclin cistron in both instances but had no effect on IRES-mediated translation of the downstream NS′ cistron (Fig. 6, lanes 3,6). Addition of wild-type eIF4A to RRL did not influence translation of these mRNAs (Fig. 6, lanes 2,5). The activity of the trans-dominant eIF4A(R362Q) mutant was confirmed by its ability to inhibit translation of luciferase mRNA and of a GUS reporter cistron linked to the EMCV IRES (data not shown). These results indicate that eIF4A (and probably eIF4F) are not required for HCV/CSFV translation. They strongly support the results obtained by reconstituting initiation reactions from purified components. They are consistent with a model for initiation in which 43S complexes bind directly to these IRESs without the involvement of eIF4A, eIF4B, or eIF4F.
Figure 6.
Dominant-negative effect of the eIF4A R362Q mutant protein on cap-mediated translation and its lack of effect on HCV and CSFV IRES-mediated translation. Reticulocyte lysate (10 μl) was preincubated alone (lanes 1,4), with eIF4A wild-type (1.2 μg) (lanes 2,5), or eIF4A mutant (1.2 μg) (lanes 3,6) for 5 min at 30°C, then incubated for 60 min at 30°C with dicistronic mRNAs (0.5 μg) as described in Materials and Methods. Translation products were analyzed by autoradiography after electrophoresis on SDS–17% polyacrylamide gel.
Structural integrity of the HCV IRES is required for stable binding of 43S complexes at the initiation codon
Disruption of HCV and CSFV IRES structure severely reduces their activity and must therefore impair one or more steps in the process of internal ribosomal entry. Initiation was analyzed on various mutant IRESs to identify these steps.
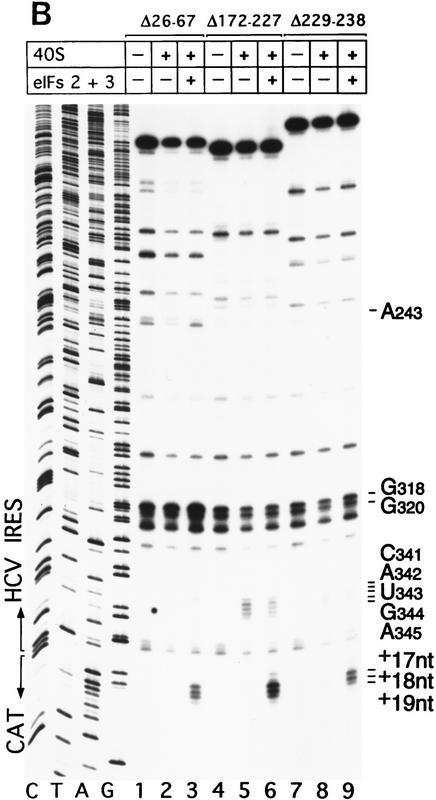
The IIa, IIIb, and IIIc hairpins contribute to HCV IRES function (Rijnbrand et al. 1995). We examined their role in binding 40S ribosomal subunits and 43S preinitiation complexes to the IRES using wild-type HCV (nucleotides 1–349)–CAT mRNA and Δ nucleotides 26–67, Δ nucleotides 172–227, and Δ nucleotides 229–238 mutant derivatives, thereof, that lack IIa, IIIb, and IIIc hairpins, respectively (Fig. 1A). The results of sucrose density gradient centrifugation showed that these three hairpins are not essential for binary complex formation. Appropriate HCV deletion mutants all bound 40S subunits strongly (Fig. 2G). Toeprint analysis of binary ribosomal complexes assembled on wild-type HCV (nucleotides 1–349)–CAT mRNA yielded stops (Fig. 7A, lane 2) that differed slightly from those obtained on wild-type HCV (nucleotides 40–372)–NS′ mRNA (Fig. 3A, lane 2). Bound 40S subunits enhanced RT stops strongly at G318 and G320 as described above, but also arrested primer extension at CAUGA341–345 (i.e., the HCV initiation codon) and at a GAG triplet 15–17 nucleotides downstream of the initiation codon. These results are summarized in Figure 1A. They show that the interaction of nucleotides downstream of the pseudoknot with 40S ribosomal subunits is influenced by the sequence of the coding region. Toeprinting of binary complexes assembled on Δ nucleotides 172–227 mutant HCV–CAT mRNA yielded stops at CAUGA341–345 and at GAG (+15–+17 nucleotides) of the same intensity as on wild-type RNA (Fig. 7A, lanes 2,5); on the Δ nucleotides 229–238 HCV mutant, corresponding stops were weaker (Fig. 7B, lane 8). The intensity of stops at G318 and G320 on these two mutant RNAs did not increase in the presence of 40S subunits. Toeprint analysis of binary complexes assembled on Δ nucleotides 26–67 HCV–CAT RNA yielded strong stops at G318 and G320 and very weak stops both at CAUGA341–345 and at GAG (+15–+17 nucleotides) (Fig. 7B, lane 2). The results of sucrose density gradient analysis indicate that the IIa, IIIb, and IIIc hairpins do not determine binding of 40S subunits to the HCV IRES; these findings are supported by the results of toeprint analysis. toeprinting also shows that these three hairpins are necessary for the wild-type pattern of interactions between the HCV IRES and the 40S subunit to occur.
Figure 7.
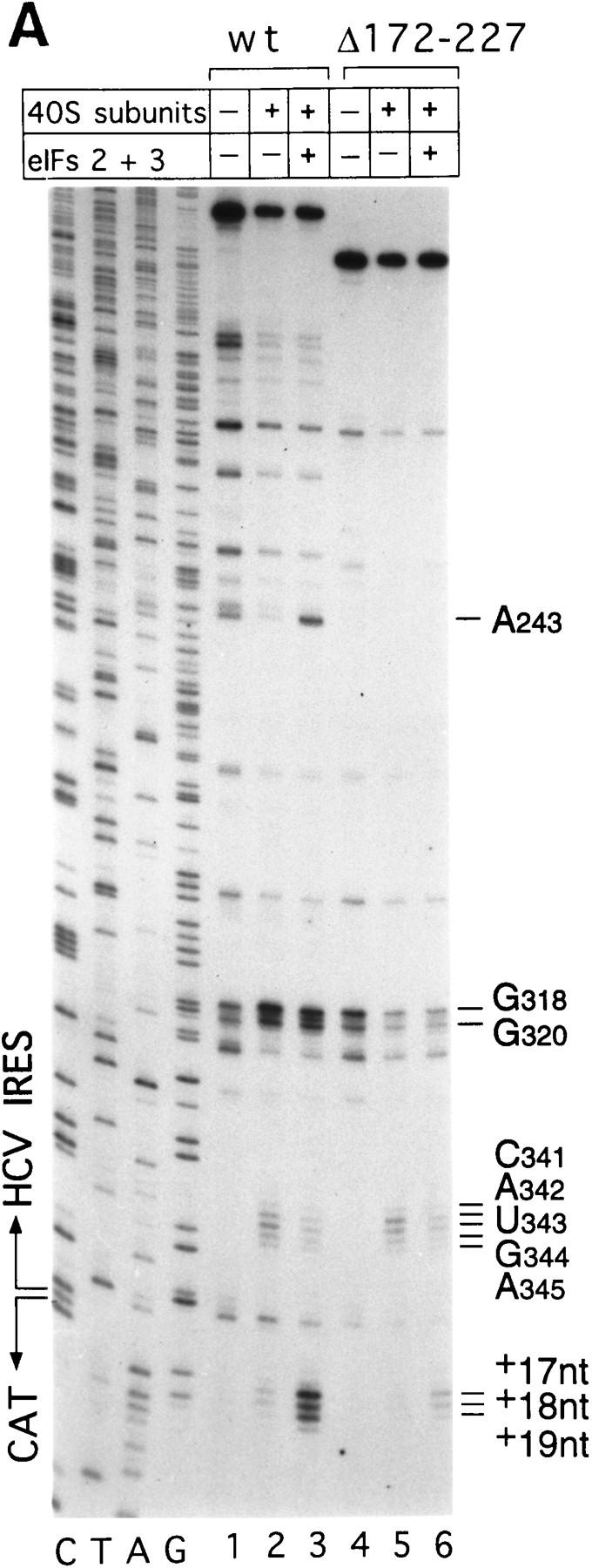
Binding of 40S subunits and assembly of 48S complexes on HCV and CSFV mutant IRESs. (A) Wild-type HCV (nucleotides 1–349)–CAT (lanes 1–3) and mutant HCV (nucleotides 1–349Δ172–227)–CAT (lanes 4–6) were incubated under standard conditions with 40S ribosomal subunits (lanes 2,3 and 5,6) and with eIF2, eIF3, Met–tRNAiMet and GMP–PNP (lanes 3,6). (B) Mutant HCV (nucleotides 1–380Δ26–67)–CAT (lanes 1–3), (nucleotides 1–380Δ172–227)–CAT (lanes 4–6) and (nucleotides 1–380Δ229–238) (lanes 7–9) RNAs were incubated with 40S subunits (lanes 2,3,5,6,8,9) and with eIF2, eIF3, Met–tRNAiMet, and GMP–PNP (lanes 3,6,9). Reaction conditions are described in Materials and Methods. A primer (5′-GCAACTGACTGAAATGCC-3′) was annealed to the CAT cistron ∼80 nucleotides downstream of the HCV initiation codon and was extended with AMV–RT. Primer extension was arrested at sites indicated on the right either as nucleotides in the HCV IRES or as positions relative to the A of the HCV initiation codon. Reference lanes C, T, A, and G depict the HCV–CAT sequence. The junction between the HCV IRES and the CAT cistron is indicated. This gel was exposed to x-ray film longer than that shown in A to show the differences in the intensity of stops on various mutant RNAs. (C) Model of the CSFV pseudoknot and adjacent nucleotides showing the substitutions present in the mutants UC325, GA357, and UC325GA357 and the deletions made in the mutants ΔA145–U148 and ΔA349–A353. In this model, helix I of the pseudoknot comprises nucleotides 129–142 and 331–346, and helix II comprises nucleotides 323–329 and 354–360. (D) CSFV (nucleotides 1–442)–NS′ RNAs containing the mutations UC325 (lanes 1–3), GA357 (lanes 4–6), UC325GA357 (lanes 7–9), and ΔA349–A353 (lanes 10–12) were incubated with 40S subunits (lanes 2,3,5,6,8,9,11,12) and with eIF2, eIF3, Met–tRNAiMet and GMP–PNP (lanes 3,6,9,12).
Toeprint analysis of 48S complexes assembled from eIF2, eIF3, Met–tRNAiMet, GMP–PNP, and 40S subunits on wild-type HCV (nucleotides 1–349)–CAT yielded stops at CAUGA341–345 that were slightly weaker, stops at G318 and G320 that were similar in intensity and stops at A243 and at GAGAA (+15–+19 nucleotides, downstream of the A of the initiation codon) that were significantly stronger than those detected on analysis of the corresponding binary IRES–40S complex (Fig. 7A, lane 3). The prominent stops at GAGAA (+15–+19 nucleotides) were not detected when Met–tRNAiMet was omitted from reactions (data not shown) and were therefore caused by 48S complexes assembled at the initiation codon. These results show that the HCV IRES undergoes a conformational rearrangement when the ternary eIF2/GTP/initiator tRNA complex binds to the binary IRES–40S complex to form a 48S preinitiation complex. The stop at A243 was attributable to bound eIF3. Under similar conditions, this stop was not detected on Δ nucleotides 172–227 HCV RNA, was very weak on Δ nucleotides 229–238 HCV RNA and had near-wild-type intensity on Δ nucleotides 26–67 mutant RNA (Fig. 7B, lanes 3,6,9). These results suggest that binding of eIF3 to this IRES requires hairpin IIIb and is enhanced strongly by hairpin IIIc.
The stops at GAGAA (+15–+19 nucleotides) caused by assembly of 48S complexes were much weaker on Δ nucleotides 172–227 mutant RNA than on wild-type HCV RNA (Fig. 7A, lanes 3,6). These stops were even weaker on Δ nucleotides 26–67 and Δ nucleotides 229–238 mutant RNAs than on Δnucleotides 172–227 mutant RNA (Fig. 7B, cf. lanes 3,6, and 9). The gel in Figure 7B that shows these data was exposed to film longer than that in Figure 7A to show the differences in intensity of stops at CAUGA341–334 and at GAGAA (+15–+19 nucleotides) between these different mutants (Fig. 7, cf. lanes 4–6 in A and B).
These results show that deletion of IIa, IIIb, or IIIc hairpins from the HCV IRES strongly impairs assembly of 48S complexes at the initiation codon. They are consistent with observations that translation of HCV mRNA is strongly impaired by deletion of any one of these hairpins (Rijnbrand et al. 1995). Sucrose density gradient analysis showed that binary ribosomal complexes assembled efficiently on these three mutant RNAs. The detailed toeprinting analysis presented here showed that despite efficient binary complex formation, the normal pattern of interaction between 40S subunits and the HCV IRES in the vicinity of the initiation codon was altered by these deletions. The result of this defect is that assembly of 48S complexes on the initiation codon was strongly impaired. Correct positioning of the 43S complex on the HCV IRES to form a 48S complex is more sensitive than binary complex formation to these structural changes. Correct positioning of the initiation codon in the ribosomal P site is therefore determined by RNA sequences within different regions of the IRES and is impaired by mutation of these sequences.
Structural integrity of the CSFV pseudoknot is required for stable binding of 43S complexes at the initiation codon
CSFV and HCV IRES elements both contain an essential pseudoknot 11–12 nucleotides upstream of the intiation codon (Wang et al. 1995; Rijnbrand et al. 1997; S.P. Fletcher and R.J. Jackson, in prep.). We examined its contribution to CSFV IRES function using three mutants, UC325, GA357, and UC325GA357, that have substitutions in helix II of the pseudoknot and one mutant, ΔA349–A353, that has a deletion in the loop connecting helices I and II. Helix I is restored in the UC325GA357 pseudorevertant. It has near-wild-type IRES activity, whereas that of all other mutants is strongly impaired (S.P. Fletcher and R.J. Jackson, in prep.). These mutations are illustrated in Figure 7C.
Subunits (40S) bound similar proportions of wild type and of these four mutant CSFV RNAs (Fig. 2H). Primer extension was arrested strongly by 40S subunits at C334 and G345 on all four mutants (Fig. 7D, lanes 2,5,8,11). The stops at UUU387–389 were much weaker on the UC325, GA357, and ΔA349–A353 mutant RNAs that had been incubated with 40S subunits (Fig. 7D, lanes 2,5,11) than at these positions on wild-type CSFV RNA (Fig. 3D, lane 2) or on the UC325GA357 pseudorevertant (Fig. 7D, lane 8) that had been incubated in identical conditions. After incubation with eIF2, eIF3, 40S ribosomal subunits, Met–tRNAiMet, and GMP–PNP, RT arrest at UGA390–392 was relatively weaker on UC325, GA357, and ΔA349–A353 mutant RNAs (Fig. 7D, lanes 3,6,12) than on wild-type CSFV RNA (Fig. 3D, lane 5) or on the UC325GA357 pseudorevertant (Fig. 7D, lane 9). A prominent stop caused by bound eIF3 appeared at A250 on all four mutant RNAs (Fig. 7D, lanes 3,6,9,12).
These results indicate that the 3′ half of the CSFV pseudoknot is not a primary determinant of 40S subunit binding to this IRES but that it influences an interaction between them that may be required for correct positioning of the initiation codon in the ribosomal P site and therefore for stable assembly of 48S ribosomal complexes at the CSFV initiation codon. Taken together with the results of analysis of the HCV deletion mutants, these observations indicate that 48S complex assembly at the initiation codon is determined by RNA elements that are dispersed throughout the linear IRES sequence.
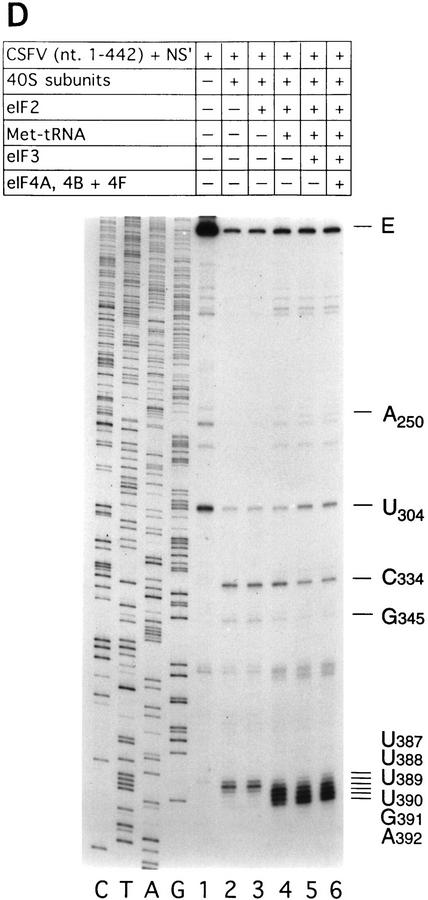
Deletion of nucleotides 145–148 abrogates binding of 40S subunits to the CSFV IRES
40S subunits bound much less strongly to CSFV nucleotides 1–321 than to CSFV nucleotides 1–442 RNA (Fig. 2I). This result indicates that an important determinant of IRES–40S subunit binding is located near the base of domain III. We deleted nucleotides 145–148 from the basal helix of this domain (Fig. 7C) to determine whether its integrity is required for binary complex formation. CSFV IRES-mediated translation of the NS′ cistron was abolished by this deletion (Fig. 8A). The difference in translation of wild-type and mutant CSFV mRNAs was not attributable to different stabilities of these templates in RRL. No significant difference in the degradation of these two templates was found after 15 min of incubation in RRL (data not shown). Ribosomal subunits (40S) bound wild-type [32P]UTP-labeled CSFV nucleotides 1–442 RNA stably; their binding to corresponding nucleotides 1–442(ΔA145–U148) mutant RNA was abolished almost totally (Fig. 2J). This result was supported strongly by the results of toeprinting analysis. Primer extension was arrested by 40S subunits at C334, G345, and UUU387–389 on wild-type CSFV RNA (Fig. 8B, lane 2), whereas RT arrest at these positions on nucleotides 1–442(ΔA145–U148) mutant RNA was insignificant (Fig. 8B, lane 5). A strong stop was detected at A250 on CSFV nucleotides 1–442(ΔA145–U148) mutant RNA incubated with GMP–PNP, Met–tRNAiMet, eIF2, eIF3, and 40S subunits (Fig. 8B, lane 5). This stop is caused by binding of eIF3 to the CSFV IRES, which is therefore not affected by deletion of nucleotides 145–148. In the same reaction, toeprinting did not yield stops at UGA390–392. This result indicates that 48S complexes are unable to form at the initiation codon on the nucleotides 1–442(ΔA145–U148) mutant IRES. The pattern of endogenous strong stops on wild-type and nucleotides 1–442(ΔA145–U148) mutant CSFV IRESs were similar, indicating that their structures did not differ grossly (Fig. 8B, lanes 1,4). cDNAs shown in Figure 8B were uniformly labeled with [α-32P]dATP and their intensity is therefore proportional to their length, whereas cDNAs shown in all other figures were end-labeled with [γ-32P]ATP and their intensities are independent of length. Taken together, these results indicate that the nucleotides 1–442(ΔA145–U148) CSFV mutant has a primary defect in binding to 40S subunits that is responsible for its inability to support assembly of 48S complexes and therefore to act as a template for translation in vitro.
Ribosomal protein S9 interacts with HCV and CSFV IRESs in binary IRES–40S subunit complexes
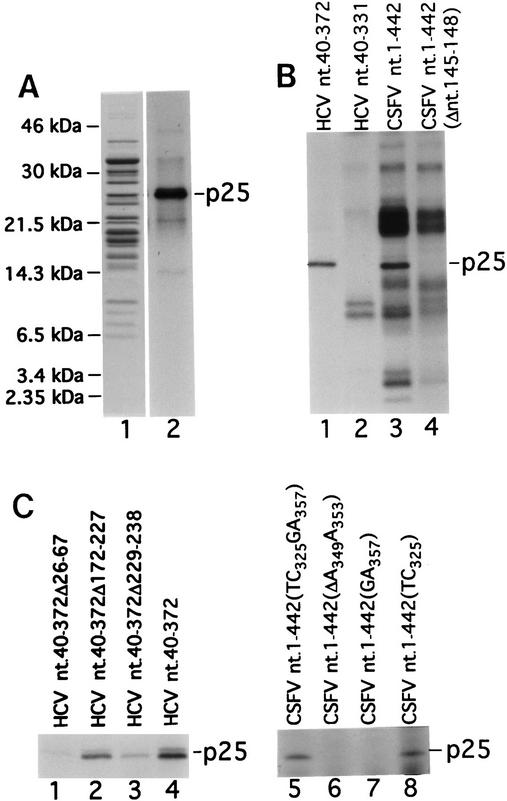
Primer extension analyses indicate that binary complexes described here are stabilized by multiple IRES–40S subunit interactions. We used UV cross-linking to determine whether specific ribosomal proteins bound to [32P]UTP-labeled HCV RNA. A single 25-kD protein (p25) was labeled after UV cross-linking binary complexes of rabbit 40S subunits and [32P]UTP-labeled HCV nucleotides 40–372 RNA (Fig. 9A, lane 2). The same protein was one of several bands that appeared after UV cross-linking binary complexes of 40S subunits and [32P]UTP-labeled CSFV nucleotides 1–442 RNA (Fig. 9B, lane 3). These additional bands also appeared after UV cross-linking CSFV nucleotides 1–442 (ΔA145–U148) RNA and 40S subunits, which do not form binary complexes (Fig. 9B, lane 4). They did not co-migrate with ribosomal proteins and are probably products of incomplete RNase digestion of UV cross-linked CSFV RNA.
Figure 9.
UV cross-linking of ribosomal protein S9 to HCV and CSFV IRESs. (A) Proteins from rabbit ribosomal 40S subunits (lane 1) or from binary complexes of rabbit 40S ribosomal subunits and [32P]UTP-labeled HCV nucleotides 40–372 RNA (lane 2) were resolved by gel electrophoresis directly (lane 1) or after UV cross-linking and RNase digestion (lane 2). The positions of molecular mass marker proteins are indicated to the left of lane 1. (B) Proteins from binary complexes of rabbit 40S ribosomal subunits and [32P]UTP-labeled HCV nucleotides 40–372 RNA (lane 1), HCV nucleotides 40–331 RNA (lane 2), CSFV nucleotides 1–442 RNA (lane 3), or CSFV nucleotides 1–442(Δ nucleotides 145–148) (lane 4) were resolved by gel electrophoresis after UV cross-linking, and RNase digestion. (C) Proteins from binary complexes of rabbit 40S ribosomal subunits and [32P]UTP-labeled HCV nucleotides 40–372Δ26–67 RNA (lane 1), HCV nucleotides 40–372Δ172–227 RNA (lane 2), HCV nucleotides 40–372Δ229–238 RNA (lane 3), HCV nucleotides 40–372 (lane 4), CSFV nucleotides 1–442 (TC325GA357) (lane 5), CSFV nucleotides 1–442 (ΔA349–A353) (lane 6), CSFV nucleotides 1–442 (GA357) (lane 7), or CSFV nucleotides 1–442 (TC325) (lane 8) were resolved by gel electrophoresis after UV cross-linking, and RNase digestion. Proteins in A (lane 1) were visualized by staining with Coomassie blue; radio-labeled proteins in all other lanes in A,B, and C were visualized by autoradiography.
p25 was not detected when [32P]UTP-labeled HCV nucleotides 40–331 RNA was cross-linked to 40S subunits (Fig. 9B, lane 2) although this RNA forms a stable binary complex with 40S subunits (Fig. 2F). p25 is therefore not a primary determinant of binary complex formation. p25 was also not detected when [32P]UTP-labeled CSFV nucleotides 1–442 (ΔA145–U148) RNA was cross-linked to 40S subunits (Fig. 9B, lane 4). This mutant RNA does not form a binary complex (Fig. 1J). The four nucleotides that are deleted in this CSFV RNA are part of a helix that is intact in HCV nucleotides 40–331 RNA. Nevertheless, these data do not indicate whether p25 makes independent contact with these IRESs or whether such contact depends on proper positioning of the RNA determined by a prior primary recognition step. CSFV nucleotides 145–148 are clearly required for primary recognition of the IRES by 40S subunits, but the effect of deleting these residues on structures elsewhere in the IRES that could determine potential independent interaction with p25 is not known.
UV cross-linking of p25 was sensitive to changes in structure of different regions of these IRESs. Labeling of p25 after UV cross-linking of binary complexes assembled on wild-type and mutant HCV IRESs was reduced by deletion of hairpins IIa and IIIc but was not significantly affected by deletion of hairpin IIIb (Fig. 9C, lanes 1–4). Mutations within the CSFV pseudoknot also affected UV cross-linking of p25. p25 became labeled much less strongly after 40S subunits were cross-linked to [32P]UTP-labeled GA357 and ΔA349–A353 mutant CSFV nucleotides 1–442 RNAs than to corresponding UC325 and UC325GA357 CSFV RNAs (Fig. 9C, lanes 5–8).
The identity of p25 was established by amino-terminal sequence analysis. The first 10 amino-acid residues of p25 were PVATRSWY(X)RK, in which (X) represents a residue whose identity could not be determined. Cysteine residues are destroyed chemically in the sequencing reaction. These residues match the amino terminus of the S9 ribosomal protein exactly (Chan et al. 1993).
Discussion
We have reconstituted internal ribosomal entry on HCV and CSFV IRESs in vitro to the stage of 48S complex formation using purified translation components to identify the necessary factors and to characterize their role and that of these two RNAs in this process. Our results show that the initiation mechanism used by HCV and CSFV IRESs differs from modes of eukaryotic initiation described previously. Some of the unusual properties of HCV and CSFV IRES-mediated initiation have parallels in the process of translation initiation in prokaryotes.
Factors and cofactors required for internal ribosomal entry
Ribosomal subunits (40S) formed stable binary complexes on HCV and CSFV IRESs in which the initiation codon is in the ribosomal P site. The processes of ribosomal binding to these IRESs, positioning of the ribosome at the initiation codon, and formation of active 80S complexes did not require the initiation factors eIF4A, eIF4B, and eIF4F and were not influenced by them. Initiation mediated by these two IRESs therefore differs fundamentally from initiation on all other eukaryotic mRNAs, including even internal initiation mediated by the EMCV IRES, which depends absolutely on one or more of these factors (Trachsel et al. 1977; Benne and Hershey 1978; Blum et al. 1989; Pause et al. 1994; Pestova et al. 1996a,b). This unexpected observation was supported strongly by the lack of effect of the eIF4A R362Q mutant on HCV and CSFV translation in RRL. This eIF4A mutant is a trans-dominant inhibitor of cap-mediated and EMCV IRES-mediated initiation (Pause et al. 1994). eIF4A, eIF4B, and eIF4F are involved in ATP-dependent attachment of 40S subunits to conventional eukaryotic mRNAs and in ATP-dependent ribosomal scanning from the binding site to the initiation codon (Merrick 1992). Initiation of HCV and CSFV translation has been shown here to differ from initiation on other eukaryotic mRNAs at both of these stages. eIF3 was also not essential for 48S complex assembly on HCV or CSFV IRESs in vitro (Fig. 3) but was absolutely required for 80S complex formation on these RNAs. Our data do not allow us to determine what role eIF3 has in 80S complex formation. It could modify the conformation of 48S complexes such that they can bind to 60S subunits, or it could be involved more directly in subunit joining. Addition of Met–tRNAiMet, eIF2, and GTP to binary IRES–40S subunit complexes was necessary and sufficient for 48S complexes to form at the initiation codon. Correct toeprints corresponding to these complexes were not detected on HCV and CSFV IRESs if Met–tRNAiMet or eIF2 were omitted from reactions or if the initiation codon was mutated. Cognate base-pairing between HCV/CSFV initiation codons and the anticodon of initiator tRNA is therefore required for assembly of stable 48S complexes.
Interactions between IRESs and translation components
Noncanonical factors are not required for internal initiation on HCV or CSFV IRESs (this study) or on the structurally distinct EMCV IRES (Pestova et al. 1996a). Internal ribosomal entry must therefore result from interactions between these IRESs and canonical components of the translational machinery. EMCV IRES function involves a stable interaction with the 4G subunit of eIF4F (Pestova et al. 1996b). eIF4F is not involved in HCV and CSFV translation, but we have identified an interaction between these two IRESs and two other essential translation components—eIF3 and the 40S ribosomal subunit. The interaction of these RNAs with eIF3 is discussed in detail elsewhere (D. Sizova, V. Kolupaeva, T. Pestova, I. N. Shatsky, and C. Hellen, in prep.).
Rabbit 40S ribosomal subunits formed stable binary complexes with HCV and CSFV IRESs in the absence of ATP that were not dissociated during reverse transcription or sucrose density gradient centrifugation. Binary complexes consisting of mRNA and the small ribosomal subunit have been identified in prokaryotes (Hartz et al. 1991) but not in eukaryotes. Binding of β-globin and EMCV mRNAs to 40S subunits is dependent on prior binding of the ternary complex (Trachsel et al. 1977; Benne and Hershey 1978; Pestova et al. 1996a). This is therefore the first report of binary complexes assembled from eukaryotic 40S ribosomal subunits and natural mRNAs.
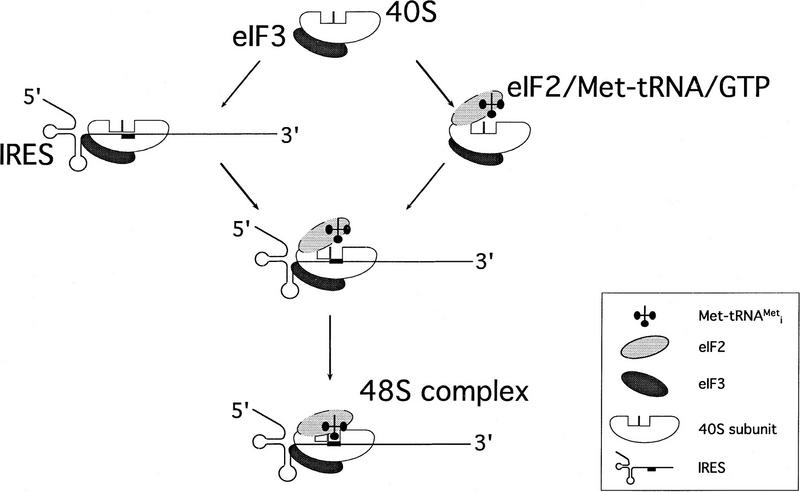
The data presented here show that it is the 40S ribosomal subunit itself that is responsible for binding of the 43S complex to the IRES to form the 48S preinitiation complex, but our data do not enable us to say in which order the IRES-containing mRNA and the ternary complex bind to the native 40S subunit under physiological conditions (Fig. 10). Base-pairing between the initiation codon and the anticodon of the initiator tRNA is a reasonable last step in assembly of 48S preinitiation complexes. This step is characterized by an increase in the intensity and a shift forward in the position of the toeprints caused by RT arrest at the leading edge of the 40S subunit. These observations suggest that mRNA downstream of the initiation codon undergoes a conformational change at this stage caused by codon–anticodon base-pairing that locks this region into place in the mRNA-binding groove of the 40S subunit, exactly as in prokaryotes (McCarthy and Brimacombe 1994).
Figure 10.
Model for the mechanism of 48S ribosomal preinitiation complex formation mediated by HCV/CSFV IRES elements. The major steps are as follows. (1) The native 40S ribosomal subunit binds directly to the IRES to form a stable binary complex in which the initiation codon is situated at the ribosomal P site. (2) The ternary eIF2/GTP/Met–tRNAiMet complex binds to the binary IRES/40S subunit complex. (3) Base-pairing between the initiation codon of the mRNA and the anticodon of the initiator tRNA causes a conformational change in the mRNA downstream of the initiation codon that locks this region into place in the mRNA-binding grove of the 40S subunit. The 40S subunit determines the interaction of the 43S complex with the IRES to form the 48S complex, but our data are not sufficient to determine in which order steps (1) and (2) occur. eIF3 is effectively a constitutive component of native 40S subunits and may stabilize ribosomal complexes by virtue of its interaction with the IRES. eIF3 is required for subsequent steps in the initiation process.
The parallels between the ribosomal complexes reported here and corresponding prokaryotic complexes (Hartz et al. 1991) are striking. Eukaryotic 48S complexes and prokaryotic ternary complexes were both detected under conditions used to toeprint binary ribosomal complexes if initiator tRNA and eIF2 (or IF2, as appropriate) were included in assembly reactions. These complexes also yielded the toeprint signals from the binary complex. This result is expected if binding of the small ribosomal subunit to mRNA is an intermediate step in initiation complex formation.
Primer extension and deletion analyses indicate that these eukaryotic binary complexes are stabilized by multiple IRES–40S subunit interactions. One contact between these IRESs and 40S subunits involves ribosomal protein S9. This protein is located on the 40S subunit at the other end of the mRNA-binding cleft from the eIF3-binding site (Lütsch et al. 1983). The results of deletion analysis suggest that nucleotides upstream of the HCV initiation codon bind to S9 and are consistent with the labeling of S9 by 5′ but not 3′ derivatised AUGUUU mRNA analogs bound to 40S subunits (Mundus et al. 1993). Labeling of S9 by HCV ΔIIa and ΔIIIc deletion mutants and by CSFV pseudoknot mutants was reduced or abolished, but these RNAs still formed stable binary IRES–40S subunit complexes. The S9–IRES interaction is therefore not the principal determinant of binary complex formation. The observation that S9 is the only ribosomal protein to be labeled significantly after UV cross-linking to the wild-type HCV IRES suggests that other interactions between it and the 40S subunit could involve 18S rRNA. Toeprint analysis of ribosomal complexes on these mutant IRESs indicated that formation of stable 48S complexes at HCV and CSFV initiation codons was impaired severely (Figs. 6 and 7). These results suggest that interaction of HCV and CSFV IRESs with S9 is either a cause or a consequence of correct alignment of their initiation codons in the ribosomal P site.
The premise that 40S subunits can bind mRNA only after binding the ternary complex is based on a failure to detect binary complexes by sucrose density gradient centrifugation or toeprinting. These methods may not reveal unstable ribosomal complexes. The possibility that 40S subunits could bind mRNA before initiator tRNA, however, has been advanced previously to explain reinitiation at a downstream AUG codon after translation of an upstream open reading frame (ORF) (Kozak 1987) and the influence of mRNA and eIF2 concentration on the relative use of tandem initiation sites in an mRNA (Dasso et al. 1990). The two eukaryotic mRNAs identified here that are bound by 40S subunits in the absence of Met–tRNAiMet or initiation factors could therefore represent one end of a spectrum of mRNAs that bind 40S subunits with different affinities. These observations indicate that 40S subunits participate actively in selection and binding of mRNA and taken together, suggest that a re-evaluation of current models for eukaryotic initiation is appropriate.
IRES function in initiation
Dissection of the initiation process mediated by the structurally distinct EMCV-like and HCV-like groups of IRESs has revealed two different mechanisms of internal ribosomal entry (Pestova et al. 1996a,b; this report). A common feature of these mechanisms is that both involve specific interactions between the IRES and canonical translation components. An efficient initiation mechanism that involves fewer translation components than that described here for HCV and CSFV is unlikely. Given the diversity of cellular and viral IRESs whose structures appear unrelated to those of either EMCV or HCV/CSFV, however, we anticipate that some IRESs exploit alternative modes of interaction with initiation factors and 40S subunits and cannot exclude the possibility that others will be found to require additional translation components.
43S complexes bound directly to the initiation codon of HCV and CSFV IRESs. The location of the ribosomal binding site is consistent with the results of mutational analyses (Honda et al. 1996; Reynolds et al. 1996; Rijnbrand et al. 1996, 1997). These results indicate that these IRESs not only attach to 43S complexes, but also that they contain determinants that direct ribosomal binding to a specific location on the IRES. Structurally distinct EMCV-like IRESs also direct ribosomal binding specifically to the initiation codon (Kaminski et al. 1990; Pilipenko et al. 1994). Even the poliovirus IRES, which is separated from the authentic initiation codon by over 150 nucleotides, requires an AUG triplet at the ribosome-binding site that can act as an initiation codon given appropriate changes in context (Pilipenko et al. 1992; Pestova et al. 1994).
The results presented here indicate that IRESs have a direct role in attaching a 43S complex to an mRNA and in directing its binding to a specific location on it. Previous models for IRES function suggested that these roles are mediated indirectly by noncanonical factors (for discussion, see Kolupaeva et al. 1996). We identified sequences located in different regions of IRESs that are recognized directly by the 40S ribosomal subunit and initiation factors such as eIF3 and eIF4F (Pestova et al. 1996a,b; this report). Mutations can abrogate IRES function without affecting one or the other of these interactions, so individually they are not sufficient to promote ribosomal binding at a defined location on an IRES. The ability to direct stable assembly of a 48S complex at a specific position is therefore probably determined by the correct spatial dispositon of multiple binding sites on the IRES. This model provides an explanation for the large size and structural complexity of IRES elements and for the functional requirement for structural integrity. It is consistent with previous observations that the sequences of a group of IRESs are conserved less strongly than their secondary structures, and that conserved residues tend to occur in unpaired loops and bulges that could participate in RNA–RNA or RNA–protein interactions (Jackson and Kaminski 1995).
Translation initiation mediated by the EMCV IRES does not require the 4E subunit of eIF4F (Pestova et al. 1996a,b) and initiation mediated by the HCV and CSFV IRESs does not require eIF4A, eIF4B, or any of the subunits of eIF4F (this report). On the basis of these results, we conclude that IRES-mediated initiation is unexpectedly simpler than cap-mediated initiation. The RNA-binding step is rate-limiting in eukaryotic initiation and the factors that mediate it are targets for regulation (Sonenberg 1996). mRNAs that contain IRESs that do not use these factors are therefore likely to be translated preferentially under conditions such as growth arrest, mitosis, or heat shock when the activities of eIF4E, eIF4B, and eIF4F are reduced. It is notable that cellular IRESs promote translation of a variety of regulatory and stress-response proteins. The observation that cap-mediated initiation is more complex than the mechanisms of IRES-mediated initiation examined to date suggests that the cap-recognition and scanning steps in initiation may be evolutionary developments that increase the mechanisms available for post-transcriptional regulation of gene expression in eukaryotes.
Materials and methods
Plasmids
The HCV plasmids pHCV(40–372).NS′, pHCV(40–354).NS′, pHCV(40–351).NS′, pHCV(40–339).NS′, and pXL.HCV(40–373).NS′ have been described (Reynolds et al. 1995). The monocistronic vectors pHCV(40–373) (AUG344 → GCG).NS′ and pHCV(40–373) (AUG344 → AAG). NS′ were made by replacing the smaller BamHI–EcoRI fragment from pHCV(40–373).NS′ by equivalent fragments from mutant bicistronic constructs (Reynolds et al. 1995). pWT–CAT contains the full HCV 5′ UTR linked to a CAT reporter cistron. pΔB–CAT, pΔE–CAT, and pΔF–CAT are identical to pWT–CAT except for deletions of HCV nucleotides 26–67, nucleotides 172–227 (hairpin IIIb), and nucleotides 229–238 (hairpin IIIc) inclusive, respectively (Rijnbrand et al. 1995).
CSFV nucleotides 1–750 cDNA (Meyers et al. 1989) was amplified by PCR and cloned into pGEM1 (Promega, Madison, WI). The resulting plasmid was linearized, digested with exonuclease III for various times, and filled-in. Residual CSFV sequences released by SalI digestion were cloned into pXLJ0 (Reynolds et al. 1995) to generate dicistronic vectors such as pXL.CSFV(1–442).NS′ in which CSFV sequences were placed between the upstream Xenopus laevis cyclin B2 cDNA and the downstream influenza NS′ cDNA. CSFV–NS′ cDNAs from these vectors were cloned into pJO (Borman and Jackson 1992) to generate the monocistronic vectors pCSFV(1–442).NS′, pCSFV(1–409).NS′, and pCSFV(1–373).NS′. Pseudoknot sequences were mutated by PCR to generate pXL.CSFV(1–442)[TC325].NS′, pXL.CSFV(1–442) [GA357].NS′, pXL.CSFV(1–442)[TC325GA357].NS′, and pXL.CSFV(1–442)[ΔA349–A353].NS′. pCSFV(1–442Δ145–148).NS′ was derived by deleting four nucleotides at the SacI site of pCSFV(1–442).NS′.
pTE1 contains EMCV nucleotides 315–1155 downstream of a T7 promoter (Evstafieva et al. 1991). The expression vectors pET(His6–eIF4A) and pET(His6-eIF4B) have been described (Pestova et al. 1996a,b).
Purification of factors and 40S ribosomal subunits
40S ribosomal subunits were prepared from RRL as described (Pestova et al. 1996a) except that after sucrose density gradient centrifugation, 40S subunits were concentrated in a Centricon S-30 (Amicon, Waltham, MA). eIF2, eIF3, and eIF4F were purified from RRL. No cross-contamination was detected by Western blotting using antibodies specific for each one of these factors. The purity and quality of factors were equal to or greater than that we described previously (Pestova et al. 1996a). Recombinant eIF4A and eIF4B were purified as described (Pestova et al. 1996a). The activities of purified initiation factors were verified using conventional assays (Merrick 1979; Pause et al. 1994).
Transcription reactions and translation assays
CSFV, HCV, and EMCV RNAs of defined length were transcribed in vitro either with or without [32P]UTP (∼3000 Ci/mmoles; ICN Radiochemicals, Irvine, CA) using wild-type or DEL 172-3 mutant T7 RNA polymerase, as appropriate, from plasmids that had been linearized at appropriate sites. CSFV plasmids were linearized by digestion with either PstI or BamHI to generate transcripts that terminated at nucleotides 321 and 442, respectively. HCV plasmids were linearized by digestion with either AccI or BamHI to generate transcripts that terminated at nucleotides 331 and 372, respectively. RNA was purified using Nuc-Trap columns (Stratagene, La Jolla, CA) as described (Pestova et al. 1991, 1996a). RNAs had specific activities of ∼300,000 to 500,000 cpm/μg. The vectors pCSFV(1–442Δ145–148).NS′, pXL.HCV(40–373).NS′, and XL.CSFV(1–442).NS′ were linearized by digestion with EcoRI prior to transcription of mRNAs that were translated in RRL (Promega) as described (Pause et al. 1994). Translation products were resolved by electrophoresis using 17% polyacrylamide gel. Gels were exposed to x-ray film overnight.
Assembly and analysis of ribosomal complexes
[35S]Methionine-labeled Met–tRNAiMet was prepared as described using rabbit tRNA (Novagen, Madison, WI) and aminoacyl-tRNA synthetase purified from Escherichia coli MRE600 (Pestova et al. 1996a). Ribosomal preinitiation complexes were assembled by incubating 1 μg of CSFV, HCV, or EMCV mRNA as appropriate, as described (Pestova et al. 1996a) except that incubation was for 3 min eIF2 (4 μg), eIF3 (10 μg), eIF4A (2 μg), eIF4B (2 μg), and eIF4F (4 μg), Met–tRNAiMet (6 pmoles) and 40S subunits (6 pmoles) were included in reaction volumes of 40 μl as indicated in the text. Ribosomal complexes were analyzed by sucrose density gradient centrifugation as described (Pestova et al. 1996a).
Ribosomal and RNP complexes were analyzed by primer extension as described (Pestova et al. 1996a), except that unless indicated, [α-32P]dATP was not present in extension reactions and primers were instead end-labeled with [γ-32P]ATP (∼6000 Ci/mmole) using T4 polynucleotide kinase. The primers 5′-CTCGTTTGCGGACATGCC-3′ and 5′-GGGATTTCTGATCTCGGCG-3′ (complementary to different parts of the NS′-coding sequence), 5′-GCAACTGACTGAAATGCC-3′ (complementary to part of the CAT-coding sequence), 5′-CGCAAGCACCCTATC-3′ (complementary to HCV nucleotides 295–309), and 5′-CCTGATAGGGTGCTGCAG-3′ (complementary to CSFV nucleotides 309–326) were used for analysis of complexes formed on CSFV–NS′, HCV–NS′, and HCV–CAT mRNAs, respectively.
80S ribosomal complexes were assembled on CSFV (nucleotides 1–442)–NS′ mRNA and β-globin mRNA (Life Technologies, Grand Island, NY) using the buffer conditions described previously (Pestova et al. 1996a) except that GMP–PNP was replaced by GTP. Reaction mixtures (100 μl) contained mRNA (2 μg), eIF2 (6 μg), eIF3 (10 μg), eIF4A (3 μg), eIF4B (2 μg), and eIF4F (3 μg), Met–tRNAiMet (8 pmoles, with specific activity 4000–8000 cpm/pmoles), 40S subunits (8 pmoles), 60S subunits (8 pmoles), and 4 μg of a 50%–70% ammonium sulphate (A.S.) RSW fraction that had been purified by elution with 0.2–0.8 m KCl from a phosphocellulose column. Incubation was for 5 min at 37°C. Ribosomal complexes were analyzed after 10%–30% sucrose density gradient centrifugation for 180 min at 39,000 rpm and 4°C using a Beckman SW41 rotor.
The same buffer conditions were used for the methionyl-puromycin synthesis reaction as described above for 80S complex formation. A 50-μl reaction mixture that contained mRNA (1 μg), eIF2 (3 μg), eIF3 (5 μg), eIF4A (1.5 μg), eIF4B (1 μg), and eIF4F (2 μg), Met–tRNAiMet (4 pmoles, with specific activity 40,000 cpm/pmole) and 40S subunits (6 pmoles) was incubated at 37°C for 5 min. Incubation was continued at 37°C for another 20 min after addition of 1 mm puromycin (Calbiochem), 60S subunits (6 pmoles) and 2 μg of the same 50%–70% ammonium sulphate (A.S.) RSW fraction as described above. The reaction mixture was then diluted with 0.2 m potassium phosphate at pH 8.0 and extracted with ethyl acetate as described (Leder and Bursztyn 1966). An aliquot of the ethyl acetate phase was counted by liquid scintillation.
UV cross-linking and protein sequencing
UV cross-linking of binary IRES–40S subunit complexes was done essentially as described previously (Pestova et al. 1991). Ribosomal proteins were resolved by tricine–SDS polyacrylamide gel electrophoresis and either dried and exposed to x-ray film or transferred to PVDF membrane for amino-terminal sequencing, using an Applied Biosystems Procise sequencer.
Acknowledgments
We thank W.C. Merrick and V.I. Agol for helpful comments during the course of this study. We thank Karen Browning for wheat germ 40S subunits, Ashkhan Haghighat and Nahum Sonenberg for eIF4A (R362Q), Dmitry Lyakhov for DEL 172-3 mutant T7 RNA polymerase, P. Bredenbeek for HCV plasmids and H.-J. Thiel for the CSFV cDNA clone that was used for all the constructions. We thank Linda Siconolfi-Baez for sequencing the S9 protein, Ernest Cuni for photography and Rodney Romain for technical assistance. This work was supported by grants from the Council for Tobacco Research (C.U.T.H.), the Wellcome Trust (R.J.J.), the Howard Hughes Medical Institute and NATO (C.U.T.H. and I.N.S.). S.P.F. gratefully acknowledges the award of a Research Studentship from the Medical Research Council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL chellen@netmail.hscbklyn.edu; FAX (718) 270-2656.
References
- Anthony DD, Merrick WC. Analysis of 40S and 80S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- Baker A-M, Draper DE. Messenger RNA recognition by fragments of ribosomal protein S4. J Biol Chem. 1995;270:22939–22945. doi: 10.1074/jbc.270.39.22939. [DOI] [PubMed] [Google Scholar]
- Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- Benne R, Brown-Luedi ML, Hershey JWB. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J Biol Chem. 1978;253:3070–3077. [PubMed] [Google Scholar]
- Blum S, Mueller M, Schmid SR, Linder P, Trachsel H. Translation in Saccharomyces cerevisiae: Initiation factor 4A-dependent cell-free system. Proc Natl Acad Sci. 1989;86:6043–6046. doi: 10.1073/pnas.86.16.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A, Jackson RJ. Initiation of translation of human rhinovirus RNA: Mapping the internal ribosome entry site. Virology. 1992;188:685–696. doi: 10.1016/0042-6822(92)90523-r. [DOI] [PubMed] [Google Scholar]
- Brown EA, Zhang H, Ping L-H, Lemon SM. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y-L, Paz V, Olvera J, Wool IG. The primary structure of rat ribosomal protein S9. Biochem Biophys Res Comm. 1993;193:106–112. doi: 10.1006/bbrc.1993.1596. [DOI] [PubMed] [Google Scholar]
- Dasso MC, Milburn SC, Hershey JWB, Jackson RJ. Selection of the 5′-proximal translation initiation site is influenced by mRNA and eIF-2 concentrations. Eur J Biochem. 1990;187:361–371. doi: 10.1111/j.1432-1033.1990.tb15313.x. [DOI] [PubMed] [Google Scholar]
- Evstafieva AG, Ugarova TY, Chernov BK, Shatsky IN. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 1991;19:665–671. doi: 10.1093/nar/19.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983;258:5804–5810. [PubMed] [Google Scholar]
- Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:1684–1689. [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Green L, Gold L. Detection of Escherichia coli ribosome binding at translation initiation sites in the absence of tRNA. J Mol Biol. 1991;218:99–105. doi: 10.1016/0022-2836(91)90876-8. [DOI] [PubMed] [Google Scholar]
- Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Howell MT, Jackson RJ. Initiation of encephalomyocarditis virus RNA translation: The authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Hellen CUT, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P, Bursztyn H. Initiation of protein synthesis II. A convenient assay for the ribosome-dependent synthesis of N-formyl-C14-methionylpuromycin. Biochem Biophys Res Comm. 1966;25:233–238. doi: 10.1016/0006-291x(66)90586-9. [DOI] [PubMed] [Google Scholar]
- Lu H-H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütsch G, Bielka H, Enzmann G, Noll F. Electron microscopic investigations on the location of rat liver ribosomal proteins S3a, 5, S6, S7, and S9 by means of antibody labeling. Biomed Biochim Acta. 1983;42:705–723. [PubMed] [Google Scholar]
- McCarthy JEG, Brimacombe R. Prokaryotic translation: The interactive pathway leading to initiation. Trends Genet. 1994;10:402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Merrick WC. Purification of protein synthesis initiation factors from rabbit reticulocytes. Methods Enzymol. 1979;60:101–108. doi: 10.1016/s0076-6879(79)60010-1. [DOI] [PubMed] [Google Scholar]
- ————— Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers G, Rumenapf T, Thiel H-J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989;171:555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- Mundus DA, Bulygin KN, Yamkovoy VI, Malygin AA, Repkova MN, Vratskikh LV, Venijaminova AG, Vladimirov SN, Karpova GG. Structural placement of the codon-anticodon interaction area in human placenta ribosomes. Affinity labeling of the 40S subunits by derivatives of oligoribo-nucleotides containing the AUG codon. Biochim Biophys Acta. 1993;1173:273–282. doi: 10.1016/0167-4781(93)90124-v. [DOI] [PubMed] [Google Scholar]
- Pause A, Méthot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT, Wimmer E. Translation of poliovirus RNA: Role of an essential cis-acting oligopyrimidine element within the 5′-nontranslated region and involvement of a cellular 57-kilodaton protein. J Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT, Wimmer E. A conserved AUG triplet in the 5′ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology. 1994;204:729–737. doi: 10.1006/viro.1994.1588. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996a;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CUT. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996b;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko EV, Gmyl AP, Maslova SV, Svitkin YV, Sinyakov AN, Agol VI. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- Pilipenko EV, Gmyl AP, Maslova SV, Belov GA, Sinyakov AN, Huang M, Brown TDK, Agol VI. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J Mol Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- Poole TL, Wang C, Popp RA, Potgieter LND, Siddiqui A, Collett MS. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ. Internal initiation of translation of hepatitis C virus RNA: The ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand R, Bredenbeek PJ, van der Straaten T, Whetter L, Inchauspé G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Letts. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- Rijnbrand R, Abbink TEM, Haasnoot PCJ, Spaan WJM, Bredenbeek PJ. The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47–56. doi: 10.1006/viro.1996.0626. [DOI] [PubMed] [Google Scholar]
- Rijnbrand R, van der Straaten T, van Rijn PA, Spaan WJM, Bredenbeek PJ. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translation control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 271–294. [Google Scholar]
- Sundkvist IC, Staehelin T. Structure and function of free 40S ribosome subunits: Characterization of initiation factors. J Mol Biol. 1975;99:401–418. doi: 10.1016/s0022-2836(75)80135-5. [DOI] [PubMed] [Google Scholar]
- Trachsel H, Erni B, Schreier MH, Staehelin T. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J Mol Biol. 1977;116:755–767. doi: 10.1016/0022-2836(77)90269-8. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosomal entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Le SY, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]