Abstract
The anomalous folding and polymerization of the β-amyloid (Aβ) peptide is thought to initiate the neurodegenerative cascade in Alzheimer's disease pathogenesis1. Aβ is predominantly a 40- or 42-amino acid peptide that is prone to self-aggregation into β-sheet-rich amyloid fibrils that are found in the cores of cerebral senile plaques in Alzheimer's disease. Increasing evidence suggests that low molecular weight, soluble Aβ multimers are more toxic than fibrillar Aβ amyloid2. The identification and quantification of low- and high-molecular weight multimeric Aβ species in brain tissue is an essential objective in Alzheimer's disease research, and the methods employed also can be applied to the identification and characterization of toxic multimers in other proteopathies3. Naturally occurring Aβ multimers can be detected by SDS-polyacrylamide gel electrophoresis followed by immunoblotting with Aβ-specific antibodies. However, the separation and detection of multimeric Aβ requires the use of highly concentrated cortical homogenates and antigen retrieval in small pore-size nitrocellulose membranes. Here we describe a technique for the preparation of clarified human cortical homogenates, separation of proteins by SDS-PAGE, and antigen-epitope retrieval/Western blotting with antibody 6E10 to the N-terminal region of the Aβ peptide. Using this protocol, we consistently detect Aβ monomers, dimers, trimers, tetramers, and higher molecular weight multimers in cortical tissue from humans with Alzheimer's pathology.
Protocol
Part 1: Preparation of clarified tissue homogenates
Dounce homogenize unfixed cortical tissue in 4 times volume of ice-cold buffer (0.1M phosphate-buffered saline [Ca++- and Mg++-free] plus 2x protease inhibitor cocktail [Santa Cruz]) with approximately 30 even pestle strokes (i.e. add 400μl buffer to 100mg tissue).
Spin homogenates for 5 minutes at 3,000g (4°C). Carefully remove clarified supernatants and store aliquots of these extracts at -80°C until use.
Determine total protein concentration (μg/μl) for clarified homogenates using a bicinchoninic acid (BCA) assay, according to the manufacturer's instructions (ThermoFisher).
Part 2: SDS-PAGE Sample preparation
With a 10 well, 10-20% Tricine gel (Invitrogen), the maximum volume that can be loaded per well is ~25μl. Total volume includes 2x SDS sample buffer and 10x reducing agent, so the maximum volume of clarified homogenate that can be loaded per well is 10μl. Clarified 20% (w/v) cortical homogenates should have a total protein concentration greater than 5 μg/μl, allowing for 50μg total protein per well. Samples with less than 5mg/ml protein will necessitate loading less total protein per well. However, high protein levels are optimal for the detection of monomeric and aggregated Aβ (50-60μg total protein is optimal).Always load the same amount of total protein per well.
For each gel, load at least one well with 10μl of a molecular weight marker such as SeeBlue Plus2 (Invitrogen). As a positive control, run 10-100ng of synthetic Aβ40 or Aβ42 peptide (diluted in 1xPBS) in another well.
Prepare reaction mixtures on ice, using freshly thawed, clarified homogenates. Vortex tubes for 5-10 seconds, heat in a dry bath at 100°C for 5 minutes, then quickly spin all samples to remove condensation in the cap.
Part 3: SDS-PAGE Gel electrophoresis
Load vortexed samples onto a 10-20% Tricine gel in the XCell Sure Lock Mini-Cell Gelbox, using Tricine SDS running buffer, according to the manufacturer's instructions (Invitrogen),
Run gel at a constant voltage of 125V for about 90 minutes. Let samples run until the 4KDa marker band is about 1 cm from the bottom of the gel.
Part 4: Transferring Proteins from Gels to Membranes
Carefully remove gels from plastic case and assemble the transfer sandwich inside the XCell II Blot Module, according to the manufacturer's instructions (Invitrogen). Pre-wet blotting pads and 0.2μm nitrocellulose membranes with Tris-Glycine transfer buffer (20% methanol), and remove any bubbles from blotting pads or the filter paper/membrane sandwich.
In the XCell Gelbox, fill the inner chamber with transfer buffer and fill the outer chamber with dIH2O (methanol exposure can wear down the plastic gelbox over time).
Run the transfer for 2-3 hours at a constant amperage of 25mA.
When the transfer is complete, deconstruct the sandwich and place the gels into dIH2O and the membranes into 1xPBS, both in square plastic petri dishes (or another appropriate container).
To visualize the efficiency of protein transfer, gels can be stained with Simply Blue SafeStain (Invitrogen), and the membranes with Ponceau S Red stain (Sigma Aldrich), both according to the manufacturer's instructions. This staining will not interfere with immunoblotting. If staining reveals inefficient protein transfer, modify transfer time in Step 3.
Part 5. Antigen-epitope retrieval & Immunoblotting
Antigen retrieval is an important step in revealing the Aβ epitopes on the membrane for antibody binding during immunoblotting. Any steamer, microwave, or water bath that maintains a constant temperature of 100°C will suffice. For antigen retrieval in a steamer, heat-seal the membrane in a heavy-duty Kapak plastic pouch filled with 1xPBS, at room temperature. Lay the pouch flat in a pre-heated steamer; once the pouch begins to expand, incubate for an additional 15 minutes. Allow the membrane to cool slowly before removing it from the pouch, to prevent excessive wrinkling.
At room temperature, carefully remove the membrane from the steaming pouch and rinse the membrane for 5 minutes in 1xPBS, followed by a 5-minute rinse in TBS-T (Tris-Buffered Saline, pH 8.0 with 0.05% Tween-20 [Sigma]), on an orbital shaker. Incubate the membrane in blocking solution (2.5% nonfat milk in TBS-T) for one hour, shaking. Without rinsing, transfer membrane to a dish or plastic pouch containing the primary antibody diluted in blocking solution (6E10 at 1:1,000 [1μg/ml] 1:5,000 [0.2μg/ml] dilution, Covance), avoiding bubbles. Incubate on an orbital shaker at room temperature for one hour and then 24-48 hours shaking at 4°C (longer incubation may give a better signal).
Rinse the membrane for 30 minutes in TBS-T (one quick rinse followed by 3 x 10 minute rinses).
Incubate the membrane in HRP-conjugated secondary antibody (Amersham ECL sheep anti-mouse, GE Healthcare), diluted at 1:10,000 in blocking solution, for 90 minutes on the shaker at room temperature.
Rinse the membrane for 30 minutes in TBS-T (one quick rinse followed by 3 x 10 minute rinses).
On the shaker, incubate the membrane in freshly-made SuperSignal West Pico Electrochemiluminescence reagent (ThermoFisher) for 5 minutes, blot off excess reagent on lint-free filter paper and place the membrane in a film cassette, between plastic sheet protectors. Blot off additional SuperSignal reagent with dust-free Kimwipes if necessary.
Expose to Kodak Biomax MR film for intervals of 30 seconds up to 30 minutes and develop in a film developer. After 5 minutes, the Aβ monomer bands will probably be saturated.
Membranes can be stripped (after rinsing in TBS-T) by shaking in Restore Plus stripping buffer for 30 minutes at room temperature and reprobed with additional antibodies if desired.
Part 6: Representative Immunoblot:
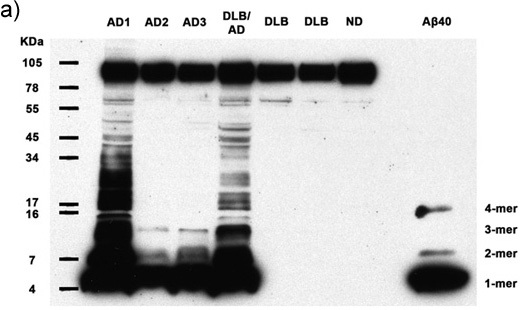
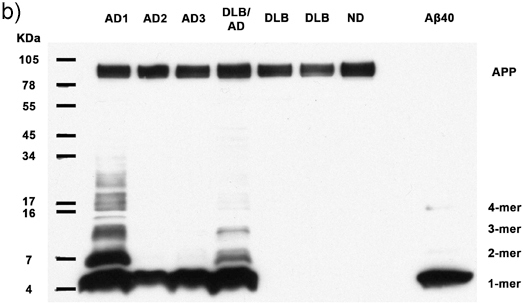
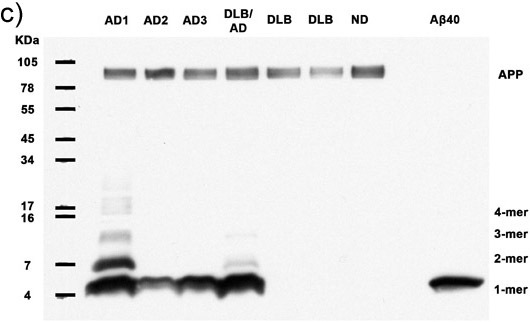 Figures 1a-c. Clarified homogenates containing 50μg total protein from 7 human subjects are analyzed for the presence of multimeric Aβ and APP. Immunoblotting with antibody 6E10 reveals Aβ monomers, dimers, trimers, tetramers, and APP (top band) in all Alzheimer's cases, as well as abundant higher molecular weight Aβ multimers in 2 AD cases. Synthetic Aβ40 confirms the identity of the lower molecular weight bands. AD: Alzheimer's disease, DLB: Dementia with Lewy Bodies, ND: Nondemented human. (a) 30 minute film exposure, (b) 5 minute film exposure, (c) 30 second film exposure.
Figures 1a-c. Clarified homogenates containing 50μg total protein from 7 human subjects are analyzed for the presence of multimeric Aβ and APP. Immunoblotting with antibody 6E10 reveals Aβ monomers, dimers, trimers, tetramers, and APP (top band) in all Alzheimer's cases, as well as abundant higher molecular weight Aβ multimers in 2 AD cases. Synthetic Aβ40 confirms the identity of the lower molecular weight bands. AD: Alzheimer's disease, DLB: Dementia with Lewy Bodies, ND: Nondemented human. (a) 30 minute film exposure, (b) 5 minute film exposure, (c) 30 second film exposure.
Discussion
Despite the importance of Aβ aggregation in the pathogenesis of Alzheimer's disease1,4,5 , few studies have described or quantified the distribution of diverse Aβ multimers in human cortical tissue samples2. Commonly used immunohistochemical techniques do not allow for the discrimination of distinct multimeric Aβ species in fixed cortical tissue. In unfixed cortical tissue homogenates, Aβ multimers can be separated and biochemically assessed using gel electrophoresis and antibody-based detection methods. However, the Aβ epitopes that are targeted may be hidden within aggregated and post-translationally modified peptide structures, preventing the detection and accurate quantification of aggregated Aβ. Using heat-induced antigen-epitope retrieval combined with SDS-PAGE and immunoblotting with an antibody to the N-terminal region of Aβ6,7, we are able to separate and detect naturally occurring Aβ multimers isolated from human brains. Distinct Aβ multimer populations in clarified tissue homogenates can then be quantified by densitometry. Additionally, the combination of gel- or membrane-extraction with Aβ immunoblotting will allow for further structural characterization of naturally occurring, post-translationally modified Aβ multimers from human tissue. It will be important to determine whether Aβ multimers in human brain are SDS-resistant, or if they are SDS-sensitive and therefore broken down into smaller aggregates by SDS denaturation under defined conditions. The characterization of diverse forms of aggregated Aβ in the human brain will facilitate the search for therapies and biomarkers for Alzheimer's disease.
Acknowledgments
Many thanks to Elaine Pranski and Carolyn Suwyn for excellent technical assistance and to Harry LeVine III, M. Paul Murphy, and Marla Gearing for important conversations. Funding was provided by RR-00165, PO1AG026423, P50AG025688, AG030539, the Woodruff Foundation and the Emory University Research Committee.
References
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell. Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Walker LC, LeVine H. The cerebral proteopathies. Neurobiol. Aging. 2000;21:559–561. doi: 10.1016/s0197-4580(00)00160-3. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Aging, amyloid, and Alzheimer's disease: a perspective in honor. of Carl Cotman. Neurochem. Res. 2003;28:1705–1713. doi: 10.1023/a:1026065122854. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Selkoe DJ. Abeta oligomers - a decade of discovery. J. Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- Swerdlow PS, Finley D, Varshavsky A. Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal. Biochem. 1986;156:147–153. doi: 10.1016/0003-2697(86)90166-1. [DOI] [PubMed] [Google Scholar]
- Ida N, Hartmann T, Pantel J, Schröder J, Zerfass R, Förstl H, Sandbrink R, Masters CL, Beyreuther K. Analysis of hetergeneous A4 peptides in human cerebrospinal fuid and blood by a newly developed sensitive Western blot assay. J. Biol. Chem. 1996;271:22908–22914. doi: 10.1074/jbc.271.37.22908. [DOI] [PubMed] [Google Scholar]


