Abstract
Background
Hepatitis C virus (HCV)–infected women—in particular, those coinfected with human immunodeficiency virus type 1 (HIV-1)—can transmit infection to their children and sex partners.
Methods
The present study was conducted to analyze the presence of HCV RNA in cervicovaginal lavage (CVL) fluid from 71 women (58 HCV/HIV-1–coinfected women and 13 HCV-infected, HIV-1–uninfected women) enrolled in the Women’s Interagency HIV Study.
Results
HCV RNA was detected (by a commercial polymerase chain reaction assay) in CVL fluid from 18 (29%) of the HIV-1–infected women and from none of the HIV-1–uninfected women (P < .05). Multivariate analysis revealed that risk factors for the presence of HCV RNA in CVL fluid were HCV viremia (odds ratio [OR], 16.81; P = .02) and HIV-1 RNA in CVL fluid (OR, 19.87; P = .02). This observation suggests local interactions between HIV-1 and HCV in the genital tract compartment. There was no correlation between HCV RNA in CVL fluid and CD4, CD8, or CD3 cell counts, HIV-1 RNA viremia, the number of leukocytes in CVL fluid, or HIV-1 therapy. Furthermore, in 3 of 5 analyzed patients who had a detectable CVL HCV RNA load, we found viral variants differing in the 5′ untranslated region that were present neither in plasma nor in peripheral-blood mononuclear cells.
Conclusions
Our observations point to the importance of the genital tract compartment, in which local HCV replication could be facilitated by local HIV-1 replication.
Hepatitis C virus (HCV) infection is common among HIV-1–infected patients, with 50%–90% being coinfected [1]. Thus, HCV coinfection has emerged as a major public health problem that contributes to significant morbidity and mortality in HIV-1–infected patients. HIV-1 coinfection accelerates the development of severe liver disease attributable to HCV [1, 2], whereas HCV coinfection has been reported to accelerate the progression of HIV-1 disease [3, 4]. Since the introduction of effective measures to screen blood and blood products for HCV, injection drug use has become the predominant mode of HCV acquisition. However, exposure via injection drugs cannot account for up to 20% of new infections [5, 6]. Several reports have suggested that HCV may be transmitted through sexual intercourse [5, 7, 8], during childbirth [9, 10], and even during casual contacts between household members [11]. The probability of mother-to-child and female-to-male transmission appears to increase in the presence of coinfection with HIV-1 [10, 12–14]. Similar findings have been reported for HIV-1–positive men who engage in high-risk sexual behaviors (such as unprotected sex) with other men [15]. However, there are conflicting results with regard to female-to-male HCV transmission [16, 17]. The mechanism of increased HCV replication in HIV-1–infected patients has been attributed to immunosuppression, as evidenced by high viral loads in patients receiving immunosuppressive drugs after transplantation [18]. However, there is also evidence suggesting that HCV replication may be directly enhanced by the presence of HIV-1 [19].
Surprisingly, despite mounting evidence for the existence of female-to-male and mother-to-child transmission of HCV, very little is known about vaginal and cervical shedding of HCV in HIV-1–positive and HIV-1–negative women. Very few studies have assessed HCV RNA in vaginal secretions. Furthermore, HCV load and quasispecies distribution in the genital tract compartment has not been analyzed previously [20–22]. This could be explained by the difficulties of obtaining cervicovaginal specimens, the methods for which have only recently been standardized in HIV-1 settings [23]. In this article, we present our findings on HCV detection, viral load, and quasispecies composition in the female genital tract using cervicovaginal lavage (CVL) fluid from HCV/HIV-1–coinfected women.
PATIENTS, MATERIALS, AND METHODS
This is a cross-sectional study nested within the Women’s Interagency HIV Study (WIHS), a prospective, multicenter effort established in August 1993 to conduct comprehensive investigations of the impact of HIV-1 infection on women. A detailed description of the WIHS cohort is available elsewhere [24]. Briefly, participants are seen every 6 months and undergo a comprehensive interview, physical and gynecological examinations, and extensive laboratory evaluations. Informed consent was obtained from all study participants or their parents or guardians, and the human experimentation guidelines of the US Department of Health and Human Services and those of the authors’ institutions were followed in the conduct of clinical research. Blood and CVL specimens were processed and stored according to a standardized WIHS protocol [24]. CVL cellular fractions were analyzed according to the study protocol and involve microscopic evaluation, round-cell staining, and measurement of the levels of hemoglobin/erythrocyte- and leukocyte-associated esterases by a commercial semiquantitative assay (Bayer Corporation). Serum, plasma, peripheral-blood mononuclear cells (PBMCs), and genital specimens were stored for each patient at each visit at a central repository monitored by BBI Biotech Research Laboratories in a state-of-the-art biological-specimen storage facility. The present study included 58 of the 113 HCV/HIV-1–coinfected women and 13 of the 23 HCV-infected, HIV-1–uninfected women enrolled at the Los Angeles WIHS site (table 1). Of these 71 women, 9 (6 of the HCV/HIV-1–coinfected women and 3 of the HCV-infected, HIV-1–uninfected women) were randomly selected for intense molecular evaluations.
Table 1.
Demographic and clinical characteristics of the hepatitis C virus (HCV)/HIV-1–coinfected women with and without HCV shedding.
| HCV in CVL fluid |
||||
|---|---|---|---|---|
| Characteristic, parameter | All | Positive | Negative | P |
| Age group | .33 | |||
| <35 years | 20 (34.5) | 5 (27.8) | 15 (37.5) | |
| ≥35 years | 38 (65.5) | 13 (72.2) | 25 (62.5) | |
| Ethnicity | .30 | |||
| White | 15 (25.9) | 2 (11.1) | 13 (32.5) | |
| Black | 23 (39.7) | 9 (50.0) | 14 (35.0) | |
| Hispanic | 18 (31.0) | 6 (33.3) | 12 (30.0) | |
| Other | 2 (3.4) | 1 (5.6) | 1 (2.5) | |
| Employed | .15 | |||
| Yes | 10 (17.2) | 1 (5.6) | 9 (22.5) | |
| No | 48 (82.8) | 17 (94.4) | 31 (77.5) | |
| No. of lifetime sex partners | .62 | |||
| 0–4 | 18 (31.0) | 6 (33.3) | 12 (30.0) | |
| 5–10 | 15 (25.9) | 3 (16.7) | 12 (30.0) | |
| 11–100 | 17 (29.3) | 5 (27.8) | 12 (30.0) | |
| >100 | 8 (13.8) | 4 (22.2) | 4 (10.0) | |
| Injection drug use | .16 | |||
| Ever | 46 (79.3) | 12 (66.7) | 34 (85.0) | |
| Never | 12 (20.7) | 6 (33.3) | 6 (15.0) | |
| Recent HAART (within previous 6 months) | .12 | |||
| Yes | 41 (70.7) | 10 (55.6) | 31 (77.5) | |
| No | 17 (29.3) | 8 (44.4) | 9 (22.5) | |
| Current CD4 cell count | .17 | |||
| Median (no. of women), cells/μL | 394 (56) | 299 (17) | 431 (39) | |
| ≤200 cells/μL | 11 (19.0) | 6 (35.3) | 5 (12.8) | |
| 201–350 cells/μL | 13 (22.4) | 5 (29.4) | 8 (20.5) | |
| 351–500 cells/μL | 14 (24.1) | 2 (11.8) | 12 (30.8) | |
| >500 cells/μL | 18 (31.0) | 4 (23.5) | 14 (35.9) | |
| Not available | 2 (3.4) | … | … | |
| Current CD8 cell count | .56 | |||
| Median (no. of women), cells/μL | 874 (56) | 790 (17) | 914 (39) | |
| ≤800 cells/μL | 25 (43.1) | 9 (52.9) | 16 (41.0) | |
| >800 cells/μL | 31 (53.4) | 8 (47.1) | 23 (59.0) | |
| Not available | 2 (3.4) | … | … | |
| Current plasma HCV RNA load | .04 | |||
| Median (no. of women), IU/mL | 941,510 (58) | 1,511,468 (18) | 387,772 (40) | |
| Undetectabl | 19 (32.8) | 2 (11.1) | 17 (42.5) | |
| ≤1,000,000 IU/mL | 12 (20.7) | 6 (33.3) | 6 (15.0) | |
| >1,000,000 IU/mL | 27 (46.6) | 10 (55.6) | 17 (42.5) | |
| Current plasma HIV-1 RNA load | .57 | |||
| Median (no. of women), copies/mL | 802 (56) | 6000 (17) | 609 (39) | |
| ≤1000 copies/mL | 30 (51.7) | 8 (47.1) | 22 (56.4) | |
| >1000 copies/mL | 26 (44.8) | 9 (52.9) | 17 (43.6) | |
| Not available | 2 (3.4) | … | … | |
NOTE. Data are no. (%) of women, unless otherwise noted. CVL, cervicovaginal lavage; HAART, highly active antiretroviral therapy.
Immunophenotyping
Three-color flow cytometry and real-time testing were performed at local sites that are certified through the Division of AIDS Quality Assurance Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Consensus protocols were used [25].
HIV-1 RNA load measurements
HIV-1 RNA loads were determined in plasma and CVL fluid by NucliSens assay (lower limit of detection, 80 copies/mL; bioMérieux) with 1 mL of CVL fluid. For HIV-1 RNA quantitation, we chose the nucleic acid sequence–based amplification (NASBA) method because of its superior performance with a wide array of clinical specimens and an extraction method that is relatively insensitive to the inhibitory substances present in vaginal and other specimen types [26]. According to the WIHS protocol, CVL specimens are processed within 6 h and are subsequently stored at −80 °C. We have performed an extensive comparison between 2 methods of detecting HIV-1 RNA in vaginal specimens collected from women enrolled in WIHS and have shown that HIV-1 RNA was readily detectable in CVL fluid from ~24% of infected women by polymerase chain reaction (PCR) and NASBA [27].
HCV RNA load measurements
Plasma specimens were tested by a commercial HCV RNA assay (lower limit of detection, 600 IU/mL; Roche Diagnostic Systems) with 100 μL of plasma. To detect and quantify HCV RNA in genital secretions, 1.0 mL of CVL fluid was ultracentrifuged at 24,000 g for 1 h; subsequently, 100 μL of the concentrated CVL pellet was used for HCV RNA extraction and detection as reported elsewhere for HIV-1 [28, 29]. We calculated HCV genome equivalents on the basis of the data published by Pawlotsky, Pawlotsky et al., and Saldanha et al. [30–32]. The sensitivity of the assay was estimated to be ~60 IU/ml for CVL testing. For the subset of 9 randomly selected women, we used an in-house qualitative HCV PCR assay that has been described elsewhere [33].
Strand-specific HCV RNA assay
The description of our strand-specific HCV RNA assay has been published elsewhere [34]. When tested on synthetic template, the assay was capable of detecting ~1 × 102−1 × 103 genomic equivalent molecules of the correct strand while nonspecifically detecting >1 × 108 genomic equivalent molecules of the incorrect strand.
Single-strand conformation polymorphism (SSCP) assay
The SSCP assay was performed as has been described elsewhere in detail [33]. Amplified sequences were compared by the SSCP assay and sequencing. The major advantages of the SSCP assay are its relative simplicity and the fact that it is less prone to sporadic artifactual polymorphism than is the cloning of PCR products and subsequent analysis of individual clones. Furthermore, this technique is able to discriminate between sequences differing by a single nucleotide substitution and to detect a minor variant admixture representing ≥3% of the whole population [35]. Because nested and seminested reverse transcriptase (RT) PCR protocols are prone to amplification artifacts, the number of cycles during the second-round amplification was kept to a minimum, to ensure the presence of a single, sharp band on the agarose gel [36].
For the subset of 9 randomly selected women, amplified sequences were compared by direct sequencing and the SSCP assay [35]. The analysis was conducted on the 5′ untranslated region. When deemed necessary, composition of the quasispecies was further analyzed by the cloning of PCR products and subsequent sequencing of individual clones. The presence of HCV RNA negative strand was determined by rTth-based strand-specific RT-PCR.
Data analysis
We assessed differences in HCV RNA detectability between the HCV/HIV-1–coinfected women and the HCV-infected, HIV-1–uninfected women by Fisher’s exact test. Further analysis was conducted among the 58 HCV/HIV-1–coinfected women. The relationships between HIV-1 RNA load in CVL fluid and plasma and between HCV RNA load in CVL fluid and plasma were analyzed by Pearson’s correlation coefficient, plotted on a log10 scale. We used logistic regression to analyze the association between detectability of HCV RNA in CVL fluid and potential predicting factors. Initially, unadjusted logistic regression analyses were performed with plasma HCV RNA load, plasma HIV-1 RNA load, CVL HIV-1 RNA load, CD4 cell count, CD8 cell count, CD3 cell count, CVL total cell count, CVL peroxidase-positive cell count, CVL color, CVL blood Chemstrip (Bayer Corporation) level, and CVL white blood cell Chemstrip level in separate models. Plasma HCV RNA load (undetectable vs. detectable or per log10 increase), plasma HIV-1 RNA load (undetectable vs. detectable; ≤1000 vs. >1000 copies/mL; or per log10 increase), CVL HIV-1 RNA load (undetectable vs. detectable; ≤150 vs. >150 copies/mL; or per log10 increase), CD4 cell count (≤350 vs. >350 cells/μL or per 100-cell/μL decrease), and CD8 cell count (≤800 vs. >800 cells/μL or per 100-cell/μL decrease) were analyzed by Pearson’s correlation coefficient in both categorical and continuous scales, as indicated above. The relationship between detectability of HCV RNA in CVL fluid and factors that were significant (P < .05) or even marginally significant (P < .1) in the unadjusted analysis were further evaluated by multivariate logistic regression analyses that adjusted for plasma HCV RNA load, CVL HIV-1 RNA load, plasma HIV-1 RNA load, and CD4 cell count. Two-sided hypotheses were tested. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated in both unadjusted and adjusted logistic regression analysis.
RESULTS
Demographic and clinical information for the 58 HCV/HIV-1–coinfected women who were and were not found to be shedding HCV in the genital tract can be seen in table 1. The majority of the women were >35 years old, were black or Hispanic, and were unemployed. More than 40% reported having >10 lifetime sex partners, and 79% reported a history of injection drug use. Seventy percent had received highly active antiretroviral therapy within the previous 6 months, 52% had HIV-1 RNA loads <1000 copies/mL, and 80% had CD4 cell counts >200 cells/μL. HCV RNA was detected in plasma from 67% (39/58) of the HCV/HIV-1–coinfected women, compared with only 46% (6/13) of the HCV-infected, HIV-1–uninfected women, and they also had higher plasma HCV RNA loads (median, 1.5 × 106 copies/mL [range 1.5 × 104 to >4.0 × 106 copies/mL] vs. 0.3 × 106 copies/mL [range, undetectable to 1.3 × 106 copies/mL]) (data not shown).
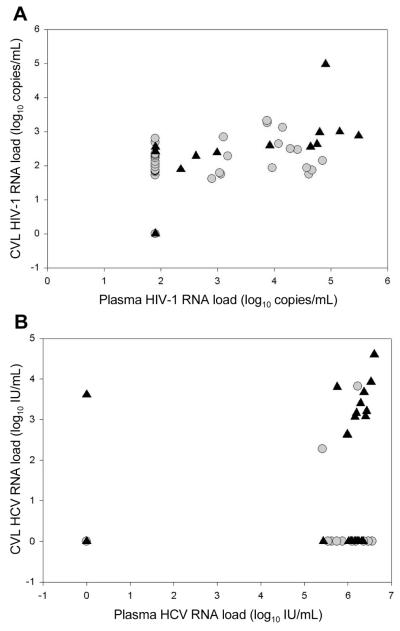
HCV RNA was detected in CVL fluid from 31% (18/58) of the HCV/HIV-1–coinfected women, although the loads were relatively low (median, 1.5 × 103 copies/mL; range, undetectable to 0.4 × 104 copies/mL); 16 of the 58 women had loads <0.8 × 103 copies/mL. None of the HCV-infected, HIV-1–uninfected women (P = .03) had detectable HCV RNA in CVL fluid when tested by a commercial HCV RNA assay (Roche Diagnostic Systems), although 2 had detectable HCV RNA by our in-house qualitative HCV PCR assay. In general, the coinfected women with detectable HCV RNA in CVL fluid were similar to those with undetectable HCV RNA in CVL fluid, except that they had higher plasma HCV RNA loads (P < .04) (table 1). The relationships between plasma and CVL HIV-1 and HCV RNA loads can be seen in figure 1A and 1B. For the relationship between log10 HIV-1 RNA load in plasma and CVL fluid, Pearson’s R = 0.50 (P = .0003); for the relationship between log10 HCV RNA load in plasma and CVL fluid, Pearson’s R = 0.33 (P = .0111).
Figure 1.
Relationships between HIV-1 and hepatitis C virus (HCV) RNA loads in plasma and detectability of HCV RNA (A) and HIV-1 RNA (B) loads in cervicovaginal lavage (CVL) fluid. In the figure, circles indicate undetectable CVL HCV (A) or HIV-1 (B) loads, and triangles indicate detectable CVL HCV (A) or HIV-1 (B) loads. For the relationship between HIV-1 RNA load in CVL fluid and plasma, Pearson’s R = 0.50 (P = .0003); for the relationship between HCV RNA load in CVL fluid and plasma, Pearson’s R = 0.33 (P = .0111).
Univariate logistic regression analysis showed possible associations between the presence of HCV RNA in CVL fluid and CD4 cell count, the presence of HCV RNA in plasma, the presence of HIV-1 RNA in CVL fluid, and blood contamination (data not shown). However, in multivariate analysis that included plasma HCV RNA load, plasma HIV-1 RNA load, CVL HIV-1 RNA fluid, and CD4 cell count, the only statistically significant predictors of HCV RNA in CVL fluid were the presence of HCV RNA in plasma (OR, 16.81 [95% CI, 1.53–185.31]; P = .02) and the presence of HIV-1 RNA in CVL fluid (OR, 19.87 [95% CI, 1.70–231.65]; P = .02) (table 2). This relationship was present only when we censored CVL HIV-1 RNA loads <150 copies/mL. We chose this censoring level because of the limited ability of the NucliSens assay to quantify HIV-1 RNA <150 copies/mL and the ability of this assay to detect viremia at this level at least 95% of the time [37, 38].
Table 2.
Determinants of hepatitis C virus (HCV) viremia in cervicovaginal lavage (CVL) fluid from the HCV/HIV-1–coinfected women.
| Women with detectable HCV RNA in CVL fluid, proportion (%) |
Unadjusted |
Adjusteda |
|||
|---|---|---|---|---|---|
| Category, parameter | OR (95% CI) | P | OR (95% CI) | P | |
| Plasma HCV RNA load | |||||
| Undetectable | 2/19 (11) | 1.00 | 1.00 | ||
| Detectable | 16/39 (41) | 5.91 (1.20–29.23) | .03 | 16.81 (1.53–185.31) | .02 |
| Per log10 increase | … | 1.36 (1.04–1.78) | .02 | … | … |
| Plasma HIV-1 RNA load | |||||
| Undetectable | 4/23 (17) | 1.00 | 1.00 | ||
| Detectable | 13/33 (39) | 3.09 (0.86–11.16) | .09 | 5.10 (0.75–34.55) | .10 |
| ≤1000 copies/mL | 8/30 (27) | 1.00 | 1.00 | ||
| >1000 copies/mL | 9/26 (35) | 1.46 (0.46–4.57) | .52 | 0.76 (0.10–5.78) | .79 |
| Per log10 increase | … | 1.62 (0.98–2.69) | .06 | … | … |
| CVL HIV-1 RNA load | |||||
| Undetectable | 2/16 (13) | 1.00 | 1.00 | ||
| Detectable | 12/34 (35) | 3.82 (0.74–19.68) | .11 | 3.80 (0.35–41.33) | .27 |
| ≤150 copies/mL | 2/22 (9) | 1.00 | 1.00 | ||
| >150 copies/mL | 12/28 (43) | 7.50 (1.46–38.47) | .02 | 19.87 (1.70–231.65) | .02 |
| Per log10 increase | … | 2.47 (0.93–6.57) | .07 | … | … |
| CD4 cell count | |||||
| >350 cells/μL | 6/32 (19) | 1.00 | 1.00 | ||
| ≤350 cells/μL | 11/24 (46) | 3.67 (1.11–12.14) | .03 | 4.11 (0.68–24.62) | .12 |
| Per 100-cell/μL decrease | … | 1.34 (1.0–1.80) | .048 | … | … |
NOTE. CI, confidence interval; OR, odds ratio.
Adjusted for the other variables in the multivariate logistic regression model, including plasma HCV RNA load, plasma HIV-1 RNA load, CVL HIV-1 RNA load, and CD4 cell count. Women with missing values were excluded.
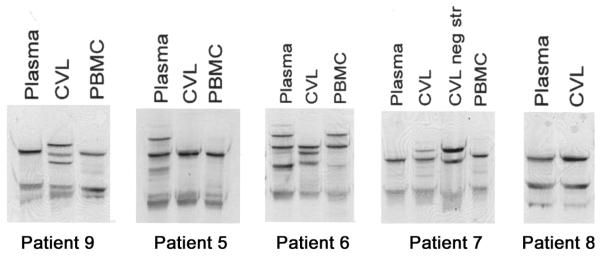
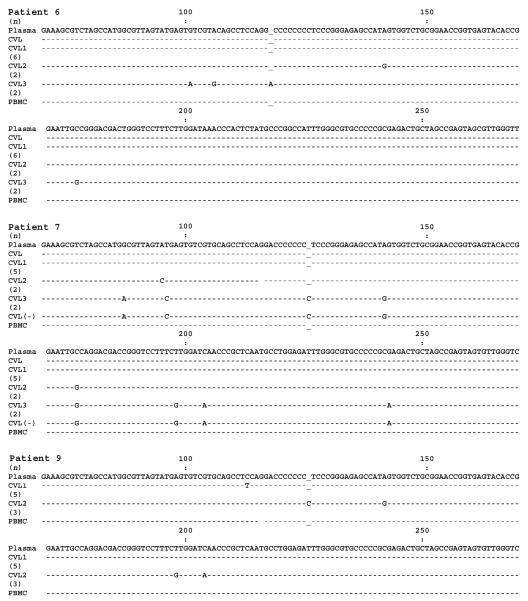
To assess for compartmentalization of HCV and genetic diversity between blood and genital HCV, a detailed analysis of plasma- and CVL-derived HCV RNA was conducted in a subset of 9 randomly selected women (6 HCV/HIV-1–coinfected women and 3 HCV-infected, HIV-1–uninfected women) (figure 2). HCV RNA was detected by a highly sensitive RT-PCR method in both plasma and CVL fluid from 5 of these women (3 of the HCV/HIV-1–coinfected women and 2 of the HCV-infected, HIV-1–uninfected women). However, HCV RNA negative strand, which is a viral replicative intermediate, was repeatedly detected in CVL fluid from only 1 of the HCV/HIV-1–coinfected women (patient 7). For 4 of the 5 women in whom HCV RNA was detected in CVL fluid (patients 5–7 and 9), the SSCP band patterns for plasma- and CVL-derived viral sequences were different, suggesting the presence of differing quasispecies variants in both of these compartments (figure 2). For all 4 patients, the PBMC-derived band pattern closely matched the plasma-derived band pattern. For patient 9, the CVL-derived band pattern was different from the plasma-derived band pattern, whereas, for patients 6 and 7, there were additional bands that were not present in the plasma-derived band pattern. For these 3 patients, viral sequences were further analyzed by direct sequencing and sequencing of cloned PCR products. CVL fluid from patient 9 was found to harbor 2 viral sequences differing from those of the plasma strain by 1 and 4 nt, respectively (figure 3). For patient 6, the dominant CVL-derived variant was identical to the plasma-derived sequence; however, cloning revealed the presence of 2 additional sequences that differed from the plasma-derived sequence (figure 3). Similarly, for patient 7, the dominant sequence variants in CVL fluid and plasma were identical, whereas 2 minor CVL-derived sequences were different; however, one of the latter was identical to the dominant negative-strand sequence in CVL fluid (figure 3). Thus, for patients 6, 7, and 9, CVL-derived HCV contained unique sequences that were present neither in plasma- nor in PBMC-derived HCV.
Figure 2.
Single-strand conformation polymorphism analysis of plasma-, cervicovaginal lavage (CVL)–, and peripheral-blood mononuclear cell (PBMC)– derived 5′ untranslated region hepatitis C virus (HCV) sequences. CVL neg str, HCV RNA negative strand in CVL fluid.
Figure 3.
Comparison of 5′ untranslated region hepatitis C virus (HCV) sequences amplified from plasma, cervicovaginal lavage (CVL) fluid, and peripheral-blood mononuclear cells (PBMCs) in patients 6, 7, and 9. In the figure, a minus sign indicates HCV RNA negative strand; “CVL” indicates the dominant variant; “CVL1,” “CVL2,” and “CVL3” indicate minor variants; and the nos. in parentheses indicate the no. of clones representing a given sequence.
DISCUSSION
To our knowledge, our study is the first to demonstrate compartmentalization of HCV in the genital tracts of HCV/HIV-1–coinfected women and possible local replication in a large proportion of HCV/HIV-1–coinfected women. We found unique but related viral variants in the blood and genital tract compartments, suggesting that HCV may replicate locally in cells shed from the genital tract. These findings have important implications for both sexual and perinatal transmission of HCV. Increased mother-to-infant and sexual HCV transmission in HCV/HIV-1–coinfected women makes it especially urgent to study and understand the dynamics of HCV in this subset of patients [1, 12–14]. Our study also suggests that, among HIV-1–infected women who are HCV viremic, there is an association between shedding of both viruses and that local control of both viruses may be impaired in those found to be shedding. This may explain the increased rate of perinatal HCV transmission to HIV-1–infected newborns and the observation that sexual transmission may be increased in coinfected patients.
Findings from our study suggest that a local HCV genital tract reservoir may exist and that this may be the source of infection for those suspected to have been infected sexually, a possibility further supported by the analysis of HCV quasispecies isolated from plasma, PBMCs, and CVL fluid. Despite the mounting evidence supporting the sexual transmission of HCV, very few reports have addressed the issue of HCV shedding in the female genital tract. In an early study, Caldwell et al. [39] reported the occasional presence of HCV RNA in multiple body fluids (including vaginal fluid collected by use of swabs), whereas, in more recent reports, Manavi et al. [21, 22] found that HCV RNA was present in cervical swabs from 8 of 22 HCV-infected women. Similar results were reported by Belec el al. [20], who detected HCV in cervicovaginal secretions from 4 of 19 women. However, none of these studies included HIV-1–positive patients. In the present study, we found that none of the HCV-infected, HIV-1–uninfected women were positive for HCV RNA in CVL fluid when tested by a commercial quantitative assay. However, both HIV-1–uninfected women in an intensely studied group were positive when tested by our sensitive in-house qualitative HCV PCR assay. These results suggest that HCV may be present at very low levels in the genital tract of some women infected with HCV but not with HIV-1. However, significant levels (>600 IU/mL) of HCV RNA in genital secretions were present only in the genital tracts of the HCV/HIV-1–coinfected women in our study. These data contrast with those published recently by Belec et al. [20, 40], who reported that up to 27% of the cellular fractions of CVL specimens from HCV-infected, HIV-1–uninfected women were HCV RNA positive. This discrepancy is most likely due to the assays used, because 2 of 3 HIV-1–uninfected women had detectable HCV RNA by our in-house assay, compared with none by a commercial assay. This may also be related to a more concentrated CVL specimen used by Belec et al. and the relatively small number of HCV-infected and HIV-1–uninfected women studied by us. The HCV/HIV-1–coinfected women in our study had rates of HCV shedding similar to those noted in the study of HIV-1–negative women by Belec et al. [40]. Additionally, inhibitors of enzymatic amplification in serum, and especially those in mucosal sites such as vaginal and oral compartments, may cause false-negative results for direct detection of HCV RNA by PCR. However, our inclusion of the ultracentrifugation step during HCV RNA extraction from CVL specimens was likely to reduce the effect of soluble PCR inhibitors. In our own experience, the ultracentrifugation step is able to eliminate the inhibitory effect in ~50% of specimens that are initially negative for HIV-1 RNA due to inhibitors of the amplification reaction (data not shown). However, it is plausible that some of the women with undetectable HCV RNA in CVL fluid may shed intermittently, and multiple sampling may yield positive results, as seen in HIV-1–infected women [27]. Thus, our data may underestimate the overall prevalence of HCV cervicovaginal shedding.
HIV-1 may locally affect HCV shedding on a molecular level or by immunologic mechanisms such as cytokines and immune activation. The latter possibility is in agreement with recently published data from Manavi et al. [21, 22] that suggest that HCV RNA in the genital tract is associated with lymphocytes and that the virus could possibly replicate there. Although high levels of HCV RNA in CVL fluid from 2 women correlated with traces of blood, the more frequently observed low-level viremia in CVL fluid of ~1000 copies/mL appears to be unrelated to blood contamination, as was shown by our multivariate analysis. Furthermore, the detection of unique CVL-derived viral variants different from those detectable either in plasma or PBMCs suggests the presence of local extrahepatic replication. This observation is in agreement with the findings of Belec et al. in HIV-1–uninfected women [40]. Their data strongly suggest an association between HCV and cells in the genital tract compartment. At present, it is unknown which cells harbor or support HCV replication in the cervix or vagina. Data from Manavi et al. point toward cervicovaginal lymphocytes as the likely source [21, 22]. Macrophages have also been reported to harbor replicating HCV in HIV-1–coinfected patients [41] and seem to play a role in mother-to-infant and sexual HIV-1 transmission [42, 43].
The mechanism by which local HIV-1 facilitates the presence of HCV in the genital tract is unclear. One possibility is concomitant infection of the same cells by both viral pathogens with subsequent enhancement of HCV replication, possibly through transactivating properties of the HIV-1 Tat protein [44]. We have recently shown that the same cell could indeed be infected by both viral pathogens [45]. Another possibility is local immune dysfunction, which would allow both viruses to replicate locally.
In summary, we have found that HCV RNA can be detected in almost 30% of HCV/HIV-1–coinfected women and that viral diversity does exist between local HCV and plasma HCV extracted from HCV/HIV-1–coinfected women. Our findings may explain a comparatively higher rate of HCV vertical transmission by HIV-1–coinfected women reported in several studies. The relationship between HIV-1 and HCV shedding is intriguing and suggests a unique local interaction between these 2 viruses in the genital tract.
Acknowledgments
The data used in this manuscript were collected by the Women’s Interagency HIV Study Collaborative Study Group, which is comprised of the following consortia or centers: the New York City/Bronx Consortium (principal investigator, Kathryn Anastos); the Brooklyn, New York, Consortium (principal investigator, Howard Minkoff); the Washington, DC, Metropolitan Consortium (principal investigator, Mary Young); the Connie Wofsy Study Consortium of Northern California (principal investigators, Ruth Greenblatt and Herminia Palacio); the Los Angeles County/Southern California Consortium (principal investigator, Alexandra Levine); the Chicago Consortium (principal investigator, Mardge Cohen); and the Data Coordinating Center (principal investigators, Alvaro Muñoz and Stephen J. Gange).
Financial support: National Institutes of Health (grants DA13760, RO1 A13760-01, and R01AI52065-01); Polish State Committee for Scientific Research (grant 2P05A 006 28 to M.R.). The Women’s Interagency HIV Study is funded by the National Institute of Allergy and Infectious Diseases, with supplemental funding from the National Cancer Institute, the National Institute of Child Health and Human Development (NICHD), the National Institute on Drug Abuse, and the National Institute of Craniofacial and Dental Research (grants U01-AI-35004, U01-AI-31834, U01-AI-34994, U01-AI-34989, U01-HD-32632 [NICHD], U01-AI-34993, U01-AI-42590, M01-RR00079, and M01-RR00083).
Footnotes
Potential conflicts of interest: none reported.
Presented in part: 9th Conference on Retroviruses and Opportunistic Infections, Seattle, February 2002; 54th annual meeting of the American Association for the Study of Liver Diseases, Boston, October 2003.
References
- 1.Eyster ME, Diamondstone LS, Lien JM, Ehmann WC, Quan S, Goedert JJ. Natural history of hepatitis C virus infection in multitransfused hemophiliacs: effect of coinfection with human immunodeficiency virus. The Multicenter Hemophilia Cohort Study. J Acquir Immune Defic Syndr. 1993;6:602–10. [PubMed] [Google Scholar]
- 2.Garcia-Samaniego J, Soriano V, Castilla J, et al. Influence of hepatitis C virus genotypes and HIV infection on histological severity of chronic hepatitis C. The Hepatitis/HIV Spanish Study Group. Am J Gastroenterol. 1997;92:1130–4. [PubMed] [Google Scholar]
- 3.Greub G, Ledergerber B, Battegay M, et al. Clinical progression, survival, and immune recovery during antiretroviral therapy in patients with HIV-1 and hepatitis C virus coinfection: the Swiss HIV Cohort Study. Lancet. 2000;356:1800–5. doi: 10.1016/s0140-6736(00)03232-3. [DOI] [PubMed] [Google Scholar]
- 4.Piroth L, Duong M, Quantin C, et al. Does hepatitis C virus co-infection accelerate clinical and immunological evolution of HIV-infected patients? AIDS. 1998;12:381–8. doi: 10.1097/00002030-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Alter H. Discovery of non-A, non-B hepatitis and identification of its etiology. Am J Med. 1999;107(Suppl):S16–20. doi: 10.1016/s0002-9343(99)00375-7. [DOI] [PubMed] [Google Scholar]
- 6.Alter MJ, Kruszon-Moran D, Nainan OV, et al. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–62. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 7.Feldman JG, Minkoff H, Landesman S, Dehovitz J. Heterosexual transmission of hepatitis C, hepatitis B, and HIV-1 in a sample of inner city women. Sex Transm Dis. 2000;27:338–42. doi: 10.1097/00007435-200007000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Halfon P, Riflet H, Renou C, Quentin Y, Cacoub P. Molecular evidence of male-to-female sexual transmission of hepatitis C virus after vaginal and anal intercourse. J Clin Microbiol. 2001;39:1204–6. doi: 10.1128/JCM.39.3.1204-1206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibb DM, Goodall RL, Dunn DT, et al. Mother-to-child transmission of hepatitis C virus: evidence for preventable peripartum transmission. Lancet. 2000;356:904–7. doi: 10.1016/s0140-6736(00)02681-7. [DOI] [PubMed] [Google Scholar]
- 10.Thomas SL, Newell ML, Peckham CS, Ades AE, Hall AJ. A review of hepatitis C virus (HCV) vertical transmission: risks of transmission to infants born to mothers with and without HCV viraemia or human immunodeficiency virus infection. Int J Epidemiol. 1998;27:108–17. doi: 10.1093/ije/27.1.108. [DOI] [PubMed] [Google Scholar]
- 11.Ackerman Z, Ackerman E, Paltiel O. Intrafamilial transmission of hepatitis C virus: a systematic review. J Viral Hepat. 2000;7:93–103. doi: 10.1046/j.1365-2893.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- 12.Eyster ME, Alter HJ, Aledort LM, Quan S, Hatzakis A, Goedert JJ. Heterosexual co-transmission of hepatitis C virus (HCV) and human immunodeficiency virus (HIV) Ann Intern Med. 1991;115:764–8. doi: 10.7326/0003-4819-115-10-764. [DOI] [PubMed] [Google Scholar]
- 13.Giovannini M, Tagger A, Ribero ML, et al. Maternal-infant transmission of hepatitis C virus and HIV infections: a possible interaction. Lancet. 1990;335:1166. doi: 10.1016/0140-6736(90)91174-9. [DOI] [PubMed] [Google Scholar]
- 14.Hershow RC, Riester KA, Lew J, et al. Increased vertical transmission of human immunodeficiency virus from hepatitis C virus–coinfected mothers. Women and Infants Transmission Study. J Infect Dis. 1997;176(Suppl):414–20. doi: 10.1086/514058. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher S. Sexual transmission of hepatitis C and early intervention. J Assoc Nurses AIDS Care. 2003;14:S87–94. doi: 10.1177/1055329003255815. [DOI] [PubMed] [Google Scholar]
- 16.Hallam NF, Fletcher ML, Read SJ, Majid AM, Kurtz JB, Rizza CR. Low risk of sexual transmission of hepatitis C virus. J Med Virol. 1993;40:251–3. doi: 10.1002/jmv.1890400315. [DOI] [PubMed] [Google Scholar]
- 17.Wyld R, Robertson JR, Brettle RP, Mellor J, Prescott L, Simmonds P. Absence of hepatitis C virus transmission but frequent transmission of HIV-1 from sexual contact with doubly-infected individuals. J Infect. 1997;35:163–6. doi: 10.1016/s0163-4453(97)91677-7. [DOI] [PubMed] [Google Scholar]
- 18.Gretch DR, Bacchi CE, Corey L, et al. Persistent hepatitis C virus infection after liver transplantation: clinical and virological features. Hepatology. 1995;22:1–9. [PubMed] [Google Scholar]
- 19.Beld M, Penning M, Lukashov V, et al. Evidence that both HIV and HIV-induced immunodeficiency enhance HCV replication among HCV seroconverters. Virology. 1998;244:504–12. doi: 10.1006/viro.1998.9130. [DOI] [PubMed] [Google Scholar]
- 20.Belec L, Legoff J, Si-Mohamed A, et al. Mucosal humoral immune response to hepatitis C virus E1/E2 surface glycoproteins and HCV shedding in saliva and cervicovaginal fluids from chronically HCV-infected patients. J Hepatol. 2003;38:833–42. doi: 10.1016/s0168-8278(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 21.Manavi M, Baghestanian M, Watkins-Riedel T, et al. Detection of hepatitis C virus (HCV) RNA in normal cervical smears of HCV-seropositive patients. Clin Infect Dis. 2002;35:966–73. doi: 10.1086/342909. [DOI] [PubMed] [Google Scholar]
- 22.Manavi M, Watkins-Riedel T, Kucera E, Czerwenka K, Hofmann H. Evidence of hepatitis C virus in cervical smears. J Infect. 1999;38:60–1. doi: 10.1016/s0163-4453(99)90038-5. [DOI] [PubMed] [Google Scholar]
- 23.Kovacs A, Chan LS, Chen ZC, et al. HIV-1 RNA in plasma and genital tract secretions in women infected with HIV-1. J Acquir Immune Defic Syndr. 1999;22:124–31. doi: 10.1097/00126334-199910010-00003. [DOI] [PubMed] [Google Scholar]
- 24.Barkan SE, Melnick SL, Preston-Martin S, et al. The Women’s Interagency HIV Study. WIHS Collaborative Study Group. Epidemiology. 1998;9:117–25. [PubMed] [Google Scholar]
- 25.Landay A, Benning L, Bremer J, et al. Correlates of immune activation marker changes in human immunodeficiency virus (HIV)–seropositive and high-risk HIV-seronegative women who use illicit drugs. J Infect Dis. 2003;188:209–18. doi: 10.1086/376509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der NJ. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bremer J, Nowicki M, Beckner S, et al. Comparison of two amplification technologies for detection and quantitation of human immunodeficiency virus type 1 RNA in the female genital tract. Division of AIDS Treatment Research Initiative 009 Study Team. J Clin Microbiol. 2000;38:2665–9. doi: 10.1128/jcm.38.7.2665-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun R, Schilling W, Jayakar H, et al. Simultaneous extraction of hepatitis C virus (HCV), hepatitis B virus, and HIV-1 from plasma and detection of HCV RNA by a reverse transcriptase-polymerase chain reaction assay designed for screening pooled units of donated blood. Transfusion. 1999;39:1111–9. doi: 10.1046/j.1537-2995.1999.39101111.x. [DOI] [PubMed] [Google Scholar]
- 29.Witt DJ, Kemper M, Stead A, Ginocchio CC, Caliendo AM. Relationship of incremental specimen volumes and enhanced detection of human immunodeficiency virus type 1 RNA with nucleic acid amplification technology. J Clin Microbiol. 2000;38:85–9. doi: 10.1128/jcm.38.1.85-89.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawlotsky JM. Diagnostic tests for hepatitis C. J Hepatol. 1999;31(Suppl 1):S71–9. doi: 10.1016/s0168-8278(99)80378-x. [DOI] [PubMed] [Google Scholar]
- 31.Pawlotsky JM, Bouvier-Alias M, Hezode C, Darthuy F, Remire J, Dhumeaux D. Standardization of hepatitis C virus RNA quantification. Hepatology. 2000;32:654–9. doi: 10.1053/jhep.2000.16603. [DOI] [PubMed] [Google Scholar]
- 32.Saldanha J, Heath A, Lelie N, Pisani G, Nubling M, Yu M. Calibration of HCV working reagents for NAT assays against the HCV international standard. The Collaborative Study Group. Vox Sang. 2000;78:217–24. doi: 10.1159/000031184. [DOI] [PubMed] [Google Scholar]
- 33.Laskus T, Radkowski M, Wang LF, Nowicki M, Rakela J. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with endstage liver disease: confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J Virol. 2000;74:1014–7. doi: 10.1128/jvi.74.2.1014-1017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C and G viruses. J Virol. 1997;71:7804–6. doi: 10.1128/jvi.71.10.7804-7806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskus T, Wilkinson J, Gallegos-Orozco JF, et al. Analysis of hepatitis C virus quasispecies transmission and evolution in patients infected through blood transfusion. Gastroenterology. 2004;127:764–76. doi: 10.1053/j.gastro.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Wang LF, Radkowski M, Vargas H, Rakela J, Laskus T. Amplification and fusion of long fragments of hepatitis C virus genome. J Virol Methods. 1997;68:217–23. doi: 10.1016/s0166-0934(97)00132-8. [DOI] [PubMed] [Google Scholar]
- 37.bioMérieux Reporting Results for NucliSens®HIV-1QT [pamphlet] 2002.
- 38.Nowicki MJ, Benning L, Bremer JW, et al. Longitudinal variability of human immunodeficiency virus type 1 RNA viral load measurements by nucleic acid sequence-based amplification and NucliSens assays in a large multicenter study. J Clin Microbiol. 2001;39:3760–3. doi: 10.1128/JCM.39.10.3760-3763.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caldwell SH, Sue M, Bowden JH, et al. Hepatitis C virus in body fluids after liver transplantation. Liver Transpl Surg. 1996;2:124–9. doi: 10.1002/lt.500020207. [DOI] [PubMed] [Google Scholar]
- 40.Belec L, Legoff J, Si-Mohamed A, et al. Cell-associated, non-replicating strand+ hepatitis C virus-RNA shedding in cervicovaginal secretions from chronically HCV-infected women. J Clin Virol. 2003;27:247–51. doi: 10.1016/s1386-6532(02)00178-6. [DOI] [PubMed] [Google Scholar]
- 41.Laskus T, Radkowski M, Piasek A, et al. Hepatitis C virus in lymphoid cells of patients coinfected with human immunodeficiency virus type 1: evidence of active replication in monocytes/macrophages and lymphocytes. J Infect Dis. 2000;181:442–8. doi: 10.1086/315283. [DOI] [PubMed] [Google Scholar]
- 42.Ometto L, Zanotto C, Maccabruni A, et al. Viral phenotype and host-cell susceptibility to HIV-1 infection as risk factors for mother-to-child HIV-1 transmission. AIDS. 1995;9:427–34. [PubMed] [Google Scholar]
- 43.van’t Wout AB, Kootstra NA, Mulder-Kampinga GA, et al. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–7. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferbeyre G, Bourdeau V, Cedergren R. Does HIV tat protein also regulate genes of other viruses present in HIV infection? Trends Biochem Sci. 1997;22:115–6. doi: 10.1016/s0968-0004(97)01011-6. [DOI] [PubMed] [Google Scholar]
- 45.Laskus T, Radkowski M, Jablonska J, et al. Human immunodeficiency virus facilitates infection/replication of hepatitis C virus in native human macrophages. Blood. 2004;103:3854–9. doi: 10.1182/blood-2003-08-2923. [DOI] [PubMed] [Google Scholar]