Abstract
The oncoprotein MDM2 binds to the activation domain of the tumor suppressor p53 and inhibits its ability to stimulate transcription. This same region of p53 is able to bind several basal transcription factors that appear to be important for the transactivation function of p53. It has therefore been suggested that MDM2 acts to inhibit p53 by concealing its activation domain from the basal machinery. Here we present data suggesting that MDM2 possesses an additional inhibitory function. Our experiments reveal that in addition to a p53-binding domain, MDM2 also contains an inhibitory domain that can directly repress basal transcription in the absence of p53. By fusing portions of MDM2 to a heterologous DNA-binding domain to allow p53-independent promoter recruitment, we have localized this inhibitory domain to a region encompassing amino acids 50–222 of MDM2. Furthermore, the function of this inhibitory domain does not require the presence of either TFIIA or the TAFs. Of the remaining basal factors, both the small subunit of TFIIE and monomeric TBP are bound by the MDM2 inhibitory domain. It is possible that MDM2 inhibits the ability of the preinitiation complex to synthesize RNA through one of these interactions. Our results are consistent with a model in which MDM2 represses p53-dependent transcription by a dual mechanism: a masking of the activation domain of p53 through a protein–protein interaction that additionally serves to recruit MDM2 to the promoter where it directly interferes with the basal transcription machinery.
Keywords: MDM2, p53, in vitro, transcriptional inhibitor, basal transcription
One of the primary biological functions of p53 is to prevent the accumulation of genomic alterations after DNA damage. To achieve this goal, p53 inhibits the growth of injured cells by two mechanisms (Haffner and Oren 1995; Ko and Prives 1996). In response to damaged DNA, p53 is capable of mediating a G1 cell cycle arrest, presumably to allow the cell time to make necessary genomic repairs. p53 can also cause the elimination of damaged cells by initiating the process of programmed cell death. The ability of p53 to induce a G1 arrest is dependent on its ability to function as a site-specific transcriptional activator (Fields and Jang 1990; Raycroft et al. 1990; Farmer et al. 1992; Kern et al. 1992; Unger et al. 1992; Zambetti et al. 1992; Attardi et al. 1996) and to induce the expression of genes whose products are themselves directly involved in arresting cell growth and proliferation (for review, see Ko and Prives 1996). This cell cycle arrest is mediated by increasing expression of the p21 protein, a cyclin-dependent kinase inhibitor (el-Deiry et al. 1993; Harper et al. 1993; Xiong et al. 1993; Dulic et al. 1994; Brugarolas et al. 1995; Deng et al. 1995). In addition, the product of another p53-response gene, GADD45, may play some role in the p53-dependent G1 arrest (Smith et al. 1994). The role of the transactivating function of p53 in the induction of apoptosis is less clear. However, the p53-response gene bax has been suggested to be involved in this process (Miyashita and Reed 1995). Taken together, these findings demonstrate the importance of the transcriptional activation properties of p53 for its ability to suppress tumor development and growth.
Because of its critical role in the control of cell growth and proliferation, the expression of p53 itself must be tightly regulated. Numerous modulators of the function of p53 have been reported, including kinases (Hupp et al. 1992; Takenaka et al. 1995), components of the ubiquitin-degradative pathway (Schneffner et al. 1993; Chowdary et al. 1994), virally encoded proteins (Bargonetti et al. 1992; Schneffner et al. 1993; Wang et al. 1994), and transcriptional inhibitors (Momand et al. 1992; Oliner et al. 1992, 1993; Yew et al. 1994). One crucial cellular regulator of p53 is the oncoprotein MDM2. Its ability to regulate p53 activity was first suggested when the mdm2 gene was found to be amplified in human sarcomas (Oliner et al. 1992) and the encoded protein was found to interact directly with p53 (Momand et al. 1992; Oliner et al. 1992). In transfection experiments, MDM2 can repress both p53-dependent transactivation of a reporter gene (Momand et al. 1992) and p53-dependent suppression of transformed cell growth (Finlay 1993). Recent studies in transgenic mice have provided a striking demonstration of the critical role MDM2 plays in p53 regulation. In these experiments, mdm2 null mice were found to possess an embryonic lethal phenotype that was completely relieved when these mdm2−/− mice were produced in a p53 null background (de Oca Luna et al. 1995; Jones et al. 1995). These results suggest that the primary defect in mdm2−/− mice is an excess of p53 activity. For these reasons, an understanding of how p53 functions to regulate growth in vivo will require that we also understand how p53 is regulated by MDM2.
Although it is clear that MDM2 is an important modulator of p53 function, the mechanism by which MDM2 inhibits p53 is not well understood. The current model for the inhibitory effects of MDM2 on p53-dependent transcription suggests that MDM2 disrupts the ability of p53 to make important contacts with the general transcription machinery through direct binding and concealment of its activation domain (Oliner et al. 1993). In support of this hypothesis, a truncated MDM2 protein, lacking the first 49 amino acids, that is no longer capable of binding p53 is not capable of inhibiting p53-dependent activation in transfection experiments (Haines et al. 1994). Furthermore, several components of the transcription machinery have been found to bind to the activation domain of p53 in a region near the MDM2 binding site (Seto et al. 1992; Chen et al. 1993; Liu et al. 1993; Truant et al. 1993; Xiao et al. 1994; Lu and Levine 1995). Together, these results suggest that the critical interaction between MDM2 and the activation domain of p53 may disrupt important interactions between p53 and the basal machinery.
Although the masking theory of p53 inhibition by MDM2 is consistent with the mutant and in vitro binding data, this mechanism has not been formally demonstrated. In addition, other transcriptional repressors that bind to transactivators are known and have been suggested to inhibit transcription via other mechanisms. For instance, both the adenovirus E1B and the cellular retinoblastoma (Rb) protein appear to contain inhibitory domains that are targeted to the appropriate promoters by interacting with p53 and E2F, respectively (Yew et al. 1994; Weintraub et al. 1995). Once at the promoter, they may inhibit additional transactivators or may directly repress the activities of the basal transcription machinery. It is possible that MDM2 may also function in a similar manner and that the interaction with p53 serves to recruit an inhibitory domain to p53-responsive promoters. In this case, MDM2 would be expected to contain a separable repression domain capable of inhibiting transcription when brought to a promoter independently of p53. This model for MDM2 inhibition is not mutually exclusive with the current, concealment model for repression by MDM2. As will be discussed later, MDM2 may repress transcription through a dual mechanism, by directly blocking p53 activation and by inhibiting activation by other transactivators bound at p53-responsive promoters.
At the present time, the specific mechanisms by which transcriptional repressors function are not well understood. Previous studies of transcriptional regulation in mammalian systems have focused primarily on transcriptional activators, and relatively few studies have addressed the properties of repressors. By studying the mechanisms by which MDM2 is capable of inhibiting p53-dependent transcription, we hope not only to understand the regulation of an important player involved in controlling cell growth but also to gain a more general insight into the functioning of transcriptional repressors.
To address the question of whether MDM2 can inhibit transcription in a p53-independent manner, we fused portions of the MDM2 protein to a heterologous DNA-binding domain to allow promoter recruitment in the absence of p53 binding. The ability of these proteins to repress transcription in both a p53-dependent and -independent manner was then tested using an MDM2-responsive, in vitro transcription system. In this way, we were able to demonstrate that MDM2 contains an inhibitory domain capable of repressing both basal and activated transcription. Furthermore, using an in vitro transcription system composed of recombinant proteins and highly purified HeLa cell nuclear fractions, we were able to identify the basal factors potentially involved in MDM2-mediated inhibition. In vitro binding assays designed to test the ability of MDM2 to interact with a minimal set of basal factors revealed a direct interaction between MDM2 and two components of the basal transcription machinery. Taken together, our data suggest that MDM2 may inhibit transcription from p53-responsive promoters by two distinct mechanisms: concealment of the activation domain of p53 and direct inhibition of the basal transcription machinery.
Results
Development of an in vitro transcription system responsive to MDM2
To dissect the mechanism by which MDM2 inhibits p53-dependent transcription, we developed an MDM2-responsive, in vitro transcription system in which to study this process. MDM2 has not been demonstrated previously to function in in vitro transcription assays, possibly because of difficulties in obtaining purified, functional, recombinant MDM2. Human MDM2 is 491 amino acids in length and is poorly expressed and largely insoluble in both Sf9 cells infected with recombinant baculoviruses and Escherichia coli (C. Thut and R. Tjian, unpubl.). In an attempt to circumvent the expression and solubility problems, we used a truncated version of MDM2 consisting of amino acid residues 1–324 [MDM2(1–324)] that had been shown previously to bind p53 (Brown et al. 1993). MDM2(1–324) was expressed and purified from E. coli by extracting proteins from lysed cell pellets under denaturing conditions. After purification and renaturation, the proteins were tested for their ability to inhibit in vitro transcription in a manner that mimics the specificity of MDM2 in transfection studies. Because MDM2 binds to the activation domain of p53 and requires this binding for its inhibitory effects, we reasoned that a hybrid protein containing the p53 activation domain fused to the Gal4 DNA-binding domain (G4–p53) would be sufficient for our study of MDM2 repression in vitro. We performed in vitro transcription reactions using a fractionated HeLa cell transcription system (Dignam et al. 1983) supplemented with purified, recombinant human TFIIA (Ranish et al. 1992) in place of the crude TFIIA-containing fraction. As expected, a template containing five Gal4 DNA-binding sites directs efficient transcriptional activation (approximately eightfold) in the presence of the G4–p53 protein (Fig. 1A, lanes 2,3). Addition of MDM2(1–324) to reactions containing G4–p53 resulted in up to a sixfold decrease in the level of transcription (Fig. 1A, lanes 4,5). Thus, our purified, renatured MDM2 protein was capable of inhibiting transcription dependent on the p53 activation domain. Next, we tested the specificity of the purified MDM2 by assaying its repressive effects on both G4–VP16-mediated and basal transcription. Previous studies performed in yeast had shown that p53 transactivation was inhibited by MDM2 but VP16 transactivation was not (Oliner et al. 1993). Likewise, in our in vitro system, transcriptional stimulation by G4–VP16 (Fig. 1A, lanes 7,8) was not inhibited by the addition of MDM2(1–324) (Fig. 1A, lanes 9,10). In addition, MDM2(1–324) did not inhibit basal transcription (Fig. 1A, lanes 11–14). These results, in combination, suggest that our simplified in vitro transcription system recapitulates the effects of MDM2 described previously in vivo.
Figure 1.
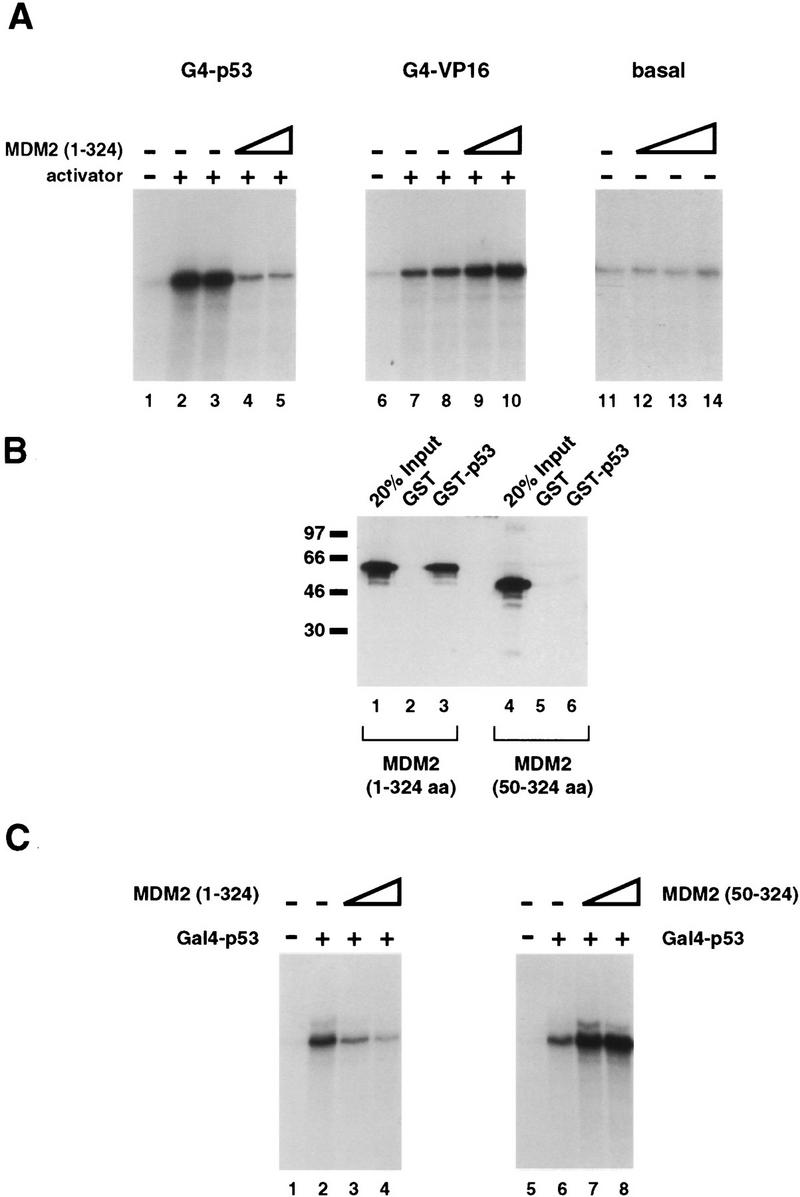
Development of an in vitro transcription system responsive to recombinant, human MDM2. (A) The first 324 amino acids of MDM2 specifically inhibit transcription dependent on the p53 activation domain in vitro. A protein containing the first 324 amino acids of human MDM2 was expressed and purified from bacteria and tested for its ability to inhibit transactivation by G4–p53, a protein consisting of the Gal4 DNA-binding domain (amino acids 1–94) fused to tandem copies of the p53 activation domain (amino acids 1–42). These proteins were assayed in an in vitro transcription system consisting of partially purified HeLa cell nuclear fractions and recombinant, purified hTFIIA using a template containing five Gal4-binding sites upstream the AdMLP. The promoter was fused to a G-less reporter, and the production of transcripts was measured by a G-less protocol. (Lane 1) The level of transcription in the absence of added activator; (lanes 2,3) reactions containing 30 ng of G4–p53. The reactions in lanes 4 and 5 contained both 30 ng of G4–p53 and 200 or 800 ng of purified, refolded MDM2(1–324), respectively. MDM2(1–324) was also tested for its ability to inhibit both basal transcription and transcription stimulated by the activation domain of VP16. (Lanes 7–10) 100 ng of a fusion protein consisting of the Gal4 DNA-binding domain (amino acids 1–147) and the activation domain of VP16 (amino acids 412–490). MDM2(1–324) protein was added to transcription reactions as follows, 200 ng (lanes 9,12), 400 ng (lane 13), or 800 ng (lanes 10,14). (B) The first 49 amino acids of MDM2 are required for an interaction with the activation domain of p53. GST (lanes 2,5) and a GST fusion protein containing amino acids 1–73 of p53 (lanes 3,6) were immobilized on glutathione resins and tested for their ability to retain soluble MDM2(1–324) (lanes 2,3) or an MDM2 protein lacking the first 49 amino acids [MDM2(50–324)] (lanes 5,6). After incubation with extracts containing the soluble MDM2 proteins, the resin and associated proteins were washed extensively, analyzed by SDS-PAGE, transferred to nitrocellulose, probed with monoclonal antibodies against MDM2 (Pharmagenics), and detected using an ECL protocol (Amersham). Lanes 1 and 4 represent 20% of the amount of MDM2 protein added to the corresponding binding reactions. (C) MDM2(50–324), which cannot interact with p53, is unable to inhibit G4–p53 dependent transcription in vitro. Using the transcription system described in A, MDM2(1–324) and MDM2(50–324) were tested for their ability to inhibit G4–p53 transactivation. Lanes 1 and 5 contain no added activator; lanes 2 and 6 contain 30 ng of G4–p53. In addition to 30 ng of G4–p53, 60 and 200 ng of MDM2(1–324) and 60 and 200 ng MDM2(50–324) were added to lanes 3 and 4 and lanes 7 and 8, respectively.
Next, we wanted to determine whether an interaction between MDM2 and the activation domain of p53 was required for repression in vitro. Prior studies indicate that the first 49 amino acids of MDM2 are required for both its ability to bind p53 and its ability to inhibit p53-dependent transcription (Haines et al. 1994). We expressed and purified a truncated MDM2 protein containing amino acid residues 50–324 [MDM2(50–324)] from E. coli, as described above, and tested its ability to function in our transcription system. As shown in Figure 1B, this protein was no longer capable of binding the activation domain of p53 as assessed by in vitro binding assays. More importantly, MDM2(50–324) was no longer able to inhibit transcription by G4–p53 (Fig. 1C, lanes 5–8) at concentrations equivalent to those at which MDM2(1–324) could efficiently repress G4–p53 transcription (Fig. 1C, lanes 1–4). Thus, the interaction between MDM2 and p53 is important for inhibition of transcription in vitro as well as in vivo.
MDM2 recruited to a promoter via a p53-independent mechanism does not require the p53 interaction domain of MDM2 to repress transcription
As mentioned previously, two models for MDM2’s mechanism of inhibition can be envisioned, both of which are consistent with the requirement for a direct, physical interaction between p53 and MDM2. First, MDM2 may conceal the activation domain of p53 through a direct interaction with this protein, thereby inhibiting basal factor contacts required for transcriptional stimulation. Second, MDM2 may contain an inhibitory domain capable of directly repressing the functions of one or more components of the basal machinery after being recruited to the promoter through an interaction with p53. If MDM2 represses transcription by the latter mechanism, it would be expected that MDM2 could also function as a repressor if targeted to a promoter via a p53-independent mechanism.
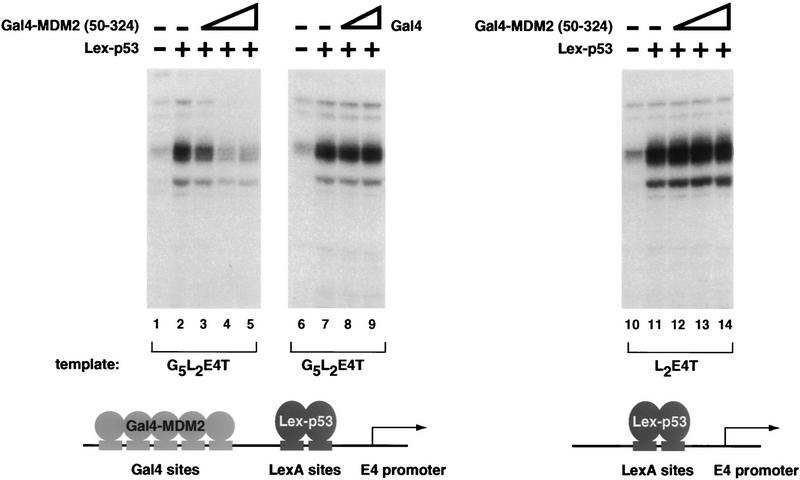
We wished to determine whether the MDM2/p53 interaction was required for MDM2 to inhibit p53-dependent transcription. To address this question, we asked whether MDM2(50–324) could inhibit the p53 activation domain when recruited to a promoter by fusion with a heterologous DNA-binding domain. A template containing five Gal4 sites upstream of two LexA sites was used to target both an MDM2/Gal4 fusion protein [G4–MDM2(50–324)] and Lex–p53 (the LexA DNA-binding domain fused to two copies of the activation domain of p53) to the E4 promoter. Lex–p53 is able to stimulate transcription from this template, as shown in Figure 2 (lanes 2,6). By adding increasing concentrations of G4–MDM2(50–324) to reactions containing Lex-p53, we were able to demonstrate that MDM2(50–324) was capable of inhibiting p53-dependent transcription once its ability to be recruited to the promoter was restored (Fig. 2, lanes 3–5). Addition of the Gal4 DNA-binding domain itself does not repress Lex–p53-dependent transcription (Fig. 2, lanes 8,9). Thus, it does not appear that simple occupancy of the Gal4 sites precludes binding of Lex–p53 and subsequent activation. Furthermore, this inhibitory effect is not attributable to a direct recruitment of G4–MDM2(50–324) by Lex–p53 because G4–MDM2(50–324) does not inhibit transcription from a template lacking Gal4-binding sites (Fig. 2, lanes 12–14). Taken together, these results demonstrate that recruitment of MDM2 to a template is sufficient to allow inhibition of p53-dependent activation.
Figure 2.
Restoring the ability of MDM2(50–324) to be targeted to a promoter enables it to inhibit p53 activation domain-dependent transactivation. To restore the ability of MDM2(50–324) to be recruited to the promoter, amino acids 50–324 of human MDM2 were fused to the Gal4 DNA-binding domain. The ability of G4–MDM2(50–324) to inhibit transcription from a template containing five Gal4 sites upstream of two LexA sites and the E4 promoter (G5L2E4T) was assayed by primer extension using the transcription system described in Fig. 1. p53 activation domain-dependent stimulation was acheived using a protein containing the LexA DNA-binding domain (amino acids 1–202) fused to two tandem copies of p53 amino acids 1–42 (Lex–p53). Lane 1 contains no added activator; lanes 2–5 have 30 ng of Lex–p53. The inhibitory properties of G4–MDM2(50–324) were tested by adding 20, 60, and 200 ng of this protein to the transcription reactions (lanes 3–5). The Gal4 DNA-binding domain alone was also tested for its ability to repress transcription from the G5L2E4T template. Lane 6 contains no added activator; lanes 7–9 contain 30 ng of Lex–p53. Lanes 8 and 9 also contain 20 and 60 ng of Gal4, respectively. The dependence of G4–MDM2(50–324) inhibition on the presence of the Gal4 sites was tested by performing the same series of reactions described for lanes 1–5 but using a template containing only two LexA sites upstream of the E4 promoter (L2E4T) (lanes 10–14).
MDM2 contains a domain that can repress basal transcription
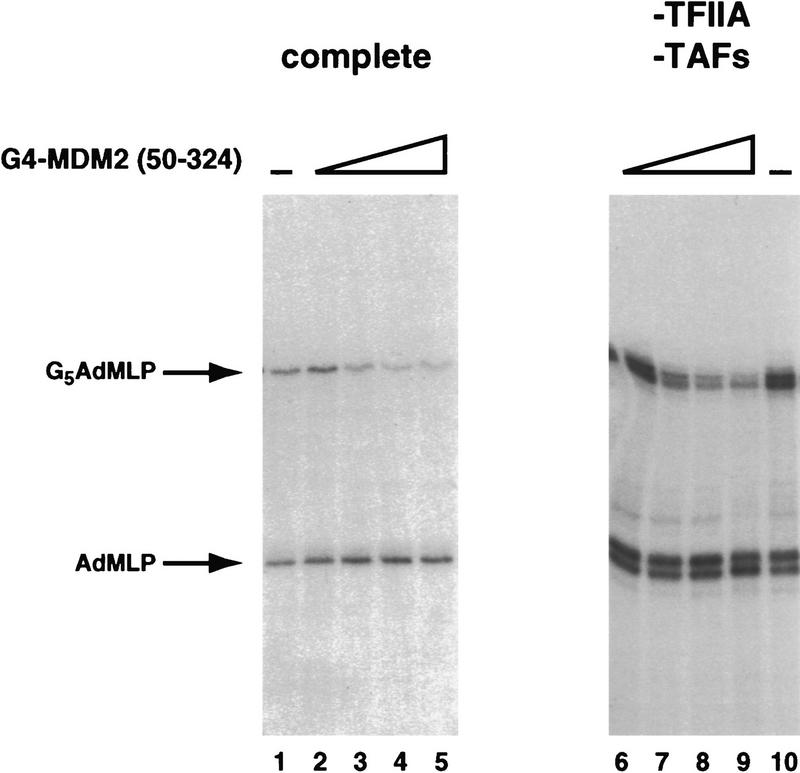
Because of the close proximity of G4–MDM2 and Lex–p53 when bound at the promoter, it was possible that G4–MDM2(50–324), although incapable of efficient in vitro binding, was still weakly interacting with the activation domain of p53 and thereby inhibiting its ability to activate transcription. For this reason, we were interested in determining whether MDM2 possessed a p53-independent inhibitory domain that could repress basal transcription directly. To this end, we tested the activity of G4–MDM2(50–324) in a fractionated HeLa cell transcription reaction. For these assays, two templates were added to each transcription reaction; one template contained five Gal4-binding sites upstream of the adenovirus major late promoter (AdMLP) to measure G4–MDM2 inhibition of transcription, and one contained only the AdMLP to serve as an internal negative control. Transcript production was assayed using a G-less protocol, and radioactive products arising from the two different templates were differentiated based on the size of their products. In this system, G4–MDM2(50–324) was capable of inhibiting basal transcription in a Gal4 site-dependent manner (Fig. 3, lanes 1–5; three- to fourfold inhibition). The G4 DNA-binding domain alone did not inhibit basal transcription in this system (data not shown). These results are consistent with the presence of an MDM2 domain capable of directly inhibiting one or more components of the basal machinery.
Figure 3.
MDM2(50–324) can inhibit basal transcription when brought to a promoter in the absence of p53, and this inhibition does not require TAFs or TFIIA. G4–MDM2(50–324) was assayed for its ability to inhibit basal transcription in a Gal4 site-dependent manner by a G-less protocol. Each reaction contains two templates that give rise to different length products: One template contains five Gal4 sites upstream of the AdMLP promoter and a G-less cassette, and one template lacks the Gal4 sites. The template giving rise to each of the two products is indicated on the left. (Lanes 1–5) The basal factors are supplied by partially purified HeLa cell nuclear fractions and recombinant hTFIIA as in Fig. 1; (lanes 6–10) purified, recombinant human TBP, TFIIB, TFIIF, TFIIE, and highly purified HeLa cell Pol II and TFIIH. Basal levels of transcription are shown in lanes 1 and 10. Lanes 2–5 and lanes 6–9 contain 7, 20, 60, and 200 ng of G4–MDM2(50–324).
TFIIA and TAFs are not required for MDM2 to inhibit basal transcription
Because MDM2 was able to repress basal transcription, we thought it likely that MDM2 was directly inhibiting the functions of one or more of the general transcription factors. To narrow the search for the basal factor component(s) inhibited by MDM2, we tested the ability of MDM2 to repress reconstituted basal transcription reactions lacking several of the general transcription factors. In the absence of an activator, it has been demonstrated that both TFIIA and the TAF [TATA-binding protein (TBP)-associated factor] components of the TFIID complex are dispensable for accurately initiated transcription from promoters containing TATA boxes. We therefore tested the ability of G4–MDM2(50–324) to inhibit transcription in a reconstituted, in vitro reaction containing recombinant TBP, TFIIB, TFIIF, and TFIIE and highly purified polymerase II (Pol II) and TFIIH. This complement of general transcription factors was capable of supporting transcription in the dual template system described previously (Fig. 3, lane 10). The purified system appears to allow transcription from an additional start site from each template, as evidenced by the appearance of a closely spaced doublet of bands. This phenomenon is likely to be the result of substituting TBP for TFIID (C. Thut and R. Tjian, unpubl.). The presence of this additional start site, however, did not affect the interpretation of the results. In this system, G4–MDM2(50–324) retained its ability to repress basal transcription (Fig. 3, lanes 6–9; upper transcript, three- to fourfold inhibition). Furthermore, this repression was dependent on the presence of Gal4 sites in the promoter (Fig. 3, lanes 6–9, lower transcript). Again, the Gal4 DNA-binding domain alone did not repress transcription (data not shown). The ability of G4–MDM2 to inhibit transcription in the absence of TFIIA and TAFs suggests that one of the remaining basal factors may be the target of the repressive activity of MDM. In addition, the purity of this MDM2-responsive transcription system argues against a mechanism of MDM2 action in which MDM2 recruits a corepressor or additional factor to mediate the observed repression.
The inhibitory domain of MDM2 lies between amino acid residues 50 and 222 and can repress p53-dependent transcription in vivo
If MDM2 does target one of the basal factors, it might be expected that MDM2 would directly interact with this factor. However, before testing the ability of MDM2 to bind the general transcription factors, we chose to more narrowly define the region of MDM2 containing the basal inhibitory domain. By this method, we hoped to limit the number of observed interactions unrelated to the inhibitory mechanism. To this end, 276 amino acids were removed from the carboxyl terminus of MDM2, and the remaining MDM2 sequences were fused to the DNA binding domain of Gal4 (G4–MDM222). Like G4–MDM2 (1–324) and G4–MDM2(50–324), G4–MDM222 was capable of inhibiting both p53-activated transcription (data not shown) and basal transcription (Fig. 4, lanes 6–9). Together, these results suggest that the basal inhibitory domain of MDM2 lies between amino acids 50 and 222.
Figure 4.
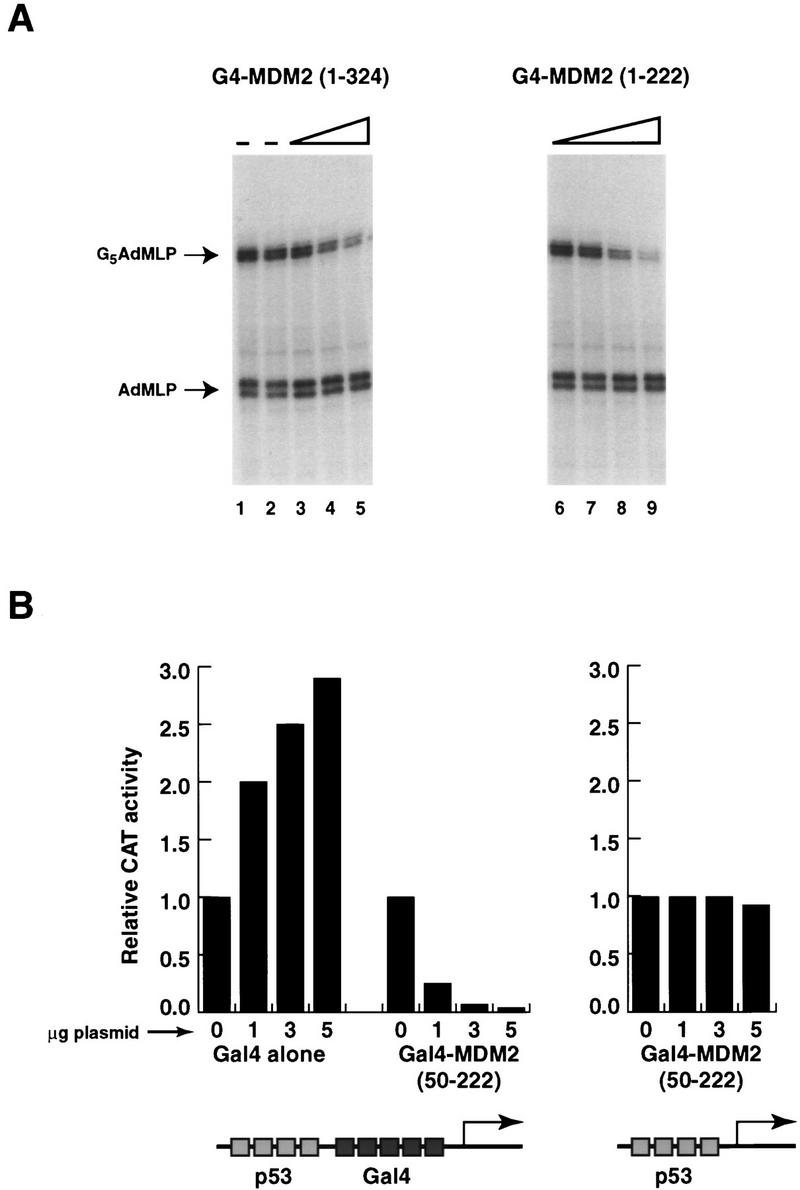
The inhibitory domain MDM2 lies between amino acid residues 50 and 222 and can repress p53-dependent transcription in vivo. (A) The first 222 amino acids of MDM2 are capable of inhibiting basal transcription. The first 222 amino acids of MDM2 were fused to the Gal4 DNA-binding domain [G4–MDM2(1–222)], and this protein was tested for its ability to repress basal transcription in a highly purified in vitro system lacking TAFs and TFIIA (see Fig. 3). The transcripts arising from the two different templates are indicated on the left. (Lanes 1,2) The basal level of transcription; (lanes 3–5) 7, 20, and 60 ng of G4–MDM2(1–324), respectively; (lanes 6–9) 7, 20, 60, and 200 ng of G4–MDM2(1–222), respectively. (B) Amino acids 50–222 of MDM2 can inhibit transcription directed by full-length p53 in transient transfections. C-33A cells were transfected with a cytomegalovirus (CMV) promoter-driven expression construct encoding full-length p53 (pC53 SN3, 0.5 μg per well) in six-well plates by a CaPO4 procedure. G4 and G4–MDM2(50–222) expression constructs were cotransfected in the amounts indicated below the graph. Two hundred nanograms of reporter plasmid [(p53)4(G4)5E1BCAT or (G4)5E1BCAT], and 500 ng of a transfection control (pCMV–lacZ) were used in each transfection. The total amount of DNA in each transfection was normalized to 7.5 μg using a CMV vector lacking an insert. All CAT activities were corrected for variations in β-galactosidase activity (a measure of transfection efficiency). Each bar represents the average of three independent experiments. In each experiment, the level of CAT activity measured in the absence of G4 or G4–MDM2(50–222) was normalized to 1 before the results were averaged.
To this point, we had identified a domain of MDM2 that was capable of inhibiting both basal and activated transcription in vitro. To determine whether this inhibitory domain was also important for the ability of MDM2 to repress transcription in vivo, we asked whether a fusion protein containing the Gal4 DNA-binding domain and amino acids 50–222 of MDM2 could inhibit full-length p53 in transiently transfected C-33A cells (a human cervical carcinoma cell line devoid of p53). In these experiments, transfection of an expression construct encoding full-length p53 resulted in a 30- to 60-fold enhancement in transcription from a template containing four p53 sites upstream of five Gal4 sites and the E1B TATA box. This promoter was fused upstream of the chloramphenicol acetyltransferase (CAT) gene, allowing promoter activity to be measured by assaying transfected cell extracts for CAT activity. Figure 4B summarizes the results of three sets of transfection experiments in which transcriptional stimulation in the presence of p53 is normalized to 1. In these experiments, cotransfection of a G4–MDM2(50–222) expression vector with the p53 expression vector and the p53/Gal4 site template significantly reduced the levels of CAT activity measured. At the highest concentration of transfected G4–MDM2 vector, the promoter activity is reduced to 5% that of p53 alone (Fig. 4B). This transcriptional inhibition is not attributable to simple occupancy of the Gal4 sites between the p53 sites and the start of transcription because expression of the Gal4 DNA-binding domain alone does not repress p53-dependent transcription (Fig. 4B), though G4-and G4–MDM2(50–222) are expressed at similar levels (data not shown). Instead, transfection of the Gal4 DNA-binding domain is slightly stimulatory, which is not unexpected as this protein has been suggested previously to contain a cryptic activation domain. Furthermore, the repression of transcription from this promoter is not attributable to a general repression of transcription resulting from G4–MDM2 overexpression because the inhibition is dependent on the presence of Gal4 sites in the template (Fig. 4B). These results demonstrate that the inhibitory domain of MDM2 identified in vitro is also capable of repressing transcription directed by full-length p53. In addition, these findings suggest that the putative repressor domain of MDM2 is capable of inhibiting transcription in a cellular setting in which the templates are likely to be assembled into chromatin and additional proteins that might interact with either p53 or MDM2 are present.
The inhibitory domain of MDM2 interacts with both the 34K subunit of TFIIE and TBP
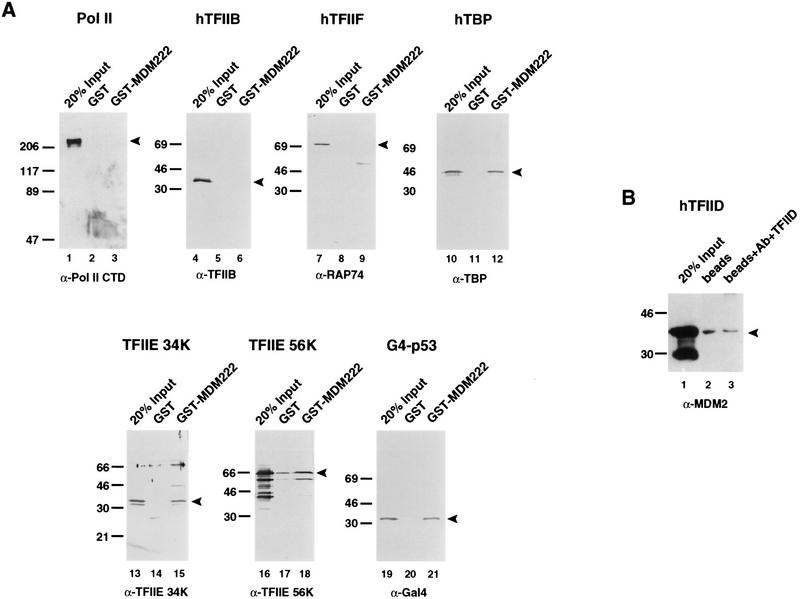
With a shortened version of the MDM2 protein containing amino acids 1–222 (MDM222), the ability of the inhibitory domain of MDM2 to bind components of the basal transcription machinery was tested. By fusing the first 222 amino acids of MDM2 to the glutathione S-transferase (GST) moiety and immobilizing this protein on a glutathione–Sepharose resin, we assessed the ability of the MDM2 inhibitory domain to interact selectively with soluble forms of the basal factors TBP, TFIIB, TFIIF, Pol II, and the 34K and 56K subunits of TFIIE. Proteins that remained bound to the GST–MDM2 resins after extensive washing were separated by SDS-PAGE, blotted to nitrocellulose, and detected using antibodies specific for these factors. As a positive control, the G4–p53 activation domain fusion was also tested for its ability to bind these resins (Fig. 5A, lanes 19–21). As shown in Figure 5A, TBP (lanes 10–12) and the 34K subunit of TFIIE (lanes 13–15) were the only basal factors that were retained to a significant extent by the GST–MDM222 beads but not the GST resin. The observed interaction between MDM2 and TBP was not unexpected and has been reported previously (Leng et al. 1995). The TFIIE 56K subunit was also retained to a small extent (Fig. 5A, lanes 16–18; <5% of the input bound), though binding to the GST resin lacking MDM2 was also detected, making it difficult to determine definitively whether TFIIE 56K and MDM2 interact specifically.
Figure 5.
(A) TBP and the small subunit of TFIIE interact with the inhibitory domain of MDM2. We next wanted to determine whether MDM2 could interact with any of the basal factors present in the minimal transcription system we had found to be capable of supporting basal repression by G4–MDM2. To this end, we fused the first 222 amino acids of MDM2 to GST (GST–MDM222), immobilized these proteins on glutathione beads, and tested their ability to specifically interact with many of the basal transcription factors: HeLa cell-purified Pol II (lanes 1–3), recombinant human (h) TFIIB (lanes 4–6), recombinant hTFIIF (lanes 7–9), recombinant hTBP (lanes 10–12), recombinant hTFIIE 34K (lanes 13–15), and recombinant hTFIIE 56K (lanes 16–18). Proteins retained by these beads were separated by SDS-PAGE, transferred to nitrocellulose, probed with antibodies against the proteins of interest (indicated below the corresponding blots), and detected using an ECL protocol (Amersham). Approximately 300–500 ng of the GST or GST–MDM222 immobilized on beads and 100–200 ng of the soluble target proteins were used in each binding reaction. The positions of the various proteins are indicated by arrowheads. (B) The inhibitory domain of MDM2 does not interact with the TFIID complex. To determine whether MDM2 can interact with TBP when assembled into the TFIID complex, we immobilized TFIID on an α-TAFII130/protein A resin and tested the ability of soluble MDM2(1–222) to be retained by this resin (lane 3). Protein A beads alone were used as a negative control (lane 2). The bound proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with α-MDM2 antibody (Pharmagenics). The arrowhead indicates the position of migration of MDM2(1–222). (Lane 1) 20% of the amount of soluble MDM2(1–222) used for each reaction.
Because TBP is notoriously “sticky” and has been reported to bind a large number of proteins, we wanted to obtain additional evidence that the interaction between TBP and MDM2 could occur in vivo. In a cellular setting, the majority of TBP is thought to exist in large multiprotein complexes, including TFIID, SL1, and TFIIIB (Goodrich and Tjian 1994a and references therein). For this reason, we tested the ability of MDM222 to interact with the TFIID complex. We linked antibodies against the TAFII130 subunit of TFIID to protein A–Sepharose beads and immobilized the TFIID complex from partially purified HeLa extracts on this resin. Next, extracts containing bacterially expressed MDM222 were tested for their ability to interact with this immobilized complex. Unlike the monomeric form of TBP, TBP incorporated into the TFIID complex was not able to interact with MDM222 (Fig. 5B, lanes 1–3). This finding suggests that an interaction between MDM2 and TBP may not occur in vivo and thus this interaction may not be relevant to the inhibitory properties of MDM2.
Discussion
In this report we provide evidence that MDM2, an oncoprotein that inhibits p53-dependent transcription, contains a previously undetected repression domain between amino acids 50 and 222 that can inhibit basal transcription in a p53-independent manner. Several mechanisms have been proposed to explain the ability of repressors to inhibit basal transcription, including direct inhibition of the activities of the general transcription machinery by the repressor (Sauer et al. 1995), modification of chromatin structure near the promoter (Bunker and Kingston 1994), and recruitment of additional factors or “corepressors” that themselves mediate transcriptional inhibition (Keleher et al. 1992; Paroush et al. 1994). We have demonstrated that MDM2 is able to inhibit basal transcription in a highly purified system devoid of chromatin. Furthermore, this system is not likely to contain potential corepressors, unless they are tightly associated with either Pol II or TFIIH. Our studies of the repression domain of MDM2 thus suggest that this domain directly inhibits one or more functions of the basal machinery. In an attempt to identify components of the basal machinery that might be targeted by MDM2, we used a combination of in vitro transcription assays and in vitro binding assays to define a subset of the basal factors sufficient to support MDM2-dependent basal inhibition and to identify potential contacts between MDM2 and the general transcription machinery. These studies led to the finding that neither TFIIA nor the TAFs are necessary for the ability of MDM2 to inhibit transcription. Of the remaining basal factors, MDM2 was found to interact with both the 34K subunit of TFIIE and monomeric TBP, suggesting that through these contacts MDM2 might inhibit their ability to function in the preinitiation complex. However, the additional finding that the inhibitory domain of MDM2 was not able to interact with TBP when in complex with TAFs leads us to speculate that the interaction between MDM2 and TBP may not be functionally relevant.
Our finding that the 34K subunit of TFIIE may be the target of the repressive activities of MDM2 was intriguing because it has been shown previously that a Drosophila repressor, Krüppel, also directly interacts with the 34K subunit of TFIIE (Sauer et al. 1995). During transcription initiation, TFIIE is thought to play a key role in the ability of the general transcription machinery to progress from the promoter-bound complex to the actively elongating complex, and repressors that target TFIIE might disrupt this transition by inhibiting one or more of the known functions of TFIIE. TFIIE appears to work in concert with the basal transcription factor TFIIH to facilitate DNA strand separation at the promoter, leading to promoter clearance and elongation (Schaeffer et al. 1993; Goodrich and Tjian 1994b; Holstege et al. 1995, 1996). TFIIE is also believed to be required for the recruitment of TFIIH (Flores et al. 1992; Maxon et al. 1994), a multisubunit complex that contains several enzymatic activities itself, including helicase, kinase, and ATPase (Sopta et al. 1989; Flores et al. 1992; Schaeffer et al. 1993; Serizawa et al. 1993; Roy et al. 1994). These enzymatic properties of TFIIH have been shown to be regulated in vitro by TFIIE (Lu et al. 1992; Ohkuma and Roeder 1994; Serizawa et al. 1994; Ohkuma et al. 1995). By binding to the small subunit of TFIIE, it is possible that MDM2 affects the ability of TFIIE to recruit TFIIH or to influence its kinase, helicase, or ATPase activities. Alternatively, TFIIE may play a more direct role in DNA strand separation or recruitment and stability of the preinitiation complex, offering other potential functions to be targeted by MDM2 (Maxon et al. 1994; Holstege et al. 1995). Future studies will address the ability of MDM2 to inhibit these specific functions of TFIIE in an attempt to determine the mechanism by which MDM2 can directly repress basal transcription.
Although the precise mechanisms by which specific repressors exert their inhibitory activities are not well known, at least three types of repressors have been proposed to exist: those that compete with activators for promoter binding sites, “quenchers” that directly inhibit the action of specific transactivators, and inhibitors that directly repress the functioning of the basal transcription machinery (Levine and Manley 1989; Johnson 1995). Much circumstantial evidence has been accumulated to suggest that MDM2 represses the transactivating functions of p53 by masking or interfering with its activation domain. This domain of p53 has been shown to interact with several components of the transcription machinery, including TAFII32 (Lu and Levine 1995; Thut et al. 1995), TAFII70 (Thut et al. 1995), TBP (Seto et al. 1992; Chen et al. 1993; Liu et al. 1993; Truant et al. 1993), and TFIIH (Xiao et al. 1994). A functional importance for these interactions has been shown in the case of TAFII32 and TAFII70 (Thut et al. 1995). Interestingly, the same mutation in the activation domain of p53 that impairs its ability to bind TAFII32 (Lu and Levine 1995; Thut et al. 1995), TAFII70 (Thut et al. 1995), and TBP (amino acids 22 and 23) also disrupts its ability to contact MDM2 (Lin et al. 1994), suggesting that MDM2 and the basal factors may interact with overlapping portions of p53. Furthermore, the recent crystal structure of the p53/MDM2 interaction interface showed that residues 22 and 23 are contained within this interface. These correlations lend further support to the hypothesis that MDM2 inhibits p53-dependent activation by disrupting interactions with components of the general transcription machinery.
Our results suggest that MDM2 can also directly inhibit the ability of the general transcription factors to potentiate mRNA synthesis. However, we do not believe that our findings exclude the possibility that MDM2 may also inhibit p53-dependent transcription by masking its activation domain from important basal factor contacts. On the contrary, the experiments described here, in conjuction with the correlation between the MDM2 and basal factor binding sites in p53’s activation domain, suggest that it is likely that MDM2 functions as both a quencher of p53 and a direct inhibitor of the basal machinery (Fig. 6). In addition, it is possible that these two mechanisms of inhibition by MDM2 complement each other and enhance the ability of MDM2 to regulate p53-responsive promoters. In support of this hypothesis, a similar dual mechanism of inhibition involving both potential masking of an activator and recruitment of a general repression domain has also been proposed to explain the inhibition of E2F-dependent transcription by Rb (Bremner et al. 1995; Sellers et al. 1995; Weintraub et al. 1995). Much like the MDM2/p53 case, an interaction between the activation domain of E2F and Rb has been shown to be important for the ability of Rb to inhibit transcription (Flemington et al. 1993; Hagemeier et al. 1993; Helin et al. 1993). In addition, Rb is capable of repressing transcription in an E2F-independent manner when fused to a heterologous DNA-binding domain, although it is not clear whether Rb targets the basal machinery or whether it serves to quench nearby activators (Bremner et al. 1995; Sellers et al. 1995; Weintraub et al. 1995). These results suggest that transcriptional inhibitors that repress transcription via multiple, distinct mechanisms are not so uncommon.
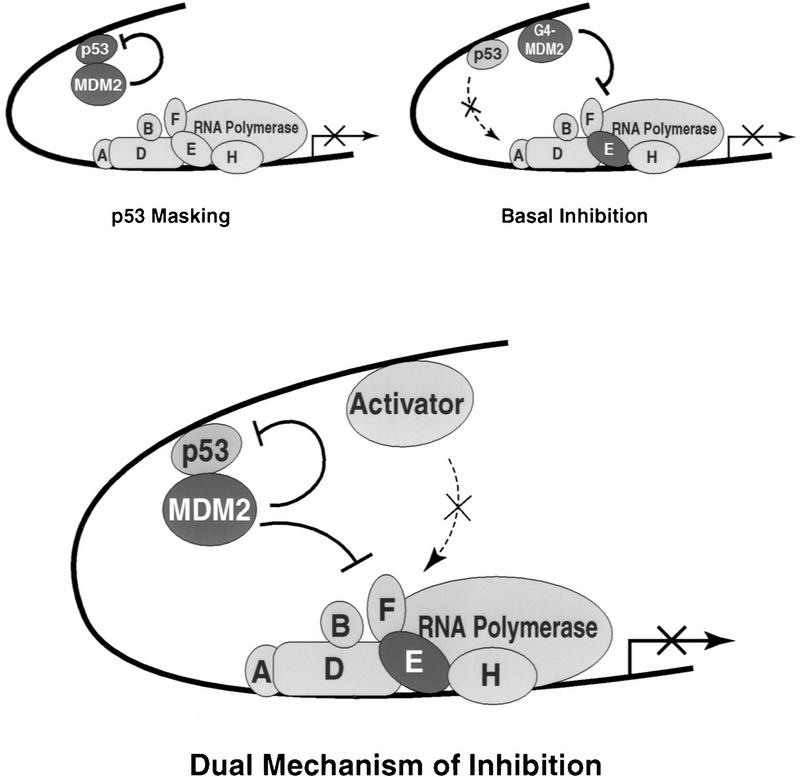
Figure 6.
A dual mechanism for inhibition of p53-responsive promoters by MDM2. The current model for inhibition of p53-dependent transcription by MDM2 suggests that by binding to the activation domain of p53, MDM2 disrupts important functional interactions between this domain and components of the basal transcription machinery. Our data suggest that MDM2 also contains a p53-independent inhibitory domain that directly represses basal transcription. The observed interaction beween the inhibitory domain of MDM2 and the 34K subunit of the basal factor TFIIE may indicate that MDM2 represses the function of this basal factor and thereby inhibits the ability of the preinitiation complex to synthesize mRNA. We propose that these inhibitory mechanisms are not mutually exclusive and may play different roles in the repressive properties of MDM2. Perhaps MDM2 simultaneously conceals the activation domain of p53 and directly represses the function of one or more components of the basal machinery to more stringently control transcription from p53-responsive promoters.
If MDM2 can inhibit the transactivating ability of p53 by directly interacting with this protein, why then should MDM2 contain an additional inhibitory domain? We believe that the answer to this question is best addressed in the context of complex eukaryotic promoters. Differential gene expression, especially in metazoans, is regulated by integrating a wide variety of positive and negative growth signals that dictate the level of transcripts arising from a particular promoter. This integration is likely to occur by the cooperative action of multiple transcriptional regulators bound at a promoter, each of which can influence the functions of the basal transcription machinery. Because most complex regulatory elements bear multiple activator binding sites, it is expected that efficient inhibition of transcription from such a promoter would require either multiple repressors affecting different sets of activators or a single repressor or silencer capable of impeding transactivation of multiple enhancer factors. The current masking model for MDM2 inhibition of p53-dependent transcription would predict that MDM2 could repress only activating signals from p53 itself but would leave the activity of other activators bound to the same promoter region largely unaffected. We suggest that the basal inhibitory domain of MDM2 functions to repress the effects of other activators bound at p53-responsive promoters (Fig. 6). In this way, MDM2 may serve to cause a more stringent regulation of p53-responsive promoters by decreasing both p53-dependent and -independent activation. If this model is correct, one would predict that MDM2 might be able to inhibit transcription stimulated by a subset of activators other than p53 if MDM2 could be recruited to the promoter in a p53-independent manner. In an attempt to test this hypothesis, we asked whether G4–MDM2 could inhibit transcription enhanced by several additional activators including Sp1, CTF (CCAAT box-binding transcription factor), and Lex–VP16. However, in none of these cases was transcription repressed by G4–MDM2 (data not shown). Although these results do not support our model, they also do not rule it out. First, transcriptional activators are likely to stimulate the activities of the basal machinery by diverse mechanisms. Some transactivators may be able to overcome the repressive effects of MDM2, perhaps by stimulating a step required for initiation that occurs downstream of the step inhibited by MDM2. For this reason, we believe that it is quite possible that MDM2 might only repress a subset of transcriptional activators and that we have not yet tested one that is appropriate. Second, basal promoters themselves are not equivalent. To alter the number of transcripts arising from a particular promoter, a transcriptional modulator must presumably affect the rate-limiting step at that promoter (e.g., basal factor recruitment, DNA strand separation, promoter clearance, etc.). This rate-limiting step will vary because of differences in the core promoter sequences and may cause some promoters to be insensitive to the effects of the inhibitory domain of MDM2. Finally, it is conceivable that in a bona fide p53-regulated promoter/enhancer, repression of p53 by MDM2 alone is sufficient to weaken the levels of transcription because of the cooperative or synergistic interplay of multiple activators that may be disrupted by repressors. Future studies using cellular, p53-responsive promoters will help to address the potential in vivo role of the inhibitory domain of MDM2 that we have described here. In addition, these studies may lead to a better understanding of the capacity of MDM2 to antagonize the biological effects of p53 and will extend our understanding of general mechanisms of transcriptional repression.
Materials and methods
Plasmids
E. coli expression vectors containing human MDM2 sequences corresponding to amino acids 1–324, 50–324, and 1–222 were constructed using a PCR-based strategy. NdeI sites were placed at the first or fiftieth codon of human MDM2, and a stop codon followed by a BamHI site was placed after amino acid 222 or 324. The PCR products and pET19b (Novagen) were then digested with NdeI and BamHI and ligated to produce the His–MDM2(1–324) and His–MDM2(50–324) vectors. His–MDM2(1–222) was constructed by placing an NdeI site at the first codon and a stop codon followed by an EspI site after amino acid 222, digesting the PCR product and pET19b with NdeI and EspI and ligating the fragments. G4–MDM2(1–324), G4–MDM2(50–324), and G4–MDM2(1–222) were engineered by ligating an NdeI (blunt)/EcoRI fragment from the corresponding His–MDM2 vectors into a pET3a-based vector containing DNA sequences encoding the first 94 amino acids of the Gal4 DNA-binding domain (J. Goodrich, unpubl.) cut with KpnI (blunt) and EcoRI. To produce G4–MDM2(50–222), the NdeI and EcoRI sites were created on either side of the MDM2 coding sequences by PCR, and digested with NdeI and EcoRI and ligated into NdeI/EcoRI-cut Gal4 vector. GST–MDM2(1–222) was constructed by digesting His–MDM2(1–222) with NdeI and EcoRI and ligating this fragment into NdeI/EcoRI-cut pGEX–2TKN (S. Ruppert, unpubl.). The (p53)4(Gal4)5E1BCAT transcription template used in transfections was produced by inserting four p53 sites contained on a HindIII–PstI fragment from pCZ PG4–lacZ (a kind gift from B. Vogelstein, Johns Hopkins Oncology Center, Baltimore, MD) into HindIII–PstI-cut G5E1BCAT (Lillie and Green 1989). Mammalian expression constructs for Gal4(1–94) and Gal4–MDM2(50–222) were constructed by transferring the NdeI–EcoRI fragments from the E. coli expression vectors containing G4 and G4–MDM2(50–222) into a derivative of pTβstops (S. Ruppert, unpubl.) to insert a Kozak sequence (NcoI site) upstream of the coding sequences. Next, a HindIII–EcoRI fragment containing the Kozak sequence and the coding regions was inserted into pcDNA3.1+ (Invitrogen) to yield pcDNA–G4 and pcDNA–G4MDM2(50–222).
Purification of recombinant proteins
The MDM2(1–324) and MDM2(50–324) proteins were expressed in E. coli (BL21 strain) by transfecting bacteria with His–MDM2(1–324) and His–MDM2(50–324). The cells were grown to an OD600 of 0.5 and induced using 0.4 mm isopropyl β-d-thiogalctopyranoside (IPTG) followed by an additional 4 hr of growth at 37°C. The cells were pelleted and sonicated, and the lysates were cleared by centrifugation. The cell pellets were then washed four times with 5 mm dithiothreitol (DTT) by dispersing the cell pellets with sonication. The washed pellets were resuspended in a small volume of guanidine buffer [6m guanidine-HCl, 20 mm Tris at pH 7.9, 10 μm ZnSO4, 10 mm β-mercaptoethanol (β-me)] and nutated at room temperature for 4 hr to extract the proteins. The extracts were cleared by centrifugation and loaded onto Ni2+ NTA resin (Qiagen) equilibrated in the same buffer. The resin was then washed with urea buffer (5 mm urea, 10 mm MES at pH 6.0, 10 μm ZnSO4, 10 mm β-me, 0.1 m NaCl) containing 35 mm imidazole. The proteins were eluted from the resin using urea buffer containing 250 mm imidazole and loaded onto a Poros HS column (PerSeptive Biosystems). The Poros HS column was eluted using a 0.1–1.0 m NaCl gradient in urea buffer, and fractions containing His–MDM2, as assessed by SDS-PAGE and Coomassie staining, were pooled. The denatured proteins were then refolded by stepwise dialysis first into guanidine buffer then into nondenaturing buffer (20 mm Tris at pH 7.9, 10 μm ZnSO4, 10% glycerol) using the steps 2 m guanidine, 1 m guanidine, 0.5 m guanidine, and 0.1 m KCl.
G4–MDM2(1–324), G4–MDM2(50–324), and G4–MDM2(1–222) were expressed and purified by a protocol similar to that for the His–MDM2 proteins. However, after extraction from the pellet, the Ni2+ affinity chromatography step was omitted and the denatured proteins were dialyzed into urea buffer and purified using a Poros HS column. The resulting protein pools were then dialyzed back into guanidine buffer and purified by size-exclusion chromatography using a Superose 12 column (Pharmacia). Renaturation followed the same procedure as that of the His–MDM2 proteins.
Lex–p53 was expressed by transforming the Lex–p53 plasmid into E. coli (BL21 strain), growing the cells to an OD600 of 0.5, inducing with 0.4 mm IPTG for 4 hr, and collecting the cells. The resulting cell pellet was resuspended in TGEM (20 mm Tris at pH 7.9, 5 mm MgCl2, 1 mm EDTA, 20% glycerol) containing 0.1 m NaCl and 0.1% NP-40, and sonicated, and the lysates were cleared by centrifugation. Lysates were then applied to anti-Flag M2 antibody resin (Kodak), nutated at 4°C for 4 hr, and eluted using Flag epitope peptide dissolved in TGEM containing 0.1 m NaCl and 0.1% NP-40.
G4–p53 was expressed as described previously (Thut et al. 1995) and purified using DEAE CL4B (Pharmacia) and Poros HS resins (PerSeptive Biosystems) eluted with linear salt gradients. GST–p53 and GST–MDM2(1–222) were expressed in BL21 cells transformed with the appropriate plasmids. Crude lysates from these cells were immobilized on glutathione–Sepharose beads (Pharmacia) by incubating the resin and cell extracts for 1 hr and washing the resin with TGEM/1.0 m NaCl. The beads were equilibrated to TGEM/0.1 m NaCl before they were used in in vitro binding assays. G4–VP16 was expressed and purified as described previously (Chasman et al. 1989).
In vitro transcription reactions
The partially purified HeLa cell extracts (Dignam et al. 1983), the recombinant TFIIA (Ranish et al. 1992), and the highly purified basal transcription factors (Goodrich and Tjian 1994b) were prepared as described previously. Transcription from the G5L2E4T and L2E4T templates (Saha et al. 1993) was assayed in the presence of partially purified HeLa cell extracts by primer extension using the E4 primer (Lillie et al. 1986). Additions of transcription factors and basal factors were carried out at 4°C in the following order: DNA template, inhibitor, activator, TBP or TFIID complex, P.5, and TFIIA. The mixtures were incubated at 30°C for 25 min, and transcription was initiated by adding nucleoside triphosphates to a final concentration of 500 μm and incubating for an additional 25 min. The reaction products were detected by primer extension, visualized by autoradiography, and quantified by PhosphorImager analysis (Fuji Photo Film Co., Ltd.). Transcription from the G5–AdML G-less (a kind gift from Michael Carey, University of California, Los Angeles) and AdML G-less (Parvin et al. 1992) templates was assayed in the presence of partially purified HeLa cell extracts or a highly purified transcription system using a G-less assay (Sawadogo and Roeder 1985). We incubated 100 ng of both templates on ice with the addition of inhibitors, activators, and basal factors in the following order: inhibitor, activator, TBP or TFIID fraction, and a mixture of the remaining basal factors. With the highly purified transcription system, the following basal factors and concentrations were used: 50 ng of Pol IIo, 20 ng of TFIIB, 30 ng of TFIIE 34K, 6 ng of TFIIE 56K, 2 ng of RAP30, 7 ng of RAP74, 0.1 μl of a TFIIH phenyl–Superose fraction (Flores et al. 1992), and 10 ng of TBP. The mixtures were incubated as before and transcription was initiated by the addition of nucleotides [500 μm ATP and CTP, 25 μm [α-32P]UTP (5 μCi per reaction)] and allowed to proceed for 25 min. After separation on a denaturing polyacrylamide gel, [α-32P]UTP incorporation was quantitated by PhosphorImager analysis.
In vitro binding assays
In vitro protein–protein interaction assays using GST fusion proteins were performed essentially as described (Goodrich et al. 1993). Approximately 0.5–1 μg of the immobilized proteins and 200–400 ng of the soluble basal factors were used in each binding reaction. The interaction assay using TFIID complexes immobilized on beads was performed by binding monoclonal antibodies against hTAFII130 to protein A–Sepharose resin (Pierce) in TGEM/0.4 m NaCl for 1 hr, washing the the unbound antibodies away with TGEM/1.0 m NaCl + 0.1% NP-40, and incubating a HeLa nuclear P1.0 fraction (Dignam et al. 1983) with the antibody beads overnight at 4°C. The beads were then washed three times with TGEM + 0.7 m NaCl and three times with TGEM + 0.1 m NaCl. The immobilized TFIID complexes were assayed for their ability to bind soluble extracts containing MDM(1–222) by a procedure similar to that used for the GST fusion protein/basal factor interactions.
Transient transfection assays
C33A cells (ATCC) were plated at a density of 3 × 105 cells per well in six-well plates (Falcon) 16–24 hr before CaPO4-mediated transfection (Sambrook et al. 1989). The medium (Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, 100 U/ml of streptomycin, and 2 mm glutamine) was replaced 18–24 hours after transfection. Twenty-four to 36 hr later, the cells were harvested, lysed, and assayed for β-galactosidase and CAT activity (Sambrook et al. 1989).
Acknowledgments
We thank Bert Vogelstein for a yeast expression vector containing the human MDM2 coding sequences and the pC53–SN3 mammalian expression vector (full-length p53), Michael Carey for the G5AdML G-less template, Jeffery Parvin for the AdML G-less template, Tom O’Brien for the Lex–p53 bacterial expression construct, Edith Wang for the anti-TAFII130 monoclonal antibodies, Mary Maxon for the anti-TFIIE antibodies, Pierre Beaurang for purified TFIIE 56K, and David King for synthesizing the Flag epitope peptide. In addition, we are grateful to members of the Tjian laboratory for their helpful suggestions and support and to Paul Kaufman, Mike Holmes, Jon Oliner, Jórk Zwicker, Gene Cutler, Grace Gill, Brian Dynlacht, Laura Attardi, and Mike Levine for critical reading of the manuscript. J.A.G. is supported by a fellowship from the Leukemia Society of America, and C.J.T. was supported by a National Science Foundation Graduate fellowship. This work was supported, in part, by a grant from the National Institutes of Health to R.T.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jlim@mendel.berkeley.edu; FAX (510) 643-9547.
References
- Attardi LD, Lowe SW, Brugarolas J, Jacks T. Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. EMBO J. 1996;15:3693–3701. [PMC free article] [PubMed] [Google Scholar]
- Bargonetti J, Reynisdottir I, Friedman PN, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes & Dev. 1992;6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- Bremner R, Cohen BL, Sopta M, Hamel PA, Ingles CJ, Gallie BL, Phillips RA. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Deb S, Munoz RM, Subler MA, Deb SP. The tumor suppressor p53 and the oncoprotein simian virus 40 T antigen bind to overlapping domains on the MDM2 protein. Mol Cell Biol. 1993;13:6849–6857. doi: 10.1128/mcb.13.11.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon G. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Bunker CA, Kingston RE. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol Cell Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman DI, Leatherwood MC, Carey M, Ptashne M. Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol Cell Biol. 1989;9:4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Farmer G, Zhu H, Prywes R, Prives C. Cooperative DNA binding of p53 with TFIID (TBP): A possible mechanism for transcriptional activation. [published erratum appears in 2652.] Genes & Dev. 1993;7:1837–1849. doi: 10.1101/gad.7.10.1837. [DOI] [PubMed] [Google Scholar]
- Chowdary DR, Dermody JJ, Jha KK, Ozer HL. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oca Luna RM, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge S, Leder P. Mice lacking p21 (CIP1/WAF1) undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. [see comments.] Nature. 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- Fields S, Jang SK. Presence of a potent transcription activation sequence in the p53 protein. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- Finlay CA. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington EK, Speck SH, Kaelin W., Jr E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Lu H, Reinberg D. Factors involved in specific transcription initiation by RNA polymerase II: Identification and characterization of factor IIH. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- Goodrich JA, Hoey T, Thut CJ, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. TBP-TAF complexes: Selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994a;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- ————— Transcription Factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994b;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Haffner R, Oren M. Biochemical properties and biological effects of p53. Curr Opin Genet Dev. 1995;5:84–90. doi: 10.1016/s0959-437x(95)90058-6. [DOI] [PubMed] [Google Scholar]
- Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines DS, Landers JE, Engle LJ, George DL. Physical and functional interaction between wild-type p53 and mdm2 proteins. Mol Cell Biol. 1994;14:1171–1178. doi: 10.1128/mcb.14.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FCP, Tantin D, Carey M, van der Vliet PC, Timmers HTM. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FCP, van der Vliet PC, Timmers HTM. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Johnson AD. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz MC, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Leng P, Brown DR, Deb S, Deb SP. Human oncoprotein MDM2 interacts with the TATA-binding protein in vitro and in vivo. Int J Oncol. 1995;6:251–259. [PubMed] [Google Scholar]
- Levine M, Manley JL. Transcripional repression of eukaryotic promoters. Cell. 1989;59:405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- Lillie JW, Green MR. Transcriptional activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Lillie J, Green M, Green MR. An adenovirus E1a protein region required for transformation and transcriptional repression. Cell. 1986;46:1043–1051. doi: 10.1016/0092-8674(86)90704-x. [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcription activation, binding to mdm-2, and the adenovirus 5 E1B 55-kD protein. Genes & Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Liu X, Miller CW, Koeffler PH, Berk AJ. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol. 1993;13:3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Levine AJ. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zawel L, Fisher L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- Maxon M, Goodrich JA, Tjian R. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: A model for promoter clearance. Genes & Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibts p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y, Roeder RG. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y, Hashimoto S, Wang CK, Horikoshi M, Roeder RG. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-α. Mol Cell Biol. 1995;15:4856–4866. doi: 10.1128/mcb.15.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Parvin JD, Timmers HTM, Sharp PA. Promoter specificity of basal transcription factors. Cell. 1992;68:1135–1144. doi: 10.1016/0092-8674(92)90084-p. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Lane WS, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- Raycroft L, Wu HY, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249:1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Schaeffer L, Humbert S, Vermeulen W, Weeda G, Egly J-M. The DNA-dependent ATPase activity associated with the class II basic transcription factor BTF2/TFIIH. J Biol Chem. 1994;269:9826–9832. [PubMed] [Google Scholar]
- Saha S, Brickman JM, Lehming N, Ptashne M. New eukaryotic transcriptional repressors. Nature. 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sauer F, Fondell JD, Ohkuma Y, Roeder RG, Jäckle H. Control of transcription by Krüppel through interactions with TFIIB and TFIIEβ. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- Sawadogo M, Roeder R. Factors involved in specific transcription by human RNA polymerase II: Analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JHJ, Chambon P, Egly J-M. DNA repair helicase: A component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Schneffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Sellers WR, Rodgers JW, Kaelin WGJ. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa H, Conaway RC, Conaway JW. Multifunctional RNA polymerase II initiation fgactor delta from rat liver: Relationship between carboxyl-terminal domain kinase, ATPase, and DNA kinase. J Biol Chem. 1993;268:17300–17308. [PubMed] [Google Scholar]
- Serizawa H, Conaway JW, Conaway RC. An oligomeric form of the large subunit of transcription factor (TF) IIE activates phosphorylation of the RNA polymerase II carboxy-terminal domain by TFIIH. J Biol Chem. 1994;269:20750–20756. [PubMed] [Google Scholar]
- Seto E, Usheva A, Zambetti GP, Momand J, Horikoshi N, Weinmann R, Levine AJ, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Chen I-T, Zhan Q, Bae I, Chen C-Y, Gilmer TM, Kastan MB, O’Conner PM, Fornace AJJ. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- Sopta M, Burton ZF, Greenblatt J. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 1989;341:410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- Takenaka I, Morin F, Seizinger BR, Kley N. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- Thut CJ, Chen J-L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- Truant R, Xiao H, Ingles CJ, Greenblatt J. Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J Biol Chem. 1993;268:2284–2287. [PubMed] [Google Scholar]
- Unger T, Nau MM, Segal S, Minna JD. p53: A transdominant regulator of transcription whose function is ablated by mutations occurring in human cancer. EMBO J. 1992;11:1383–1390. doi: 10.1002/j.1460-2075.1992.tb05183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub SJ, Chow KNB, Luo RX, Zhang SH, He S, Dean DC. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier JL, Triezenberg SJ, Reinberg D, Flores O, Ingles CJ, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Science. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- Yew PR, Liu X, Berk AJ. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes & Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- Zambetti GP, Bargonetti J, Walker D, Prives C, Levine AJ. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes & Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]