Abstract
Homothallic strains of Saccharomyces cerevisiae can change mating type as often as every generation by replacing the allele at the MAT locus with a copy of mating type information present at one of two storage loci, HML and HMR, located on either end of chromosome III. Selection of the appropriate donor locus is dictated by a mating type-specific repressor protein, α2p: Cells containing α2p select HMR, whereas those lacking α2p select HML. As a repressor protein, α2p binds to DNA cooperatively with the transcriptional activator Mcm1p. Here we show that two α2p/Mcm1p-binding sites, DPS1 and DPS2, control donor selection. DPS1 and DPS2 are located ∼30 kb from the left arm of chromosome III, well removed from HML, HMR, and MAT. Precise deletion of only DPS1 and DPS2 results in random selection of donor loci and in a cells without affecting selection in α cells. Reciprocally, deletion of only the α2p binding segments in each of these two sites results in selection of the wrong donor loci in α cells without affecting preference in a cells. These results suggest that Mcm1p, bound to these two sites in the absence of α2p, activates HML as donor. Binding of α2p blocks the ability of Mcm1p bound to DPS1 and DPS2 to activate HML, resulting in default selection of HMR as donor. DPS1 and DPS2 also regulate expression of several noncoding RNAs, although deletion of at least one of these RNA loci does not affect donor preference. This suggests that transcriptional activation, rather than transcription of a specific product, is the initiating event in activating the left arm of chromosome III for donor selection.
Keywords: S. cerevisiae, mating type interconversion, repression protein α2p, transcriptional activation, donor selection, recombinational enhancer
Mating type interconversion in yeast provides a striking example of developmentally regulated interactions between distant regions of the genome. Most laboratory strains of Saccharomyces exhibit one of two haploid mating types, a or α, dictated by the particular allele, a or α, present at a single locus, MAT, on chromosome III. However, those strains that carry the HO (homothallic) gene do not exhibit a stable mating type but, rather, change mating type as often as every generation (for review, see Herskowitz et al. 1992). The product of the HO gene, a double-stranded DNA endonuclease, initiates this process of mating type interconversion by generating a double-strand break uniquely at a specific site within the MAT locus (Strathern et al. 1982; Kostriken et al. 1983; Kostriken and Heffron 1984). The cell then repairs this double-strand break by a gene conversion event, inserting at MAT a copy of the DNA present at one of two repositories of mating type information, known as the HML and HMR silent mating type loci. Each of these loci, located at the opposite ends of chromosome III, contains a complete but transcriptionally inactive copy of one mating type allele, with HML normally carrying the α allele and HMR the a allele. Thus, replacement of the a allele at MAT with a copy of the a allele resident at HMR leads to a switch in mating type (cf. Fig. 1).
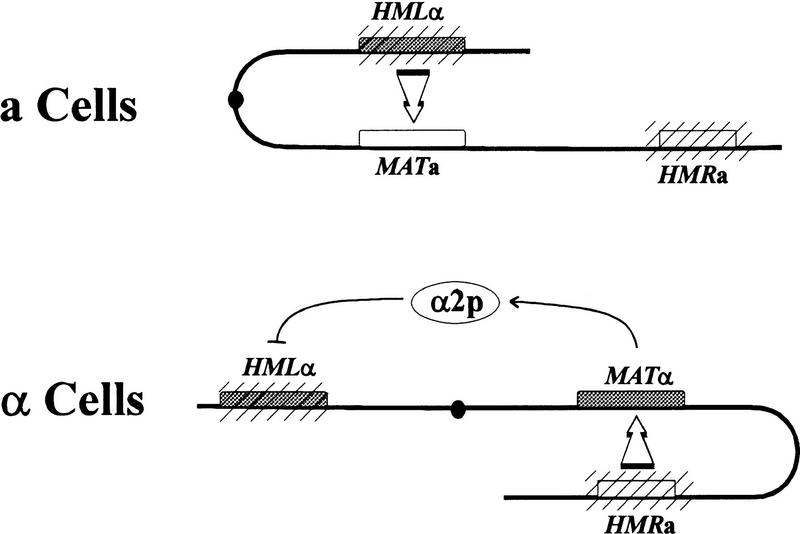
Figure 1.
Donor preference during mating type interconversion in Saccharomyces cerevisiae. Diagram of chromosome III indicating the cell-dependent bias in donor preference during mating type interconversion. Mating type loci are indicated by boxes, with filled boxes representing α mating alleles and open boxes representing a mating alleles. The transcriptionally silent mating type loci, HML and HMR, are denoted by hatching, and the centromere is designated by the filled circle. In a cells (top), HML serves as donor in ∼80% of switching events. In α cells (bottom), the product of the MATα2 locus, α2p, blocks HML from serving as donor, and as a consequence, HMR serves as donor in ∼95% of the switching events.
The process of mating type interconversion is tightly regulated (Herskowitz et al. 1992). Restricted expression of HO ensures that switching occurs only during G1 and only in cells that have budded at least once (Nasmyth 1983; Breeden and Nasymth 1985; Bobola et al. 1996; Sil and Herskowitz 1996). In addition, because HO expression is repressed in diploid cells, switching only occurs in haploids (Jensen and Herskowitz 1984). Finally, cell type dictates which of the silent loci, HML or HMR, will serve as donor following cleavage of the MAT locus. a Cells predominantly use HML as donor, whereas α cells most often use HMR as donor (Herskowitz et al. 1992). Given the disposition of alleles between the two donor loci, this process of donor preference ensures that most HO-initiated events result in a change of mating type, rather than a futile replacement of the mating type allele at MAT with the same allele. This regulation requires that the cell be able to promote selective interaction among loci positioned over an entire chromosome.
The fact that the cell exhibits donor preference in mating type switching indicates that the cell can distinguish HML from HMR in a cell type-specific manner. Cell type-specific recognition of the two loci is dictated by α2p, one of the two transcriptional regulatory proteins encoded by the MATα allele. Cells expressing α2p select HMR as donor, whereas cells lacking α2p select HML as donor, regardless of the presence or absence of other mating type regulatory proteins (Jensen and Herskowitz 1984; Tanaka et al. 1984; Wu et al. 1996; Szeto and Broach 1997). In haploid cells, α2p functions as a repressor of genes, such as STE2 and MFA1, that are responsible for conferring an a mating phenotype to the cell (generically designated a-specific genes). Repression results from the binding of a heteromeric complex, composed of two molecules of α2 protein and two molecules of the constitutively expressed MADS box protein, Mcm1p, to operator sites upstream of the regulated genes (Keleher et al. 1988, 1989; Passmore et al. 1989). These binding sites consist of a 16-bp dyad symmetrical Mcm1p-binding region bracketed by inversely oriented 9-bp α2p-binding segments (Sauer et al. 1988; Keleher et al. 1989). Repression requires recruitment of the Tup1p/Ssn6p general repression complex through direct interaction between α2p and Tup1p (Keleher et al. 1992; Komachi et al. 1994). We have recently shown that α2p dictates donor preference as a complex with Mcm1p and through recruitment of Tup1p/Ssn6p (Szeto and Broach 1997). Thus, the same components and interactions required for transcriptional repression by α2p are also essential for cell type-specific donor prefernce.
The particular features of HML or HMR that the cell uses to distinguish between them during mating type interconversion have begun to be clarified. Our previous studies demonstrated that neither the alleles at the donor loci, the structure of these loci, nor any feature near HML, HMR, or MAT provides a marker used by the cell to select one donor locus versus the other during interconversion (Weiler and Broach 1992). Nonetheless, because donor preference is maintained even when MAT is moved to a different chromosome, the two donor loci reside in intrinsically distinct chromosomal domains that control accessibility of the loci in a cell type-specific manner (Weiler and Broach 1992). This observation can be appreciated in the context of recent results from Wu and Haber (1995), who suggested that the mechanism underlying cell-specific donor preference involves enhanced recombinogenicity of the left arm of chromosome III in a cells versus α cells. In particular, they observed that the frequency of mitotic recombination between heteroalleles of LEU2, where one allele was inserted at HML and the other at MAT, was 20- to 30-fold higher in a cells than in α cells. They also observed that HML remained the preferred donor in a cells when situated at several locations within 40 kb of the left arm of chromosome III but not when positioned further to the right. Thus, Wu and Haber suggested that enhanced mitotic recombination potential of the left arm of chromosome III in a cells renders HML the preferred donor, whereas this enhanced recombination potential is lost in α cells. In the absence of activated recombination, HMR is the default donor for as yet undefined reasons. Recently, Wu and Haber (1996) identified a small region on the left arm of chromosome III significantly removed from MAT or either donor loci that is required to activate enhanced recombination in a cells. They have designated this region a recombination enhancer.
In this report we present results further defining the cis-acting region on chromosome III responsible for cell type-specific selection of donor locus during mating type interconversion. We confirmed the existence of the recombinational enhancer on chromosome III and demonstrated that the activity of this enhancer is dependent on two α2p/Mcm1p-binding sites within this region. We show further that these sites regulate expression of several a-specific RNA transcripts from this domain. Because deletion of the genomic region encoding one of these RNAs does not alter donor preference, transcriptional activation—rather than any particular product of transcription—may be sufficient to stimulate recombination. These results suggest that yeast use a novel mechanism to activate one chomosome arm for donor accessibility during mating type interconversion.
Results
A cis-acting site-specifying donor preference resides on the left arm of chromosome III
Because a yeast cell selects one mating type donor locus in preference to the other as a function of cell type, the cell is capable of distinguishing HML from HMR. Because our previous studies indicated that features of chromosome III used by the cell to distinguish HML from HMR during mating type interconversion are well removed from either locus, we developed a method to systematically survey chromosome III to identify such cis-acting sites. We created a series of inversions within chromosome III, by the procedure described in Materials and Methods, and then tested strains carrying these inversions for donor preference during normal mating type interconversion. Assuming, for example, that a site on the left arm of chromosome III dictates selection of HML in a cells, then an inversion with a breakpoint between HML and this site would yield a strain in which the site is now positioned near HMR rather than HML. Accordingly, in a cells carrying this chromosome, HMR would become the preferred donor. In contrast, if the breakpoint leaves the site associated with HML, then HML would remain the preferred donor in a cells carrying the inverted chromosome.
Strains carrying different chromosome III inversions were tested for donor preference in both a and α cells, as described in Materials and Methods, and a summary of this analysis is provided in Figure 2 adjacent to a schematic of each chromosome tested. In the first series of experiments, we held the breakpoint within the right arm of chromosome III constant and varied the position of the breakpoint within the left arm of chromosome III. As evident from these results, proper donor preference in a cells depends on a region located between 24 and 60 kb on the left arm. That is, the preferred donor in a cells is always the locus located immediately distal to the 24- to 60-kb domain. A parallel but reciprocal correlation is evident for donor preference in α cells: The preferred donor locus in α cells is the one removed from the 24- to 60-kb domain. The most straightforward interpretation of these results, in light of the observations from Wu and Haber (1996), is that a single cis-acting site resides between 24 and 60 kb on the left arm of chromosome III and that this site activates the region around it for recombination in a cells but not in α cells. Data presented below are consistent with this hypothesis.
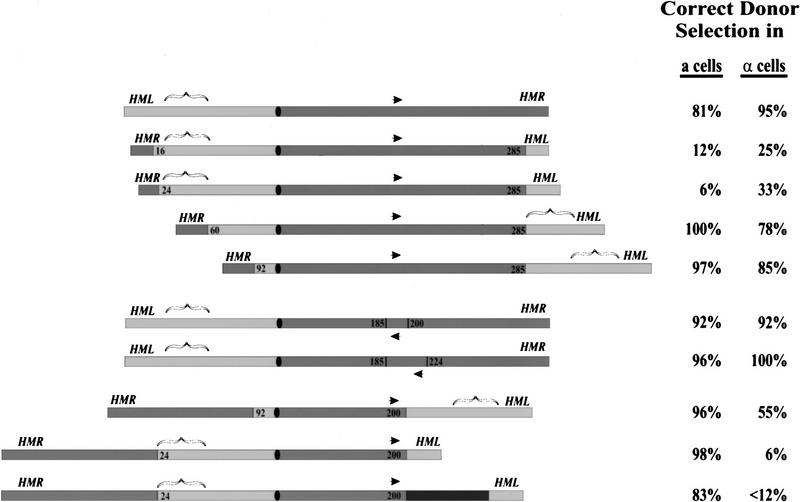
Figure 2.
Chromosome inversions reveal a cis-acting site on the left arm of chromosome III required for donor preference. (Left) Diagram of the structure of chromosome III in selected inversion strains. The location of the donor loci, HML and HMR, are indicated, as is the position of the centromere (filled oval) and the MAT locus (arrow). Sequences from the left arm are shaded light gray, and those from the right are shaded dark gray. The positions of the inversion endpoint (in kb from the left arm of the chromosome) are designated on each inversion chromosome. The bracket denotes the region, determined from this analysis, that apparently dictates donor preference. The black segment in the last construct corresponds to 25 kb of Caulobacter cresentus DNA inserted at the inversion breakpoint. (Right) The proportion of switching events in which the cell selected the appropriate locus (HML for a strains, HMR for α strains), determined as described in Materials and Methods. Each value represents analysis of at least 25 individual spore clones. Switching events in which the correct donor locus was not selected include those in which the opposite donor was preferentially selected and those in which no preferential donor locus could be determined.
The additional chromosome III rearrangements are in agreement with the above results, although they indicate the existence of additional features of the system. In a second set of experiments, we examined the effect of inverting the MAT locus. As evident in Figure 2, neither a local inversion of MAT nor a larger pericentric inversion including MAT alters donor preference in a cells. This is consistent with the model proposed above and with previous results from our laboratory indicating that MAT did not serve as a reference point for donor selection (Weiler and Broach 1992). The reduced efficiency of donor selection in α cells carrying the pericentric inversion and the selection of HML in both cell types in the last two constructs suggests that additional features besides the site between 24 and 60 kb contribute to donor selection. The implications of these results on donor selection are explored in the Discussion.
Two α2p/Mcm1p-binding sites reside in the donor preference locus on chromosome III
As noted in the introductory section, cell type-specific donor preference is established by the activity of α2p in conjunction with Mcm1p. Accordingly, we examined the 24- to 60-kb region of chromosome III for the presence of sites with homology to known α2p/Mcm1p-binding sites. We found that two such sites reside in this interval, centered at positions 29,209 and 30,595, both of which were noteworthy for several reasons. First, unlike the six known α2p/Mcm1p sites characterized previously (Zhong and Vershon 1997), neither of these two sites lies immediately upstream of an open coding region (cf. Figure 5, below). Second, both of these sites conform to the consensus α2p/Mcm1p-binding motif, deduced from the six previously characterized sites, better than any other sequence within the yeast genome. Finally, these two sites lie in or near the region defined by Wu and Haber (1996) to be involved in donor preference. Thus, these two sites resemble bona fide α2p/Mcm1p-binding sites, are not associated with expression of a protein product, and reside in a region required for donor preference.
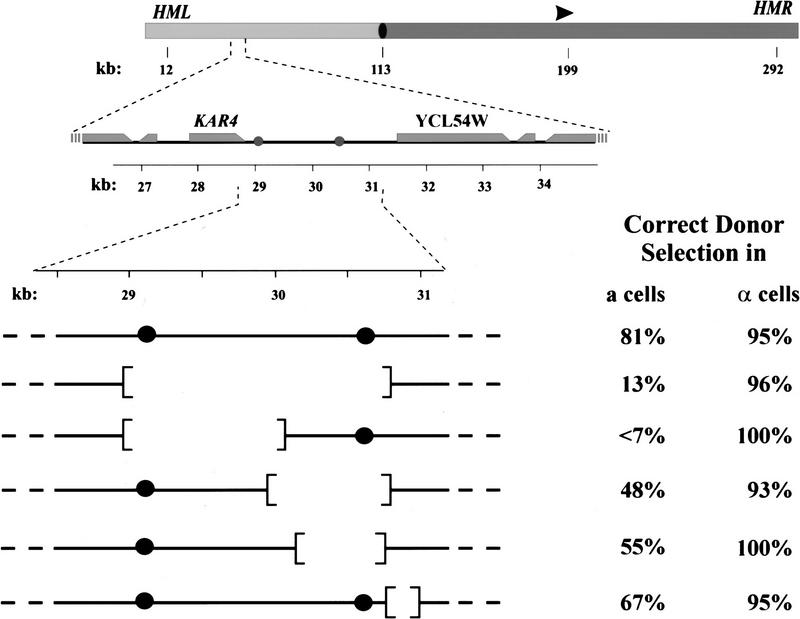
Figure 5.
α2p/Mcm1p-binding sites on the left arm of chromosome III dictate donor preference in a cells. (Top) A diagram of chromosome III is shown which indicates the positions of HML, HMR, MAT (arrowhead), the centromere (filled oval), the left arm (light gray) and the right arm (dark gray). The positions of the three mating type loci and the centromere are specified (in kb from the left end of the chromosome). (Middle) The region from 26 to 35 kb of chromosome III is expanded to show the positions of the two α2p/Mcm1p-binding sites (shaded circles) and the adjacent open reading frames (shaded rectangles, taper at the 3′ end of the reading frame). (Bottom) The segment of chromosome III from 29.5 to 31 kb present in the wild-type (first line) and in several deletion strains, indicating the locations of the two α2p/Mcm1p sites (•) and the regions deleted in each strain. To the right of each chromosome segment is indicated the proportion of switching events in which the cell selected the appropriate locus (HML for a strains; HMR for α strains), determined as described in Materials and Methods.
Most α2p/Mcm1p-binding sites are present upstream (∼200 bp) of the transcriptional start site of a-specific genes. In the absence of α2p, Mcm1p binds to these sites and functions as a transcriptional activator. In the presence of α2p, α2p and Mcm1p bind cooperatively to the site and repress transcription. To test whether the sequences at 29.2 and 30.6 kb are authentic α2p/Mcm1p-binding sites, we performed several in vitro and in vivo binding experiments. For in vitro binding assays we synthesized and cloned fragments corresponding to the region from 29,194 to 29,229 (designated DPS1) and to the region from 30,578 to 30,613 (designated DPS2). Each of the cloned fragments was isolated, labeled, and incubated with varying amounts of purified intact α2p and constant amount of the purified amino terminal domain (amino acids 1–96) of Mcm1p (Mead et al. 1996). The proportion of fragment bound by the proteins was then determined by electrophoresis on nondenaturing polyacrylamide gels. As a control, we also tested binding of the equivalent DNA fragment corresponding to the α2p/Mcm1p-binding site from the STE6 promoter, a known α2p/Mcm1p-binding site. The results of this analysis, presented in Figure 3, indicate that α2p and Mcm1p bind individually to both DPS1 and DPS2 with the same relative affinities as they show for the STE6 operator sequence. In addition, they exhibit the same cooperativity in binding to both DPS1 and DPS2 as they do with the STE6 operator.
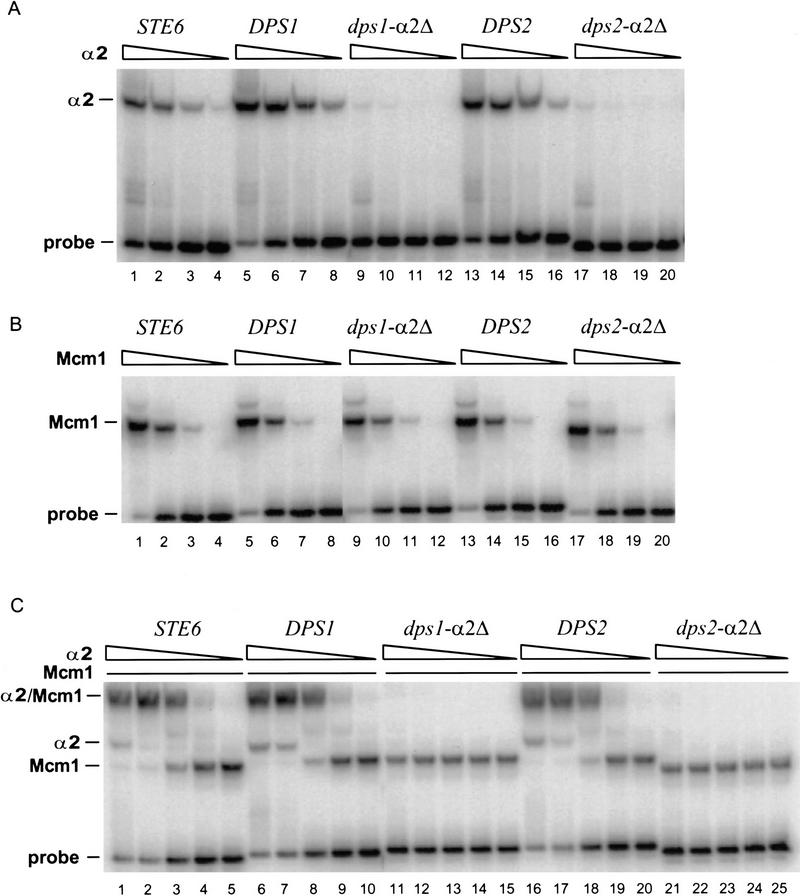
Figure 3.
DPS1 and DPS2 bind α2p and Mcm1p in vitro. (A) Binding of α2p to the indicated α2p/Mcm1p sites (sequences are shown in Table 1) as determined by EMSAs (described in Materials and Methods). Purified α2p was added to the binding reactions at 4.0 × 10−7 m (lanes 1,5,9,13,17), 8.0 × 10−8 m (lanes 2,6,10,14,18), 1.6 × m 10−8 m (lanes 3,7,11,15,19), or 3.2 × 10−9 m (lanes 4,8,12,16,20). (B) Binding of Mcm1p to the indicated site. Purified Mcm1p1-96 fragment was added to the binding reactions at 1.7 × 10−8 m (lanes 1,5,9,13,17), 1.7 × 10−9 m (lanes 2,6,10,14,18), 1.7 × 10−10 m (lanes 3,7,11,15,19), or 1.7 × 10−11 m (lanes 4,8,12,16,20). (C) Binding of α2p and Mcm1p to the indicated site. Pruified α2p was added to the binding reactions at 8.0 × 10−8 m (lanes 1,6,11,16,21), 1.6 × 10−8 m (lanes 2,7,12,17,22), 3.2 × 10−9 m (lanes 3,8,13, 18,23), 6.4 × 10−10 m (lanes 4,9,14,19,24), or 1.3 × 10−10 m (lanes 5,10,15,20,25).
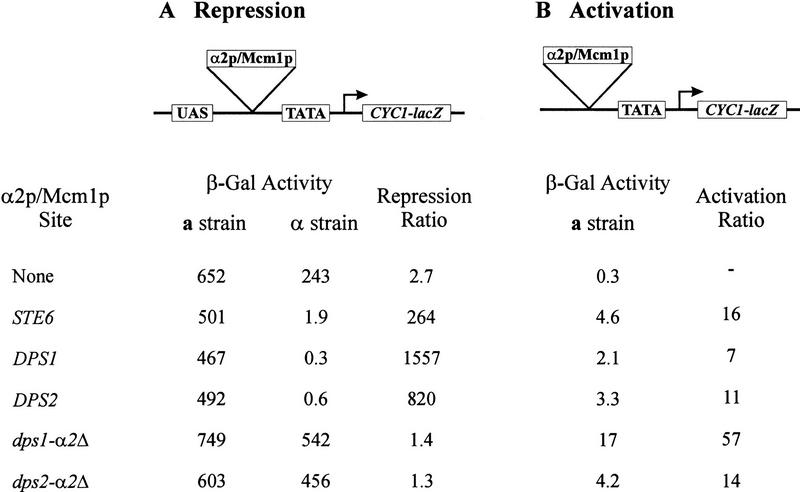
To confirm that the binding affinities exhibited by DPS1 and DPS2 were physiologically relevant, we examined whether these sites would exert transcriptional repression in vivo in the presence of α2p and promote transcriptional activation in the absence of α2p. To test for α2p-mediated repression activity, we inserted DPS1 or DPS2 between the upstream activating sequence CYC1 (USACYC1) and the TATA element upstream of a CYC1–lacZ reporter gene (Acton et al. 1997). The plasmid constructs were then transformed into isogenic a and α strains, and the expression of the reporter gene determined by assaying β-galactosidase in individual transformants. The extent of repression was then calculated as the ratio of expression in the α strains carrying the test plasmid to that from the a strain carrying the same plasmid. As evident from the results presented in Figure 4, both DPS1 and DPS2 are as effective in exerting α2p-dependent repression as is the operator from STE6. We also tested whether these sites would exhibit transcriptional activation in the absence of α2p. Accordingly, we deleted the UASCYC1 from each of the plasmids used in the repression assay and transformed the resulting plasmids into an a strain. The level of β-galactosidase elicited by the test plasmid compared to that of a comparable plasmid lacking any UAS elements provided an indication of the transcriptional activation capacity of the α2p/Mcm1p sites. As shown in Figure 4, both DPS1 and DPS2 possess weak UAS activity, but the level of activation is comparable to that of the STE6 operator. Thus, we conclude that DPS1 and DPS2 are capable of binding Mcm1p and α2p with affinities comparable to other α2p/Mcm1p-binding sites in the cell and that they are capable of exerting Mcm1p-mediated transcriptional activation and α2p/Mcm1p-mediated repression in vivo.
Figure 4.
DPS1 and DPS2 can mediate cell type-specific transcriptional activation and repression in vivo. The α2p/Mcm1p binding sites listed on the left (sequences shown in Table 1) were inserted into the assay vectors indicated at the top and tested for repression activity (A) or activation activity (B), as described in Materials and Methods. (A) β-Galactosidase levels were determined on three independent transformants of the indicated plasmid in strain EG123 (a strain) and strain 246.1.1 (α strain). Values are the average of the three determinations. Repression ratio is the activity obtained in the a strain containing the indicated plasmid divided by the activity obtained in the α strain containing the same plasmid. (B) β-Galactosidase levels were determined on three independent transformants of the indicated plasmid in strain EG123 (a strain). Activation ratio was calculated by dividing the β-galactosidase activity obtained with the plasmid containing the indicated site by that obtained with the plasmid lacking any site.
DPS1 and DPS2 regulate donor selection
To test whether DPS1 and DPS2 affect donor preference, we created strains with various deletions spanning these sites and then examined donor preference, during normal switching events in strains carrying these deletions. The results of this analysis are summarized in Figure 5. The largest deletion examined removed both α2p/Mcm1p-binding sites while retaining the open coding regions flanking the sites. Haploid strains carrying this deletion were viable, indicating that no essential functions resided within the deleted sequences. This deletion had a dramatic effect on donor preference: α cells carrying the deletion exhibited completely normal donor preference—selecting HMR in 95% of the switching events—but a cells carrying the deletion exhibited inverted preference, selecting HMR rather than HML in most of the switching events. Further analysis indicated that both sites contributed to normal switching patterns. A deletion encompassing DPS1 while retaining DPS2 yielded a defect in switching in a cells equivalent to that seen with strains carrying the deletion removing both sites. A deletion spanning DPS2 also reduced efficiency of donor selection in a cells, although not as dramatically as did deletion of the region spanning DPS1. Thus, the region encompassing two α2p/Mcm1p-binding sites is required for proper donor preference in a cells, although deletion of this region does not affect switching in α cells.
Given that α2p is the principal trans-acting determinant of donor preference, deletion of the region encompassing the α2p/Mcm1p-binding sites DPS1 and DPS2 had an unexpected effect on donor preference. That is, deletions spanning this region reversed donor preference in a cells, in which α2p was not present, but had no effect on donor preference in α cells, in which α2p was present. One explanation for this apparent conundrum is that these sites play an active role in promoting selection of HML in a cells and that this function is repressed by the binding of α2p to the sites. This model is consistent with our observation (Szeto and Broach 1997) that donor selection in the presence of α2p—a preference for HMR over HML—is the same as that obtained by deletion of the sites.
To test this hypothesis, we created several specific alleles of DPS1 and DPS2 and then tested their effect on donor preference in a and α cells. We first precisely deleted either or both α2p/Mcm1p-binding sites (cf. Table 1) and then examined donor preference in the resulting three strains. As evident from Table 2, deletion of DPS1 alone had a small but significant effect on donor preference in a cells and a minor effect on donor preference in α cells. Deletion of DPS2 had no detectable effect on donor preference in either cell type. Deletion of both DPS1 and DPS2, leaving the remainder of this region intact, rendered donor preference completely random in a cells while causing little or no effect on donor preference in α cells. Thus, we conclude that the two α2p/Mcm1p sites act redundantly to promote selection of HML in a cells. We note that deletion of only DPS1 and DPS2 does not yield as severe a donor preference phenotype as does deletion of the entire region spanning the two sites. This suggests that the recombination enhancer consists of more than just these two sites, a hypothesis elaborated in the Discussion.
Table 1.
α2p/Mcm1p sites and plasmids used in this study
Uppercase letters represent the sequences of the genomic α2p/Mcm1p sites from the loci listed on the left that were cloned into the plasmids listed on the right to test for transcriptional repression (repress) and transcriptional activation (activate activity), results from which are presented in Fig. 4. Lowercase letters represent nucleotides that were added to facilitate cloning into the plasmid. Underlined sequence represents the Mcm1p-binding domain at each site; the italicized sequence represents the α2p-binding domains. The sequences of the mutant alleles of DPS1 and DPS2 are also shown, indicating the altered (bold) or deleted (space) base pairs. The mutation on the left side of dps1 created a ClaI restriction site; the mutation on the right side of dps1 created a BstBI restriction site; the mutation on the left side of dps2 created a PvuII restriction site; the mutation on the right side of dps2 created an EagI site. These restriction sites were used to verify the presence of the mutant alleles in every spore clone used in donor preference analysis.
Table 2.
α2p/Mcm1 sites mediate donor preference
| Strain
|
Allelea
|
Donor selectionb
|
|||
|---|---|---|---|---|---|
| in a cells
|
in α cells
|
||||
|
HML
|
HMR
|
HMR
|
HML
|
||
| Y2201 | wild type | 81% (152) | 13% (24) | 95% (239) | 2% (6) |
| Y2388 | dps1Δ | 68% (13) | 26% (5) | 84% (16) | 11% (2) |
| Y2389 | dps2Δ | 86% (18) | 10% (2) | 100% (28) | 0% (0) |
| Y2390 | dps1Δ dps2Δ | 30% (7) | 26% (6) | 75% (46) | 7% (4) |
| Y2391 | dps1–α2Δ | 95% (21) | 0% (0) | 70% (28) | 23% (9) |
| Y2392 | dps2–α2Δ | 87% (26) | 3% (1) | 90% (18) | 5% (1) |
| Y2393 | dps1–α2Δ dps2–α2Δ | 79% (30) | 5% (2) | 23% (9) | 55% (22) |
| Y2394 | dps1Δ–dps2Δ [DPS1 DPS2] | 40% (4) | 50% (5) | 61% (17) | 4% (1) |
(dps1Δ) Substitution of the sequence GGATCC for the 31 bp of chromosome III from position 29,194 to 29,224; (dps2Δ) substitution of the sequence GGATCC for the 31 bp of chromosome III from position 30,580 to 30,610; (dps1–α2Δ, dps2–α2Δ) sequences shown in Table 1. Strain Y2394 consists of strain Y2390 containing a centromeric plasmid carrying the 5.33-kb BamHI fragment spanning DPS1 and DPS2.
Donor preference was determined as described in Materials and Methods by examining the predominant allele at MAT in colonies derived from individual spores of the indicated cell type with the designated inversion obtained following dissection of the indicated strain. Numbers in parentheses are the absolute number of spore clones that had selected the designated donor during mating type interconversion. Only those colonies that gave a single predominant MAT allele are listed. The remainder for each strain represents colonies in which both donor alleles were represented essentially equally in the spore clone.
In a second series of experiments, we examined the effects on donor preference of alleles of DPS1 and DPS2 in which we had ablated both α2p-binding regions at each locus while leaving the Mcm1p-binding region intact. As a control for these experiments, we first tested these alleles for the ability to bind Mcm1p and α2p in vitro and promote transcriptional activation and repression in vivo. As evident from the gel shift assays shown in Figure 3, both sites were capable of binding Mcm1p with the same affinity as that of the corresponding wild-type allele. However, both mutant sites exhibited at least a 100-fold reduction in their ability to bind α2p. These results were confirmed in the in vivo transcriptional assay. As shown in Figure 4, both mutant alleles promoted transcriptional activation of the CYC1 reporter with the same or better efficiency than did the corresponding wild-type alleles or the cognate site from STE6. However, neither of the alleles exhibited any α-specific repression activity. Thus, both mutant alleles retained their ability to promote transcriptional activation but could not exert α-specific transcriptional repression.
The donor preference during mating type interconversion of strains carrying either or both of these mutant alleles is presented in Table 2. Introduction of either mutant allele alone had no appreciable effect on donor preference in either a cells or α cells. However, strains carrying these mutant alleles at both sites exhibited normal donor preference in a cells but now showed an inverted donor selection in α cells. This suggests that α2p binding to either site restricts selection of HML. This result is the reciprocal of that observed above for strains in which the sites are completely deleted. From these mutant studies, we conclude that DPS1 and DPS2 play an active role in donor selection in a cells and that binding of α2p renders these sites inactive for donor selection.
DPS1 and DPS2 regulate a-specific transcripts
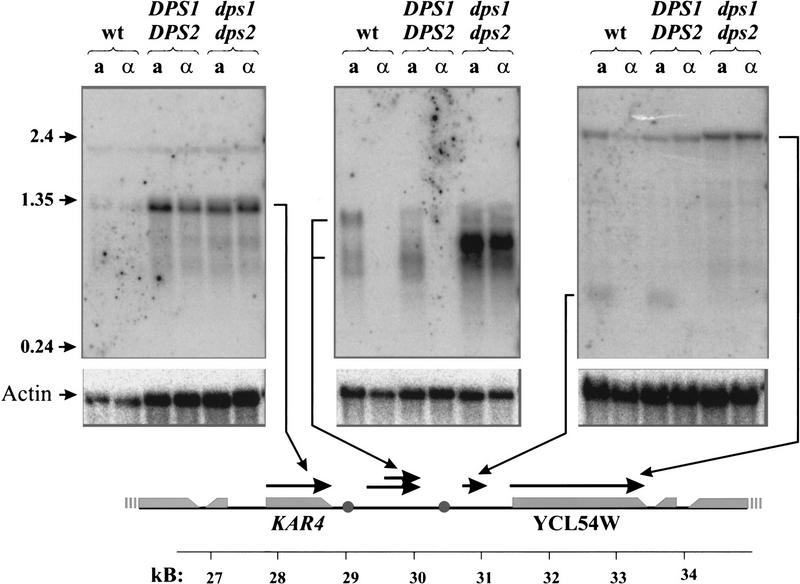
Given the strict correlation in the structural requirements of α2p/Mcm1 for both donor preference and transcriptional repression, we examined whether DPS1 or DPS2 regulates transcription within the region of the recombination enhancer. We isolated total RNA from two different pairs of isogenic a and α strains containing an intact recombination enhancer and from a and α strains deleted specifically for DPS1 and DPS2. Gel-fractionated RNA was probed with DNA corresponding to different regions across the recombination enhancer domain, the results of which are presented in Figure 6. As evident from these results, the two genes flanking the enhancer region, KAR4 and YCL54W, are transcribed at essentially equal levels in both a and α cells. However, several transcripts that derive from the enhancer region exhibit an a-cell-specific pattern of expression. The orientation and initiation sites of these transcripts were determined by RNase protection and primer extension assays (data not shown) and are indicated on the schematic diagram at the bottom of Figure 6. Transcription of a 0.3-kb species initiates at a site 250 bp to the right of DPS2, occurs only in a cells, and is dependent on the presence of DPS1 and/or DPS2. Transcription from the region lying between DPS1 and DPS2 is somewhat more complex. a-Specific trancripts ranging in size from 0.3 to 1.1 kb that derive from this region are evident in two different strains, although the relative amounts of the different species differs in two different strains. Because strains carrying deletions of DPS1 and DPS2 contain transcripts over this region not seen in DPS1+DPS2+ strains, we cannot readily determine whether the a-specific transcripts are dependent on these two sites. None of the a-specific transcripts encompasses an extended open coding region, and all of these transcripts are at low abundance: By quantitative RNase protection assays, none of the transcipts has a steady-state level higher than that of the α2 mRNA (data not shown).
Figure 6.
Transcription within the recombination enhancer. Samples (20μg) of total RNA isolated from the indicated strains were fractionated on formaldehyde–agarose gels, transferred to nylon membranes, and hybridized with 32P-labeled probes corresponding to positions 26,740–30,145 (left), 29,963–30,803 (middle),or 30,803–31,955 (right) of chromosome III. Orientation and precise positions of RNA species homologous to these fragments were determined by RNase protection assays and are indicated at the bottom on a diagram of a 26- to 35-kb segment of chromosome III [(shaded circles) DPS1 and DPS2; (shaded rectangles) open coding regions, taper at the 3′ end of the reading frame]. (Bottom panels) Hybridization filters were stripped and reprobed with DNA specific for actin mRNA. Strains: (wt) 14a (ahis1) and 17α (αhis1); DPS1 DPS2, a and α spore clones of a ho::URA3 derivative of strain Y2201; dps1 dps2, a and α spore clones of an ho::URA3 derivative of strain Y2390.
The recombination enhancer acts in cis
The fact that specific RNA species are transcribed from the recombination enhancer raises the possibility that these species could be diffusible elements of the donor preference machinery. Accordingly, we asked whether specific mutations of the recombination enhancer could be complemented in trans. As noted in Table 2, a cells deleted specifically for DPS1 and DPS2 select a donor locus during mating type interconversion essentially randomly. In addition, these cells show reduced expression of at least one of the a-specific RNA species transcribed from this region. We introduced into this strain a centromeric plasmid carrying a fragment spanning DPS1, DPS2, and the two a-specific RNA-transcribed regions. As evident from the data in Table 2, the presence of the plasmid did not correct the donor preference defect resulting from deletion of the two α2p/Mcm1p sites. Thus, if any of these two RNA species are involved in donor selection, they apparently cannot be provided in trans.
Discussion
The nature of the donor preference locus
We have shown in this study that two α2p/Mcm1p-binding sites play critical roles in the function of the recombination enhancer on chromosome III responsible for donor preference during mating type interconversion. Specific deletion of both of these sites renders donor preference in a cells completely random while having no effect on donor preference in α cells. In contrast, inactivation of the α2p-binding domains from these loci, which alleviates α2p-mediated repression while retaining the transcriptional activation potential of the sites, has the opposite effect: Donor preference is normal in a cells but completely inverted in α cells. These results indicate that in the absence of α2p these two sites redundantly play a positive role in promoting enhanced recombination. The presence of α2p abrogates this enhanced recombination potential. Because α2p had no effect on the frequency of selecting HMR in α cells carrying the enhancer near HMR only (data not shown), we can conclude that α2p bound to the enhancer does not promulgate a negative recombination potential. Rather, the enhancer elevates the recombination potential of the surrounding chromosomal domain when α2p is absent but does not affect the intrinsic recombination potential of a region when α2p is present.
Although the α2p/Mcm1p-binding sites are necessary for proper donor selection, they are not sufficient. In molecular dissections of the enhancer region performed by Wu and Haber (1996), a segment just encompassing DPS1 was insufficient to impart donor preference activity. Rather, reasonable activity required not only DPS1 but also the centromere-proximal 500-bp region adjacent to DPS1. In addition, we find that precise deletion of both DPS1 and DPS2 does not yield as severe a phenotype as does deletion of the entire domain spanning both DPS1 and DPS2: The former deletion yields random selection of donor sites while the latter deletion yields a complete bias for HMR as donor in a cells. From these observations, we conclude that the donor preference locus, or recombination enhancer, consists of two elements: a cell type-regulated transcriptional enhancer (which can be provided either by DPS1 or DPS2), and a second element of unknown function. The fact that deletion of the entire locus was a more severe phenotype than deletion of DPS1 and DPS2 alone suggests either that this second element has intrinsic activity in the absence of a transcriptional enhancer or that some other transcriptional activation element resides in this region. As discussed below, this second element could be the coding domain for a cis-acting RNA or, more likely, the entry site for a recombination-promoting complex capable of diffusion along the chromosome.
Why is HMR the default donor locus?
Although the presence of a cell type-specific recombination enhancer on the left arm of chromosome III accounts for preferential selection of HML in a strains, the reason for the preference for HMR in α strains is not as clear. As noted above, because deleting the recombination enhancer or placing it near HMR has no effect on donor preference in α cells (data not shown), we conclude that the enhancer does not act to repress selection of HML in α cells. This would suggest that in the absence of activation of HML for recombination, HMR is intrinsically more recombinogenic during mating type interconversion than is HML. This intrinsically higher recombinational potential of HMR could be attributable to the local chromosomal context of HMR or to its position relative to MAT. Because previous studies have documented that HMR is the preferred donor in α cells even when MAT resides on a separate chromosome, local context plays a role in selection of HMR (Weiler and Broach 1992; Wu et al. 1996). However, as noted from the data in this study, the position of HMR relative to MAT also contributes to the intrinsic donor preference in α cells. In the normal chromosomal context, MAT is twice as far from HML as from HMR. In strains carrying a chromosome III inversion in which this ratio is inverted, donor preference in α cells becomes random (Fig. 2, line 8), and in strains carrying an inversion chromosome that places MAT much nearer HML than HMR, HML is the preferred donor in α cells (Fig. 2, line 9). In these inversions, the 100-kb chromosomal context in which HMR is embedded remains unchanged from that in wild-type cells, indicating that local context does not override relative position. Thus, we conclude that both relative position and local context collaborate to establish HMR as the preferred donor in the absence of activation of the recombination enhancer.
Models for the role of α2p in donor preference
We can envision two classes of models that account for the role in donor preference of α2p and the two α2p/Mcm1p-binding sites within the recombination enhancer on chromosome III. These are diagrammed on Figure 7. The first model suggests that DPS1 and DPS2 regulate transcription of one or more RNAs that act in cis, spreading outward along the chromosome from their site of synthesis to enhance the recombinational potential of DNA with which they are associated. How such an RNA or RNAs would promote recombination is not known, although RNA as a recombination enhancer is not unprecedented (Kotani et al. 1996; Yoon et al. 1996). Several precedents exist for cis-acting RNAs that profoundly affect the behavior of the associated chromosome. Xist RNA, transcribed from the XIC locus on the inactive X chromosome, coats the inactive X chromosome and is required for its inactivation (Brown et al. 1992; Rastan 1994; Penny et al. 1996). Similarly, genes encoding structural RNAs of unknown functional reside within all known regions of mammalian genomes known to be subject to parental imprinting, circumstantially implicating these RNAs in chromosomal imprinting. An attractive feature of this model is that it readily accounts for the strict correlation between the roles of α2p in transcriptional regulation and donor preference.
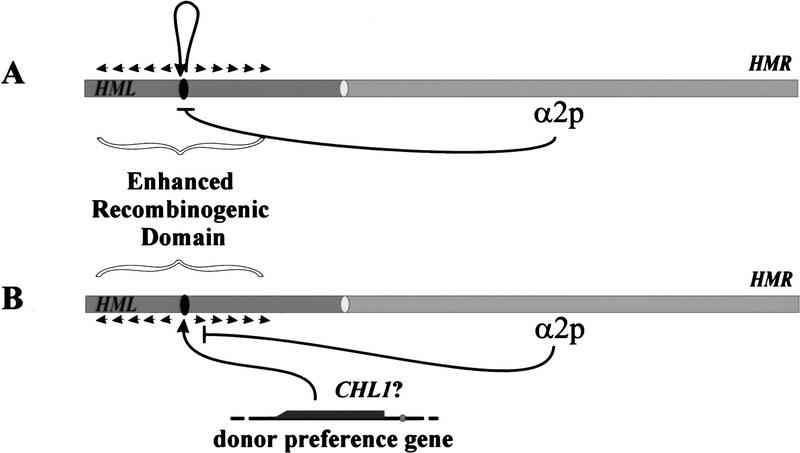
Figure 7.
Two models for the role of α2p in donor preference. (A) Cis-acting RNAs. One or more RNA species from the recombination enhancer domain expressed in a cells associates with the left arm of chromosome III to promote enhancer recombination potential of the underlying DNA. Repression of expression of these RNAs by α2p abrogates the enhanced recombination potential of the left arm in α cells. (B) Loading recombination proteins. In a cells, the recombination enhancer provides a loading site for one or more proteins, possibly Chl1p, that spread outward along the left arm of chromosome III and promote enhanced recombination. Alteration of the chromatin structure of the enhancer domain by α2p precludes access of the recombination proteins to the loading site to preclude enhanced recombination in α cells.
The second model postulates that the recombination enhancer serves as an entry site for loading a protein or complex onto chromosome III, which then spreads outward along the chromosome from the site to promote enhanced recombination in the region surrounding the site. One candidate for such a protein is the product of the CHL1 locus. Deletion of this gene renders donor preference random in a cells but has no effect on donor preference in α cells (Weiler et al. 1995). The function of the Chl1p in donor preference is unknown, but the protein exhibits significant homology to Rad3p, a DNA helicase (Gerring et al. 1990). Thus, we might postulate that Chl1p would enhance recombination by promoting strand exchange when present on the chromatin in the vicinity of a donor locus.
In this model we presume that the entry site for the recombination complex would be the second element of the recombination enhancer discussed above. Accessibility of this site would be dependent of the transcriptional activation potential of DPS1 and DPS2: In a cells DPS1 and DPS2 would function as activators to render the site accessible to the complex, and in α cells DPS1 and DPS2 would function as repressors to make the site refractory. Transcription of the locus in a cells could be required for opening the chromatin to allow entry of the complex. Alternatively, such transcription could simply be coincidental, because of the fact that activating the chromatin structure for entry of the complex could render the local region accessible to the basal transcriptional apparatus. In either event, binding of α2p to DPS1 or DPS2 would preclude entry of the complex by repressing transcription or by converting the local chromatin structure into a “closed” configuration. In either case, this would likely involve a Tup1p/Ssn6p-mediated organization of phased nucleosomes emanating outward from DPS1 and DPS2 (Cooper et al. 1994; Szeto and Broach 1997).
Correlation between selective recombination and transcriptional activation has also been noted in hematopoietic lineages. In heavy-chain class switching in B cells, transcription occurs in the 5′ region of one of the heavy-chain constant domains prior to selection of that particular domain for recombination. Ultimately, the transcriptionally activated constant domain is joined to the previously matured variable domain to generate a new heavy chain (Yancopoulos et al. 1986; Jung et al. 1993; Zhang et al. 1993). Thus, transcription marks the prospective recombination site. Similarly, the recombination machinery—Rag1, Rag2, Ku, and so forth—responsible for rearrangements of immunoglobulin genes in B-cell lineages is the same as that required for rearrangement of T-cell receptor genes in T-cell lineages. Nonetheless, only immunoglobulin genes are rearranged in B cells and only T-cell receptor genes are rearranged in T cells. In both cell types, transcription occurs in the region subject to recombination but not in the region repressed for recombination (Stanhope-Baker et al. 1996). In neither of these cases is the functional relationship between transcription and recombination known: Transcription may serve to activate the locus or domain for the recombination event or it may be a by-product of “opening” the chromatin over this region to allow recombination. The experimental accessibility of the yeast donor preference locus may allow resolution of the nature of the correlation in at least this one case.
Materials and methods
Strains and plasmids
All of the strains used in this study were derived from strain LS255, a ciro derivative (Rose and Broach 1990) of strain Y2201 (Szeto and Broach 1997) and are listed in Table 3. Inversions were induced using the Flp1p recombinase and Flp1p recognition target (FRT) sites from the yeast 2μ circle plasmid. Plasmids pKSW42 and pKSW43 were used to introduce FRT sites in inverted orientation into chromosome III at the endpoints of the desired inversions. Plasmid pKSW42 is a derivative of pUC18 containing a 674-bp HindIII–BamHI spanning FRT and a 1106-bp HindIII–AvaI frament containing the URA3 gene. Plamid pKSW43 is a derivative of pUC18 with the same FRT site and a 1764-bpBamHI fragment carrying the HIS3 gene. Approximately 1-kb DNA segments corresponding to the endpoints of the desired inversion were obtained by PCR amplification and cloned separately into plamsids pKSW32 and pKSW43. The resulting plasmids were digested with a restriction enzyme unique to the inserted chromosomal III segment, and both transformed into strain LS255. Resulting transformants bearing the correct insertions as determined by Southern hybridization were then transformed with YEp51–GAL–FLP1 (Rose and Broach 1990), and individual isolates were cultured briefly in galactose-containing medium and plated on YEPD (2% yeast extract, 1% Bacto-peptone, 2% glucose). Isolates that had lost the YEp51–GAL–FLP1 plamid were screened by Southern hybridization for those that carried the inverted chromosome, and isolates with the correct structure were retained. Strains Y2381 and Y2382 were derived from strains Y2379 and Y2380 by transforming them to leucine prototrophy with plasmid pLS41, targeted by homologous recombination to the FRT site at the inversion breakpoint. Plasmid pLS41 consists of the cosmid vector pLAFR1 carrying the yeast LEU2 and FRT genes and 25-kb Caulobacter crescentus DNA.
Table 3.
Strains used in this study
| Strain
|
Genotypea
|
|---|---|
| LS255 | hmlα1α2Δ–inc/hmlα1α2Δ–inc MATa/MATαhmra1Δ101–inc/hmra1Δ101–inc HO/HO leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 ura3-52/ura3-52 trp1-289/trp1-289 |
| Y2365 | LS255, MATa/MATα–inv(16–285 kb) |
| Y2366 | LS255, MATa–inv(16–285 kb)/MATα |
| Y2367 | LS255, MATa/MATα–inv(24–285 kb) |
| Y2368 | LS255, MATa–inv(24–285 kb)/MATα |
| Y2369 | LS255, MATa/MATα–inv(60–285 kb) |
| Y2370 | LS255, MATa–inv(60–285 kb)/MATα |
| Y2371 | LS255, MATa/MATα–inv(92–285 kb) |
| Y2372 | LS255, MATa–inv(92–285 kb)/MATα |
| Y2373 | LS255, MATa/MATα–inv(185–200 kb) |
| Y2374 | LS255, MATa–inv(185–200 kb)/MATα |
| Y2375 | LS255, MATa/MATα–inv(185–224 kb) |
| Y2376 | LS255, MATa–inv(185–224 kb)/MATα |
| Y2377 | LS255, MATa/MATα–inv(92–200 kb) |
| Y2378 | LS255, MATa–inv(92–200 kb)/MATα |
| Y2379 | LS255, MATa/MATα–inv(24–200 kb) |
| Y2380 | LS255, MATa–inv(24–200 kb)/MATα |
| Y2381 | LS255, MATa/MATα–inv(24–200 kb)∷pLS41 |
| Y2382 | LS255, MATa–inv(24–200 kb)∷pLS41/MATα |
| Y2383 | LS255, (dps1 dps2)Δ28961–30803/DPS1 DPS2 |
| Y2384 | LS255, dps1Δ28961–30183/DPS1 |
| Y2385 | LS255, dps2Δ29963–30803/DPS2 |
| Y2386 | LS255, dps2Δ30145–30739/DPS2 |
| Y2387 | LS255, DPS2Δ30739–30997/DPS2 |
| Y2388 | LS255, dps1Δ/DPS1 |
| Y2389 | LS255, dps2Δ/DPS2 |
| Y2390 | LS255, dps1Δ dps2Δ/DPS1 DPS2 |
| Y2391 | LS255, dps1-α2Δ/DPS1 |
| Y2392 | LS255, dps2-α2Δ/DPS2 |
| Y2393 | LS255, dps1-α2Δ dps2-α2Δ/DPS1 DPS2 |
| Y2394 | LS255, dps1Δ dps2Δ/DPS1 DPS2 [YCp–DPS1–DPS2] |
| 246.1.1 | MATα trp1 leu2 ura3 his4 |
| EG123 | MATa trp1 leu2 ura3 his4 |
| GY12 | MATa his4-917 lys2-128 leu2 ura3-52 nrp2-1 |
Strains Y2365–Y2382 each contain an inversion within the indicated chromosome III homolog with the indicated inversion endpoints. Chromosome III in strains Y2381 and Y2382 carries 25 kb of C. crescentus DNA inserted at the inversion breakpoint between HML and MAT. Deletion alleles in strains Y2388–Y2394 are described in the footnote to Table 2.
Strains containing specific deletion of the recombination enhancer region of chromosome III were constructed by two-step replacements in strain Y2201. Large deletions were constructed in vitro by appropriate restriction digestion of a genomic fragment spanning the recombination enhancer from 28,542 to 31,609 and replacement of the deleted sequences with a 1.7-kb BamHI fragment spanning the yeast HIS3 gene. Deletion strains were constructed by transforming strain Y2201 to His+ with EcoRI-digested DNA from the deletion/insertion plasmid. Smaller deletions of DPS1 and DPS2 were constructed by oligonucleotide mutagenesis of the genomic DNA corresponding to position 28,542–31,069 of chromosome III, cloned into plasmid pALTER (Promega), using the Altered Sites mutagenesis protocol as described by the manufacturer. The sequence of each mutation was designed to create a novel restriction site to facilitate subsequent identification of the mutation in yeast. All mutations were confirmed by PCR and restriction site analysis and then cloned into plasmid YIp5. Plamids carrying the desired mutations were digested appropriately to target insertion into chromosome III and then used to transform strain Y2201 to uracil prototrophy. Ura+ transformants were purified and plated onto SC plates [0.67% Difco yeast nitrogen base, 2% glucose, amino acids, and nucleotide bases as specified (Rose et al. 1990)] containing 5′-fluoro-orotic acid (5-FOA) (1.0 mg/ml). 5-FOA-resistant colonies were purified and then screened by Southern hybridization for those carrying the appropriate mutation(s).
Strains for analysis of cell type-specific transcription of the enhancer region were obtained by transforming diploid strains of the desired DPS1 DPS2 genotype to Ura+ with plasmid pKSW72 cleaved with Eco47III. This plamid contains an allele of ho truncated at both the 5′ and 3′ ends and a copy of URA3 (Weiler et al. 1995). The resulting diploid strain, heterozygous for the dps allele and for the ho::URA3 allele, was sporulated and the spore clones tested by PCR analysis to determine the dps allele and by marker analysis for the ho::URA3. Strains of the appropriate genotype were retained.
The levels of α2p/Mcm1p-mediated repression and Mcm1p-mediated activation by wild-type and mutant α2p/Mcm1p-binding sites were measured by expression of a heterologous CYC1–lacZ reporter plasmid. Plasmids containing the wild-type and mutant sites were constructed by inserting double-stranded oligonucleotides of the sites with TCGA overhangs into the XhoI site between the TATA and UAS elements in the CYC1–lacZ promoter of plasmid pTBA23 (Mead et al. 1996). Plasmids containing single oligonucleotide inserts were screened by restriction digests and verified by sequence analysis. To measure Mcm1p-mediated activation, the CYC1UAS elements in the CYC1–lacZ promoter of these plasmids were deleted as described (Acton et al. 1997).
Donor preference assays
Tetrad dissection and determination of spore mating type were performed as described previously (Weiler and Broach 1992). The donor preference of the first offspring of an a or α spore to switch mating type was inferred by determining the predominant mating type cassette at the MAT locus in cells of the colony originating from that spore (Szeto and Broach 1997). Because both donor loci of the strains used in these assays contained inc mutations, the initial mating type switch precludes any subsequent switches in the progeny of the switched cell. Accordingly, the colony originating from a single spore should consist predominately of identical progeny from the first switching event, most often a switch carried out by the mother cell in the second generation. Determination of the predominant mating type allele resident at MAT in spore colonies was accomplished by PCR analysis of the MAT locus using primers that yielded different sized fragments for all four possible alleles—those initially resident at MAT and those derived from the silent loci. Colonies predominantly exhibiting mata1δ101 allele were scored as selecting HMR as donor, colonies predominantly exhibiting matα1α2 were scored as selecting HML as donor, and colonies that failed to exhibit one predominant allele were scored as ambiguous. These latter colonies resulted either from diminished switching efficeincy, in which a significant number of cells carrying the original wild-type allele resident at MAT are present in the colony, or from random donor preference, in which both donor loci are represented at MAT in cells in the colony. For spores originating from a parent heterozygous for a chromosome III inversion or mutation, spore clones were analyzed for the presence of the markers or by PCR analysis. Only colonies carrying the mutation or inversion were included in the analysis.
Repression/activation assays
To measure α2p/Mcm1p-mediated repression, pTBA23-derived plasmids containing individual α2p/Mcm1p sites were transformed into yeast strains 246.1.1 and EG123 (Sicilano and Tatchell 1984), and the resultant transformants assayed for β-galactosidase activity (Goutte and Johnson 1988). The level of repression was determined by comparing the level of β-galactosidase expression observed in strain 246.1.1 carrying a test plasmid to that of strain EG123 carrying the same plasmid. To measure Mcm1p-mediated activation, plasmids carrying the site of interest and lacking the CYC1 UAS were transformed into the yeast strain GY12 and individual transformants assayed for β-galactosidase activity (Acton et al. 1997). Activation levels were determined by comparing β-galactosidase levels in transformants carrying the test plasmid to that of the same strain carrying a control plasmid lacking an α2p/Mcm1p site. Assays were performed with three independent transformants for each plasmid, and the β-galactosidase units were averaged. Standard deviations were within 20%.
Electrophoretic mobility shift assays
The relative α2p/Mcm1p DNA-binding affinities for the natural and mutant α2p/Mcm1p sites were determined by electrophoretic mobility shift assays (EMSAs), as described (Zhong and Vershon 1997). Labeled DNA fragments used in EMSAs were synthesized by PCR amplification with labeled primers and purified by electrophoresis on a 10% native polyacrylamide gel. Labeled DNA fragments were incubated with purified α2 and Mcm11–96 proteins at the concentrations given in the legend to Figure 3 at room temperature for 3 hr. Both α2 and Mcm11–96 proteins were purified as described previously and were >90% pure as judged by Coomassie-stained SDS-polyacrylamide (Vershon et al. 1993; Vershon and Johnson 1995). All binding reactions were electrophoresed through 0.5× TBE/4.5% native polyacylamide gels at 200 V for 2–3 hr. Gels were dried, exposed to phosphor screens, and images were scanned on a Molecular Dynamics PhorphorImager model 425.
Northern analysis
Total RNA was prepared as described from yeast cells in exponential phase in YEPD, fractionated on agarose gels containing 8% formaldehyde, transferred to Hybond nylon membrane in high salt, and probed with the indicated DNA fragments labeled by random primer extension. Washed filters were exposed to phosphor screens, and images were scanned on a Molecular Dynamics PhosphorImager model 425.
Acknowledgments
The research presented in this report was supported by grants from the National Institutes of Health to J.R.B. and A.K.V.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jbroach@molecular.princeton.edu; FAX (609) 258-6175.
References
- Acton TB, Zhong H, Vershon AK. DNA-binding specificity of Mcm1: Operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol Cell Biol. 1997;17:1881–1889. doi: 10.1128/mcb.17.4.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching in mother cells. Cell. 1996;85:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Breeden L, Nasymth KA. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:525–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Roth SY, Simpson RT. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes & Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- Gerring SL, Spencer F, Hieter P. The CHL1 (CTF1) gene product of Saccharomyces cervisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte C, Johnson AD. a1 protein alters the DNA specificity of α2 repressor. Cell. 1988;52:875–882. doi: 10.1016/0092-8674(88)90429-1. [DOI] [PubMed] [Google Scholar]
- Herskowitz I, Rine J, Strathern J. Mating-type determination and mating type interconversion in Saccharomyces cerevisiae. In: Jones EW, Pringle JR, Broach JR, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 583–656. [Google Scholar]
- Jensen RE, Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harbor Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- Keleher CA, Goutte C, Johnson AD. The yeast cell-type specific repressor α2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988;53:927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Keleher CA, Passmore S, Johnson AD. Yeast repressor α2 binds to its operator cooperatively with yeast protein Mcm1. Mol Cell Biol. 1989;9:5228–5230. doi: 10.1128/mcb.9.11.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Komachi K, Redd MJ, Wolberger C. The WD repeats of Tup1 interact with the homeo domain protein α2. Genes & Dev. 1994;8:2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- Kostriken R, Heffron F. The product of the HO gene is a nuclease: Purification and characterization of the enzyme. Cold Spring Harbor Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- Kostriken R, Strathern JN, Klar AJS, Hicks JB, Heffron F. A site-specific endonuclease essential for mating type switching in Saccharomyces cerevisiae. Cell. 1983;35:167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Kotani H, Germann MW, Andrus A, Vinayak R, Mullah B, Kmiec EB. RNA facilitates RecA-mediated DNA pairing and strand transfer between molecules bearing limited regions of homology. Mol & Gen Genet. 1996;250:626–634. doi: 10.1007/BF02174450. [DOI] [PubMed] [Google Scholar]
- Mead J, Zhong H, Acton TB, Vershon AK. Yeast α2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins in higher eukaryotes. Mol Cell Biol. 1996;16:2135–2143. doi: 10.1128/mcb.16.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth KA. Molecular analysis of a cell lineage. Nature. 1983;302:670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- Passmore S, Elble R, Tye BK. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes & Dev. 1989;3:921–935. doi: 10.1101/gad.3.7.921. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Rastan S. X chromosome inactivation and the Xist gene. Curr Opin Genet Dev. 1994;4:292–297. doi: 10.1016/s0959-437x(05)80056-5. [DOI] [PubMed] [Google Scholar]
- Rose AB, Broach JR. Propagation and expression of cloned genes in yeast: 2-μm circle-based vectors. Methods Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sauer RT, Smith DL, Johnson AD. Flexibility of the yeast α2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes & Dev. 1988;2:807–816. doi: 10.1101/gad.2.7.807. [DOI] [PubMed] [Google Scholar]
- Siciliano PG, Tatchell K. Transcription and regulatory signals at the mating-type locus in yeast. Cell. 1984;37:969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Sil A, Herskowitz I. Identification of an asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- Strathern JN, Klar AJS, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31:183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Szeto L, Broach JR. Role of α2 protein in donor locus selection during mating type interconversion. Mol Cell Biol. 1997;17:751–759. doi: 10.1128/mcb.17.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Oshima T, Araki H, Harashima S, Oshima Y. Mating type control in Saccharomyces cerevisiae: A frameshift mutation at the common DNA sequence, X, of the HMLα locus. Mol Cell Biol. 1984;4:203–212. doi: 10.1128/mcb.4.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon AK, Johnson AD. A short, disordered protein region mediates interaction between the homeodomain of the yeast α2 protein and the MCM1 protein. Cell. 1993;72:105–112. doi: 10.1016/0092-8674(93)90054-t. [DOI] [PubMed] [Google Scholar]
- Vershon AK, Jin Y, Johnson AD. A homeo domain protein lacking specific side chains of helix 3 can still bind DNA and direct transcriptional repression. Genes & Dev. 1995;9:182–192. doi: 10.1101/gad.9.2.182. [DOI] [PubMed] [Google Scholar]
- Weiler KS, Broach JR. Donor locus selection during Saccharomyces cerevisiae mating type interconversion responds to distant regulatory signals. Genetics. 1992;132:929–942. doi: 10.1093/genetics/132.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Szeto L, Broach JR. Mutations affecting donor preference during mating type interconversion in Saccharomyces cerevisiae. Genetics. 1995;139:1495–1510. doi: 10.1093/genetics/139.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Haber JE. MATa donor preference in yeast mating type switching: Activation of a large chromosomal region for recombination. Genes & Dev. 1995;9:1922–1932. doi: 10.1101/gad.9.15.1922. [DOI] [PubMed] [Google Scholar]
- ————— A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- Wu X, Moore JK, Haber JE. Mechanism of MATα donor preference during mating-type switching of Saccharomyces. Mol Cell Biol. 1996;16:657–668. doi: 10.1128/mcb.16.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, DePinho RA, Zimmerman KA, Lutzker SG, Rosenberg N, Alt FW. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Cole-Strauss A, Kmiec EB. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA.DNA oligonucleotide. Proc Natl Acad Sci. 1996;93:2071–2076. doi: 10.1073/pnas.93.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bottaro A, Li S, Stewart V, Alt FW. A selective defect in IgG2b switching as a result of targeted mutation of the I gamma 2b promoter and exon. EMBO J. 1993;12:3529–3537. doi: 10.1002/j.1460-2075.1993.tb06027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Vershon AK. The yeast homeodomain protein MATα2 shows extended DNA binding specificity in complex with Mcm1. J Biol Chem. 1997;272:8402–8409. doi: 10.1074/jbc.272.13.8402. [DOI] [PubMed] [Google Scholar]