Abstract
Cellular responses to mechanical perturbation are vital to cell physiology. In particular, migrating cells have been shown to sense substrate stiffness and alter cell morphology and speed. Zyxin is a focal adhesion protein that responds to external mechanical forces; however, the mechanisms of zyxin recruitment at force-bearing sites are unknown. Using force-sensing microfabricated substrates, we simultaneously measured traction force and zyxin recruitment at force-bearing sites. GFP-tagged zyxin accumulates at force-bearing sites at the leading edge, but not at the trailing edge, of migrating epithelial cells. Zyxin recruitment at force-bearing sites depends on Rho-kinase and myosin II activation, suggesting that zyxin responds not only to the externally applied force, as previously shown, but also to the internally generated actin-myosin force. Zyxin in turn recruits vasodilator-stimulated phosphoprotein, a regulator of actin assembly, to force-bearing sites. To dissect the domains of zyxin that are essential for this unique force-dependent accumulation, we generated two zyxin truncation mutants: one lacking the LIM domain (ΔLIM) and one containing only the LIM domain with all three LIM motifs (LIM). GFP-tagged ΔLIM does not localize to the force-bearing sites, but GFP-tagged zyxin LIM-domain is sufficient for the recruitment to and dynamics at force-bearing focal adhesions. Furthermore, one or two LIM motifs are not sufficient for force-dependent accumulation, suggesting that all three LIM motifs are required. Therefore, the LIM domain of zyxin recruits zyxin to force-bearing sites at the leading edge of migrating cells.
Introduction
Mammalian cells are exposed to various types of mechanical stimuli from their environment and neighboring cells. In response to these external forces, cells alter their morphology, migration, and function. For example, in a stiff three-dimensional matrix, mammary epithelial cells develop an invasive phenotype (1), and fibroblasts migrate faster on a stiff substrate than on a soft one (2). Furthermore, stem cells differentiate into specific lineages according to the substrate stiffness (3,4). These observations highlight the importance of a cell's response to mechanical cues for directing its fate and functions. However, the exact molecular mechanisms by which cells sense and respond to mechanical stimuli remain elusive.
The adhesion complex is an integral component of the transmission of mechanical stimuli. Of interest, the application of external forces changes the localization of the focal adhesion protein zyxin. For example, when adherent cells are stretched by pulling on the underlying flexible substrate, zyxin proteins accumulate to actin stress fibers (5) and enhance actin assembly at focal adhesions (6), whereas zyxin-deficient cells fail to respond to external strain (7). Moreover, when cells are prodded in the vicinity of actin stress fibers, zyxin reversibly accumulates at the perturbed fibers (8,9). These findings suggest that zyxin is a mechanosensing protein and an ideal candidate for regulating mechano-induced cell signaling.
Zyxin binds to α-actinin, an actin cross-linking protein (10) and Enabled (Ena)/vasodilator-stimulated phosphoprotein (VASP) proteins with actin anticapping activity (11) at its N-terminus region. Previous studies have shown that zyxin recruits VASP to focal adhesions (12) and force-bearing cadherin junctions (13). Mislocalization of VASP and mammalian Ena is observed when the overexpressed zyxin LIM domain displaces the endogenous zyxin from focal adhesions (6,14). The zyxin LIM domain consists of three LIM motifs in the C-terminus of zyxin, and the LIM domain alone is sufficient for focal adhesion localization (14). The LIM motif has structures similar to that of zinc fingers, which binds DNA, and some LIM proteins localize to the nucleus and have been shown to play a transcriptional role (15,16). The functional diversity of LIM proteins suggests that the LIM domain plays a unique role in various cellular processes. However, the precise functions of the zyxin LIM domain remain unclear.
To study the role of zyxin as a mechanosensor, we analyzed the accumulation of proteins at force-bearing sites using a microfabricated force-sensing substrate and confocal live-cell microscopy. Here, we show that zyxin accumulated in a Rho-kinase and myosin II-mediated, force-dependent manner at the leading edge, but not at the trailing edge, of migrating cells. Zyxin in turn recruited VASP to these force-bearing sites. We isolated the role of LIM domains by creating zyxin constructs without LIM domains (ΔLIM) and with only LIM domains (LIM). The zyxin-ΔLIM proteins were unresponsive to traction force, whereas the LIM domain responded to traction force with localization and dynamics similar to those observed for the zyxin-GFP proteins. Of the three LIM motifs in the zyxin LIM domain, one or two alone did not accumulate at force-bearing sites. Thus, we propose that all three LIM motifs are required for force-dependent accumulation of zyxin.
Materials and Methods
Cell culture and reagents
Madin-Darby canine kidney (MDCK) GII cells were cultured in low-glucose Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Cells were fixed in phosphate-buffered saline (PBS) containing 3% paraformaldehyde, 0.3% Triton X-100, for 10 min and stained with Alexa-568 phalloidin (Invitrogen, Carlsbad, CA), VASP antibody (BD Biosciences, San Jose, CA), zyxin 4302 (Epitomics, Burlingame, CA), or zyxin B71 antibody (12). The GFP-tagged full-length human zyxin, zyxin-ΔLIM (1-383AA), zyxin-LIM (338-572AA), LIM1 (377-443AA), LIM2 (437-503AA), LIM3 (496-572AA), LIM12 (377-503AA), LIM23 (437-572AA), or VASP plasmids were transfected into MDCK cells, and G418 antibiotic-resistant stable cells were subcloned to homogeneity. Zyxin knockdown cell lines were generated and characterized with the use of pSuper.gfp plasmids (Oligoengine, Seattle, WA) as described previously (13). The canine specific zyxin shRNA sequence in the stable zyxin knockdown clone used in this study is 5′-GACAAGAACTTCCACATGA-3′, and the endogenous zyxin level was <5% of normal cells (13). We previously tested these cell lines for off-targeting effects using a scrambled sequence (5′-CACATAAGGTCCATACGAA-3′) and found no side-effects (13).
Live-cell confocal microscopy and fluorescence recovery after photobleaching
All samples were imaged on a Zeiss Axio Observer equipped with a Yokogawa spinning disk confocal system, a 40× objective, 488 and 561 nm solid-state lasers, and a CoolSNAP HQ camera. The microscope system was controlled and automated by Slidebook software (Intelligent Imaging Innovations, Denver, CO). Live cells were imaged on glass-bottom dishes (MatTek, Ashland, MA) in a temperature-controlled chamber at 37°C. The GFP accumulation on a single adhesion site was selectively photobleached with the use of a photo-ablation system (Intelligent Imaging Innovation, Denver, CO) and imaged every 3 s for 5 min. The GFP intensity was quantified with ImageJ and analyzed in Microsoft Excel. The normalized GFP intensity was curve-fitted with a single exponential function to determine the half time and the degree of fluorescence intensity recovery.
Fabrication of micropillar arrays
Micropillar arrays were fabricated as described previously (17). The micropillar master was etched with deep reactive ion etcher and had dimensions of 2 μm in pillar diameter, 6 μm in height, and 4 μm in pitch (AdvancedMEMS, Berkeley, CA; see Fig. S5 A in the Supporting Material). The bright-field (BF) images of collapsed pillars were used to quantify the actual height and diameter (Fig. S5 B). A droplet of 100 μg/mL fibronectin (BD Biosciences, San Jose, CA) solution spiked with rhodamine fibronectin (Cytoskeleton, Denver, CO) in a 1:5 ratio was deposited on the pillar tips. After incubation for 10 min, the pillars were immersed in PBS with 1% bovine serum albumin and 0.05% Triton X-100 for 1 h. The pillars were then washed with PBS and the growth media. This approach created a fibronectin mesh on the pillar array (Fig. S5 C and Fig. S6), making it easier for the cells to adhere and spread. As a result of reorganization of fibronectin by migrating cells, the some cells were able to deflect distant pillars using fibronectin bundles (Fig. S6, arrowhead). We avoided this phantom deflection in our analysis.
Quantification of traction force and protein accumulation
We calculated the traction force based on the displacement of pillar tips from the original positions (Fig. S7 A). We measured the elasticity of polydimethylsiloxane (PDMS) using an Instron tensile tester (Norwood, MA), and used the pillar stiffness (2.5 MPa) to convert the pillar displacements to traction force. To quantify the GFP intensity, the pillar regions of interest (ROIs) were shifted half a pillar diameter in the direction of deflection and placed on the GFP image (Fig. S7 B). Within each ROI, each thresholded region was normalized to the corresponding background intensity (Fig. S7 C).
Results and Discussion
Zyxin is recruited to force-bearing sites
To examine the mechanosensing ability of zyxin, we simultaneously analyzed the accumulation of zyxin-GFP expressed in MDCK epithelial cells and the corresponding traction force using microfabricated force-sensing substrates. The force sensor is based on arrays of closely packed elastic pillars that deflect as membrane extensions make contact and pull (see Materials and Methods) (4,17,18). The tips of the pillars were coated with fibronectin to facilitate cell adhesion and spreading (see Materials and Methods). Zyxin-GFP expressing cells seeded on a pillar array rapidly spread out and extended lamellipodia with focal adhesion-like zyxin positive puncta at the tips of force-bearing pillars (Fig. 1 A and Movie S1). These zyxin-GFP puncta accumulated at the side of the pillars proximal to the cell body and oriented in the direction of the generated force (Fig. 1 A).
Figure 1.
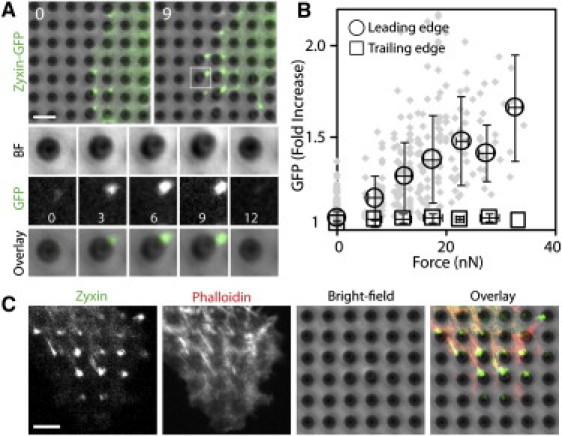
Zyxin is recruited to force-bearing sites. (A) Zyxin-GFP accumulated as puncta at the force-bearing pillars of a migrating cell. The white square region is shown as a montage of an individual pillar. BF, bright-field. Time in minutes. (B) Quantification of traction force and zyxin accumulation at the leading edge (gray diamonds, ncell = 15; open circles are the average values) and trailing edge (gray squares, ncell = 9). The trailing edge was defined as the tail end of persistently migrating cells. Error bars are the standard deviation (SD). (C) Immunofluorescence staining of endogenous zyxin and phalloidin in WT cells. Scale bars: 5 μm.
Previous studies showed that zyxin accumulates in the sites of externally applied forces (5,6,8), and, with the addition of Rho-kinase inhibitor, zyxin dissipates from focal adhesions (19). Although the Rho-kinase and myosin II inhibitors are assumed to reduce the traction force of migrating cells, some cells can still migrate in the presence of such inhibitors (20). Therefore, we explicitly analyzed the changes in traction force and zyxin accumulation in the presence of Y27632 (a Rho-kinase inhibitor) or blebbistatin (a myosin II inhibitor). The generation of traction force and the zyxin recruitment at force-bearing sites depended on Rho-kinase and myosin II activity, as both Y27632 and blebbistatin simultaneously decreased the traction force and zyxin accumulation (Fig. S1). This is consistent with a previous finding that the zyxin dynamics is regulated by Rho-kinase (19), and blebbistatin-dependent zyxin accumulation demonstrates that the effector of Rho-kinase that regulates the force-dependent zyxin recruitment is likely myosin II.
At the leading edge of migrating cells, the accumulation of zyxin-GFP correlated temporally with the magnitude of pillar deflection and therefore the amount of traction force (Fig. 1 A). At the trailing edge of migrating cells, however, these zyxin puncta were clearly absent (Fig. S2). The quantification of pillar deflection and zyxin accumulation shows that as traction force increased, more zyxin accumulated relative to the surrounding region at the leading edge (Fig. 1 B). At the trailing edge, however, increased zyxin accumulation was not observed (Fig. 1 B, square, and Fig. S2). Therefore, zyxin is a marker of traction force at the leading edge of migrating cells.
Our experimental results indicate that force-dependent zyxin accumulation occurs at the leading edge, but not at the trailing edge, of migrating cells. One possible explanation for the differential zyxin recruitment is that the force-generation mechanisms are highly distinctive at the leading and trailing edges. At the leading edge, actin polymerization and myosin contraction mediate retrograde flow that drives lamellipodia protrusions and force generation (21,22). On the other hand, the trailing edge has greater myosin II accumulation and thus greater acto-myosin contraction (23). These varying force-generation mechanisms imply that each edge also has distinctive molecular compositions. Because zyxin only accumulates at the leading edge, it does not indiscriminately localize to force-bearing sites; rather, it has an ability to distinguish between these distinct adhesion sites.
To analyze the colocalization of zyxin and the actin network, we labeled the migrating wild-type (WT) cells on the pillar array with a zyxin antibody and phalloidin. The immunofluorescence staining showed a localization of endogenous zyxin similar to that observed for zyxin-GFP, confirming that zyxin-GFP is a faithful marker for endogenous zyxin localization (Fig. 1 C). Zyxin-positive focal complexes at the tips of lamellipodia were not observed, consistent with previous observations that zyxin is absent at the lamellipodia tips (24). Phalloidin-labeled actin bundles colocalized with the bright, mature puncta located farther away from the leading edge (Fig. 1 C). Smaller, nascent zyxin puncta were present near the tips of lamellipodia, whereas phalloidin-positive actin bundles were absent (Fig. 1 C), suggesting that formation of these puncta precedes the assembly of the actin bundle network.
Zyxin recruits VASP to the force-bearing sites
The actin assembly is required for lamellipodia extension and retrograde flow, and VASP regulates actin assembly by its anticapping activity (25). Because zyxin contains a proline-rich VASP-binding domain (11), we analyzed force-dependent accumulation of VASP using the force-sensitive substrate and cells stably expressing GFP-tagged VASP. Similarly to force-dependent localization of zyxin, GFP-VASP accumulated at the tips of force-bearing pillars as distinct puncta (Fig. 2 A and Movie S2). The intensity of the VASP puncta increased as the migrating cell exerted force against the pillar, and decreased as the migrating cell released the pillar (Fig. 2 A). Zyxin-GFP and GFP-VASP showed quantitatively similar force-dependent accumulation (Fig. 2 B), suggesting a direct physical interaction between zyxin and VASP at force-bearing sites of migrating cells.
Figure 2.
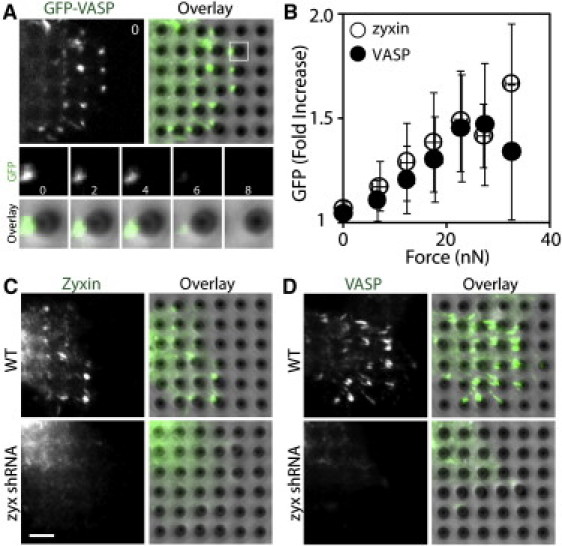
Zyxin recruits VASP to force-bearing sites. (A) GFP-VASP accumulated as puncta at the force-bearing pillars of a migrating cell. The white square region is shown as a montage of an individual pillar. Time in minutes. (B) Quantification of traction force and VASP accumulation (filled circle, ncell = 19). The zyxin-GFP values (open circle) from Fig. 1B are included as a reference. Error bars are the SD. (C) Immunofluorescence staining of zyxin in WT and zyxin-knockdown cells. (D) Immunofluorescence staining of VASP in WT and zyxin-knockdown cells. Scale bar: 5 μm.
To test whether zyxin is required for VASP recruitment to the force-bearing sites, we analyzed VASP localization in WT and zyxin knockdown cells. The zyxin knockdown cell lines were generated by stable expression of canine zyxin specific shRNA and characterized in our previous publication (13). In WT cells, both zyxin and VASP accumulated at force-bearing sites (Fig. 2, C and D). In a stable zyxin knockdown clone (Fig. 3 B) (13), endogenous VASP did not localize to force-bearing pillars, even though the zyxin-deficient cells exerted traction force against the pillars (see below). Our analysis demonstrates that zyxin accumulation at force-bearing sites precedes and is required for VASP recruitment at force-bearing pillars.
Figure 3.
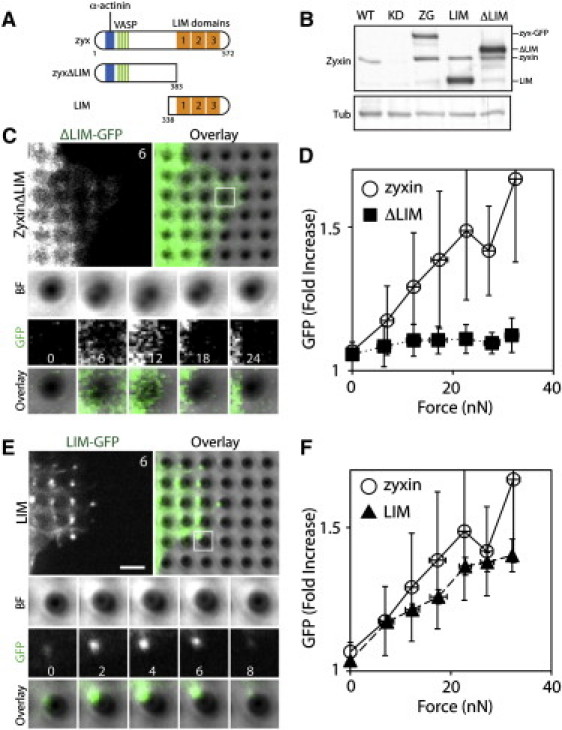
LIM domain of zyxin is sufficient for localization at force-bearing sites. (A) Schematic of full-length zyxin, truncated zyxin lacking LIM domains (zyxΔLIM), and truncated zyxin containing only LIM domains (LIM). (B) Western blots of cells expressing zyxin and its mutants. (C) Unlike zyxin-GFP (open circle), ΔLIM-GFP did not accumulate at force-bearing pillars. The white square region is shown as a montage of an individual pillar. (D) The intensity of ΔLIM-GFP (filled square, ncell = 10) did not correlate with traction force. (E) LIM-GFP accumulated as puncta at the force-bearing pillars. The white square region is shown as a montage of an individual pillar. (F) In similarity to zyxin-GFP (open circle), the intensity of LIM-GFP (filled triangle, ncell = 20) correlated positively with traction force. Scale bar: 5 μm. Error bars are the SD. Time in minutes.
Zyxin has been suggested to play an integral role in generating traction force by recruiting VASP to force-bearing sites. Surprisingly, we found that migrating zyxin knockdown cells exerted traction forces similar to those of WT cells (Fig. 2, C and D, and Fig. S3). In addition, these zyxin knockdown and WT cells were shown to migrate at similar speeds on a collagen- or fibronectin-coated glass coverslip (13). These results differ slightly from those of a previous study, in which zyxin null fibroblasts were shown to migrate faster on a fibronectin-coated substrate (12). Zyxin null fibroblast cells retain the ability to exert traction force with similar magnitudes as zyxin null fibroblasts rescued with exogenous zyxin, except that traction forces of high magnitudes were observed less frequently in zyxin null cells (9). This difference in force generation by zyxin-deficient cells may be due to the substrate stiffness, given that substrate stiffness can alter cell morphology and migration speed (2). On a soft polyacrylamide substrate, zyxin accumulation is inversely correlated to traction force generation (26), suggesting that matrix stiffness may play a critical role in zyxin localization at force-bearing sites. Because the pillar geometry defines the pillar rigidity, which in turn changes the cytoskeletal contractility (4) and cell stiffness (27), this experimental approach may reveal the stiffness-dependent zyxin dynamics. Alternatively, because the force-anchoring points are limited to the pillar tips in our substrates, the geometric difference between flat and pillar-based substrates may be an important factor in how zyxin-deficient cells generate traction forces. Despite the differences in experimental methods, these studies suggest that, in the absence of zyxin, migrating cells can generate sufficient force to migrate in both flat and pillar-based substrates.
The LIM domain of zyxin is sufficient for force-dependent recruitment
To identify the domain of zyxin that is required for force-dependent recruitment, we constructed zyxin mutants with the truncated LIM-domain (ΔLIM-GFP) and with only the LIM domains (LIM-GFP) (Fig. 3 A), and expressed these mutants in epithelial cells (Fig. 3 B). Both mutant-expressing cells generated traction forces similarly to zyxin-GFP-expressing cells without any defects (see the similar magnitude of traction force generated by the normal and mutant expressing cells in Fig. 3, D and F). Although ΔLIM-GFP-expressing cells generated traction forces, ΔLIM-GFP proteins did not accumulate at force-bearing sites (Fig. 3 C and Movie S3). The intensity of ΔLIM-GFP remained diffused in the lamellipodia that extended at the force-bearing pillars (Fig. 3 C). Quantification of the GFP intensity and force shows that ΔLIM-GFP intensity did not correlate with traction force (Fig. 3 D). In contrast, LIM-GFP accumulated at the force-bearing pillars as puncta, positively responding to the pillar deflection (Fig. 3 E and Movie S4). Quantification of the LIM-GFP intensity and traction force generated by the migrating cells demonstrates that LIM-GFP and zyxin-GFP had similar rates of increase in intensity (Fig. 3 F). Thus, the LIM domain of zyxin is sufficient for responding to the traction force generated by migrating cells.
To test whether endogenous zyxin is required for the LIM domain recruitment at force-bearing sites, we transfected the LIM-GFP into zyxin knockdown cells. Despite the significantly reduced level of zyxin in these cells, LIM-GFP robustly colocalized with deflected pillars similarly to LIM-GFP expressed in WT cells (Fig. 4, A and B, and Movie S5) and at focal adhesions (Fig. 4 C). Although there is a slight decline in the GFP-LIM intensity at a higher traction force (Fig. 4 B), the difference is within the overlapping distributions. Because the zyxin deficiency reduces actin assembly at focal adhesions (6,13), these data suggest that LIM recruitment to force-bearing sites does not require endogenous zyxin or zyxin-mediated actin assembly.
Figure 4.
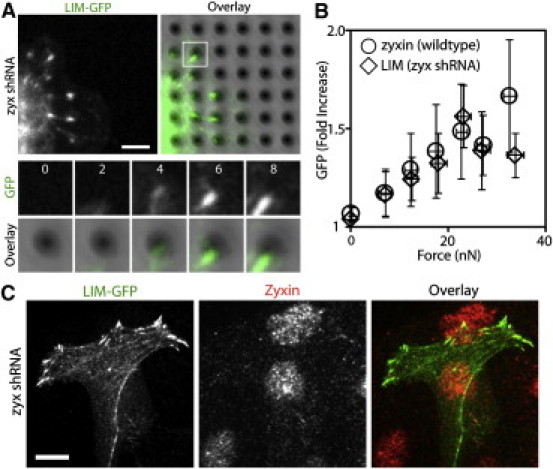
Zyxin LIM domain in the absence of endogenous zyxin is recruited to force-bearing sites. (A) In the zyxin-knockdown cells, zyxin LIM-GFP accumulated at force-bearing sites. The white square region is shown as a montage of an individual pillar. Time in minutes. Scale bar: 5 μm. (B) Quantification of traction force and LIM-GFP accumulation (open diamonds, ncell = 8). The zyxin-GFP values (open circle) from Fig. 1B are included as a reference. Error bars are the SD. (C) In the zyxin-knockdown cells, zyxin LIM-GFP accumulated at focal adhesions. Scale bar: 10 μm.
The zyxin LIM domain consists of three LIM repeats, with each LIM motif containing two zinc-binding residues that form a tandem zinc finger topology (15). To determine which LIM motifs are essential for recruitment to force-bearing sites, we further dissected the zyxin LIM domain into individual LIM motifs (LIM1, LIM2, and LIM3) or truncated LIM motifs (LIM12 and LIM23), and tested their ability to localize at force-bearing sites. The individual LIM motifs primarily localized to cytoplasm, and in some cells the individual LIM motifs accumulated at cell-cell contacts (A. Steele and S. Yamada, unpublished data). However, none of these LIM motifs accumulated at focal adhesions (Fig. S4 A), which is consistent with previous observations of focal adhesion targeting by LIM mutants, albeit in the presence of VASP- and α-actinin-binding domains (14). In addition, the single LIM motifs failed to accumulate at force-bearing sites (Fig. 5, A and B), and similar results were obtained with truncated LIM mutants (Fig. 5, C and D). Significantly, the truncated LIM mutants localized to focal adhesions (Fig. 5 E and Fig. S4 B), suggesting the presence of distinct adhesive complexes at focal adhesions established on a coverslip and force-bearing sites observed using force-sensing pillar arrays. These data suggest that the individual or truncated LIM motifs are not sufficient for force-dependent accumulation, and that zyxin recruitment requires all three LIM motifs. Of interest, many LIM proteins contain multiple LIM repeats (15), suggesting that the LIM motif may require multiple repeats for proper protein interactions.
Figure 5.
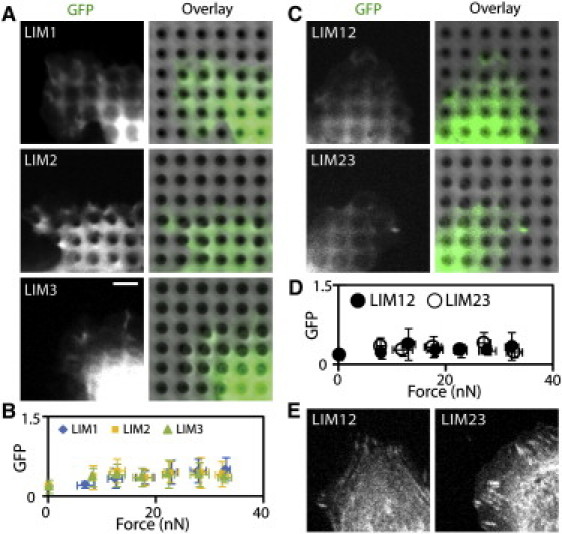
Truncated LIM mutants are not recruited to force-bearing sites. (A) Individual zyxin LIM motifs do not localize to force-bearing sites. Scale bar: 5 μm. (B) Quantification of traction force and GFP-tagged LIM1, LIM2, or LIM3 accumulation (diamonds: LIM1, ncell = 9; square: LIM2, ncell = 10; triangles: LIM3, ncell = 12). (C) Truncated zyxin LIM domains do not localize to force-bearing sites. Scale bar: 5 μm. (D) Quantification of traction force and GFP-tagged LIM12 or LIM23 accumulation (filled circles: LIM12, ncell = 10; closed circles: LIM23, ncell = 11). (E) GFP-tagged LIM12 and LIM23 localized to focal adhesions, albeit less robustly than the zyxin LIM domain containing all three LIM motifs (see also Fig. S4).
The zyxin LIM domain recapitulates zyxin dynamics at force-bearing sites
Although the zyxin LIM domain is sufficient for localizing at force-bearing sites, zyxin's binding partners (e.g., α-actinin and Ena/VASP) may stabilize zyxin proteins at force-bearing sites. To analyze the dynamics of zyxin at the force-bearing site, we selectively photobleached GFP-tagged full-length zyxin or LIM domain puncta that accumulated at the force-bearing pillar. After these puncta were photobleached, both zyxin-GFP and LIM-GFP recovered rapidly (Fig. 6 A). Quantification of the GFP intensity change after photobleaching reveals that the intensity of zyxin-GFP and LIM-GFP fully recovered to the prephotobleaching level at the same rate (Fig. 6 B). The intensity recoveries of zyxin- and LIM-GFP yielded statistically identical averages for percentage of recovery (Fig. 6 C) and half-time of recovery (Fig. 6 D). These results are consistent with the full-length zyxin protein dynamics previously observed at the focal adhesions of bovine endothelial cells (19). The observation of similar dynamics in intensity recovery at force-bearing pillars suggests that the LIM domain alone, without additional support from α-actinin or Ena/VASP, can stabilize zyxin to the force-bearing sites.
Figure 6.
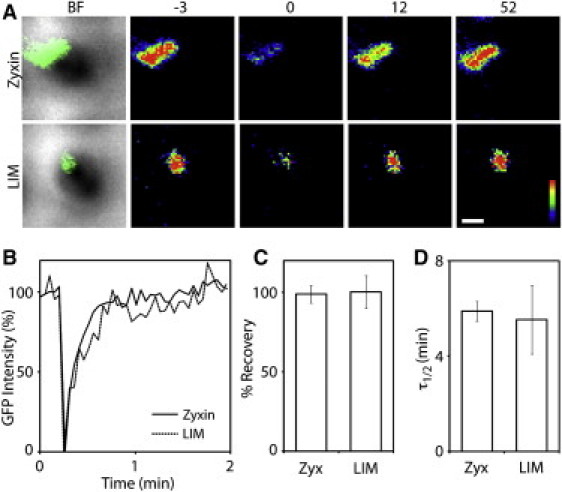
Dynamics of the LIM domain is similar to that of zyxin at force-bearing sites. (A) A montage of individual force-bearing pillars with zyxin-GFP or LIM-GFP puncta. GFP intensity was pseudo-colored to represent highest intensity as red (or white in print version) and lowest intensity as black. Time in seconds. Scale bar: 1 μm. (B) Quantification of the GFP intensities of zyxin and LIM domain. (C) The average fluorescence recovery of zyxin-GFP (ncell = 18 and traction force, mean ± SE = 14.3 ± 5.7 nN) and LIM-GFP (ncell = 14 and traction force, mean ± SE = 23.9 ± 7.0 nN). (D) The average half-time of recovery of zyxin-GFP and LIM-GFP. Error bars are the mean ± SE.
Currently, the only known binding partners of zyxin LIM domain are cysteine-rich protein (CRP), which is thought to be involved in muscle differentiation (28,29); synemin, a member of an intermediate protein family (30); and p130cas (31), which is thought to undergo a force-dependent conformational change (32). CRP binds to the first LIM motif of zyxin (33), yet the LIM1 of zyxin alone does not localize to force-bearing junctions (Fig. 5 A). Unlike other zyxin-binding proteins, synamin does not localize to focal adhesions; instead, the localization pattern of synamin is similar to that of vimentin (30). In addition, synamin-depleted HeLa cells have proper zyxin localization at focal adhesions (30). Furthermore, in p130cas null cells, zyxin localizes to focal adhesions but not at focal complexes, suggesting that p130cas is not essential for zyxin recruitment at focal adhesions (31). On the basis of these findings, we can conclude that none of these zyxin-interacting proteins are responsible for zyxin LIM recruitment at force-bearing sites.
Our results indicate that zyxin recruitment to force-bearing sites requires only the LIM domain of zyxin, and does not require the N-terminus of zyxin containing VASP- and α-actinin-binding sites. This in turn suggests that the interactions between zyxin and VASP or α-actinin are not required for force-dependent recruitment. Moreover, by truncating the LIM domain into one or two LIM motifs, we were able to show that these LIM truncated mutants do not accumulate at force-bearing sites, which suggests that all three LIM motifs are required for proper force-dependent accumulation. The combined mutational and traction force analyses demonstrate for the first time (to our knowledge) that the zyxin LIM domain, but not individual LIM motifs, is sufficient for force-dependent recruitment of zyxin.
Because zyxin can shuttle between the cytoplasm and nucleus (34), zyxin may regulate cell function in a force-dependent manner, and the ability to sense mechanical force may be an integral part of gene expression regulation. Although we do not yet know the precise molecular interactions of the zyxin LIM domain at force-bearing sites, the identification of the LIM domain as the module for force-dependent recruitment demonstrates that force-sensitive interactions may be a key feature of other proteins that contain the LIM domain.
Acknowledgments
We thank Dr. Mary Beckerle (University of Utah) for sharing the zyxin B71 antibody, Dr. Juergen Wehland (Gesellschaft für Biotechnologische Forschung, Germany) for the zyxin-GFP plasmid, Dr. Hiroaki Hirata (Nagoya University) for the zyxin-LIM plasmid, Dr. Frank Gertler (MIT) for the GFP-VASP plasmid, and Dr. Grant Sumida for a critical reading of the manuscript.
This work was supported by a Beckman Young Investigator Award (S.Y.), a Hellman Family New Faculty Award (S.Y.), a EUREKA Award from the National Institutes of Health (S.Y.), the University of California Cancer Research Coordinating Committee (S.Y.), an Achievement Rewards for College Scientists Award (T.N.), and a grant from the Training Program in Biomolecular Technology, National Institutes of Health (T32-GM08799 to T.N.).
Footnotes
Thuc-Nghi Nguyen's present address is Allen Institute for Brain Science, Seattle, Washington.
Supporting Material
References
- 1.Paszek M.J., Zahir N., Weaver V.M. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Lo C.M., Wang H.B., Wang Y.L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engler A.J., Sen S., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Fu J., Wang Y.K., Chen C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshigi M., Hoffman L.M., Beckerle M.C. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 2005;171:209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirata H., Tatsumi H., Sokabe M. Mechanical forces facilitate actin polymerization at focal adhesions in a zyxin-dependent manner. J. Cell Sci. 2008;121:2795–2804. doi: 10.1242/jcs.030320. [DOI] [PubMed] [Google Scholar]
- 7.Ngu H., Feng Y., Yin F.C. Effect of focal adhesion proteins on endothelial cell adhesion, motility and orientation response to cyclic strain. Ann. Biomed. Eng. 2010;38:208–222. doi: 10.1007/s10439-009-9826-7. [DOI] [PubMed] [Google Scholar]
- 8.Colombelli J., Besser A., Stelzer E.H. Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J. Cell Sci. 2009;122:1665–1679. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- 9.Smith M.A., Blankman E., Beckerle M.C. A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Dev. Cell. 2010;19:365–376. doi: 10.1016/j.devcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford A.W., Michelsen J.W., Beckerle M.C. An interaction between zyxin and α-actinin. J. Cell Biol. 1992;116:1381–1393. doi: 10.1083/jcb.116.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drees B., Friederich E., Golsteyn R.M. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J. Biol. Chem. 2000;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman L.M., Jensen C.C., Beckerle M.C. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J. Cell Biol. 2006;172:771–782. doi: 10.1083/jcb.200512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen T.N., Uemura A., Yamada S. Zyxin-mediated actin assembly is required for efficient wound closure. J. Biol. Chem. 2010;285:35439–35445. doi: 10.1074/jbc.M110.119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nix D.A., Fradelizi J., Beckerle M.C. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- 15.Kadrmas J.L., Beckerle M.C. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Gilmore T.D. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta. 2003;1593:115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- 17.Yang M.T., Fu J., Chen C.S. Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity. Nat. Protoc. 2011;6:187–213. doi: 10.1038/nprot.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan J.L., Tien J., Chen C.S. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lele T.P., Pendse J., Ingber D.E. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J. Cell. Physiol. 2006;207:187–194. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- 20.Straight A.F., Cheung A., Mitchison T.J. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 21.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y.L. Flux at focal adhesions: slippage clutch, mechanical gauge, or signal depot. Sci. STKE. 2007;2007:pe10. doi: 10.1126/stke.3772007pe10. [DOI] [PubMed] [Google Scholar]
- 23.Conti M.A., Adelstein R.S. Nonmuscle myosin II moves in new directions. J. Cell Sci. 2008;121:11–18. doi: 10.1242/jcs.007112. [DOI] [PubMed] [Google Scholar]
- 24.Zaidel-Bar R., Ballestrem C., Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 25.Bear J.E., Gertler F.B. Ena/VASP: towards resolving a pointed controversy at the barbed end. J. Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beningo K.A., Dembo M., Wang Y.L. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 2001;153:881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tee S.Y., Fu J., Janmey P.A. Cell shape and substrate rigidity both regulate cell stiffness. Biophys. J. 2011;100:L25–L27. doi: 10.1016/j.bpj.2010.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crawford A.W., Pino J.D., Beckerle M.C. Biochemical and molecular characterization of the chicken cysteine-rich protein, a developmentally regulated LIM-domain protein that is associated with the actin cytoskeleton. J. Cell Biol. 1994;124:117–127. doi: 10.1083/jcb.124.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadler I., Crawford A.W., Beckerle M.C. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J. Cell Biol. 1992;119:1573–1587. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun N., Huiatt T.W., Robson R.M. Synemin interacts with the LIM domain protein zyxin and is essential for cell adhesion and migration. Exp. Cell Res. 2010;316:491–505. doi: 10.1016/j.yexcr.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Yi J., Kloeker S., Beckerle M.C. Members of the zyxin family of LIM proteins interact with members of the p130Cas family of signal transducers. J. Biol. Chem. 2002;277:9580–9589. doi: 10.1074/jbc.M106922200. [DOI] [PubMed] [Google Scholar]
- 32.Sawada Y., Tamada M., Sheetz M.P. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmeichel K.L., Beckerle M.C. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 34.Nix D.A., Beckerle M.C. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


