Abstract
Apical membrane antigen 1 (AMA1) is essential for malaria parasite invasion of erythrocytes and is therefore an attractive target for drug development. Peptides that bind AMA1 have been identified from random peptide libraries expressed on the surface of phage. Of these, R1, which binds to a hydrophobic ligand binding site on AMA1, was a particularly potent inhibitor of parasite invasion of erythrocytes in vitro. The solution structure of R1 contains a turn-like conformation between residues 5–10. Here the importance of residues in this turn-like structure for binding to AMA1 was examined by site-directed mutagenesis and NMR spectroscopy. The peptide was expressed as a fusion protein following replacement of Met16 by Leu in order to accommodate cyanogen bromide cleavage. This modified peptide (R2) displayed the same affinity for AMA1 as R1, showing that the identity of the side chain at position 16 was not critical for binding. Substitution of Phe5, Pro7, Leu8, and Phe9 with alanine led to significant (7.5- to > 350-fold) decreases in affinity for AMA1. Comparison of backbone amide and CαH chemical shifts for these R2 analogues with corresponding values for R2 showed no significant changes, with the exception of R2(P7A), where slightly larger differences were observed, particularly for residues flanking position 7. The absence of significant changes in the secondary chemical shifts suggests that these mutations had little effect on the solution conformation of R2. The identification of a non-polar region of these peptides containing residues essential for AMA1 binding establishes a basis for the design of anti-malarial drugs based on R1 mimetics.
Keywords: apical membrane antigen 1, malaria, Plasmodium falciparum, peptide expression, NMR, structure, dynamics
INTRODUCTION
Malaria caused by infection with the protozoan parasite Plasmodium falciparum remains one of the world’s most serious public health problems. Young children who have not developed protective adaptive immune responses to the parasite are particularly susceptible to developing life-threatening disease, with more than 1 million dying from malaria each year.1 Vaccination is a particularly cost-effective means of controlling infectious diseases but no vaccine is yet available for malaria.2 Proteins on the surface of the merozoite stage of the parasite are attractive candidates for inclusion in a vaccine directed against the asexual blood-stage parasites, which cause the symptoms of malaria, as they are accessible to plasma antibodies that block parasite invasion of host cells. Plasmodium falciparum apical membrane antigen-1 (PfAMA1) is one such molecule. Immunization with AMA1 induced protection in both simian and rodent models of the human disease.3–6 The disulfide-bond stabilized conformation of the ectodomain was essential for protection, as the antigen was not an effective vaccine after reduction and alkylation.4,5
AMA1 is an 83-kDa antigen synthesized by mature blood-stages of P. falciparum and is located initially in micronemes, secretory apical organelles of sporozoites and merozoites.2,7 Prior to merozoite invasion of host erythrocytes, AMA1 is processed into a 66-kDa product and released onto the merozoite surface.8,9 AMA1 appears to be important for reorientation of the merozoite on the erythrocyte surface prior to invasion.10 Recent evidence indicates that AMA1 forms a complex with several rhoptry neck proteins11–13 as part of the moving junction that propels the merozoite into the erythrocyte.14–17 Its importance is emphasized by the fact that it has not been possible to obtain targeted disruptions of the AMA1 gene that knocked out its function.18
Substantial sequence identity exists among AMA1 from different Plasmodium species7,19–22 and the 16 Cys residues, which form eight intramolecular disulfide bonds23,24 in the ectodomain, are conserved in all sequences. The structures of individual domains of PfAMA1 have been determined in solution,24,25 and crystal structures have been reported for AMA1 from P. vivax 26 and for the first two domains of PfAMA1, alone 27 and in complex with inhibitory antibodies.28,29 A striking feature of the PfAMA1 crystal structure is a large hydrophobic cleft on the surface of AMA1 created by the interaction of domains I and II.27 This cleft is the site of interaction with several inhibitory antibodies, including the monoclonal 1F928 and single-domain shark antibodies, the extended CDR3 loop of which penetrates deep into the AMA1 hydrophobic cleft.29
As a consequence of the critical role of AMA1 in invasion, molecules that bind to and neutralize its activity are of interest not only as tools to probe its structure and function, but also as potential leads in the development of antimalarial therapeutics.30 Peptides that bind to both recombinant and parasite-derived AMA1 have been identified from random peptide libraries expressed on the surface of phage.31–33 Of these peptides, R1 displayed a high binding affinity for AMA1 and was able to inhibit merozoite invasion of malaria parasites cultured in vitro.33 Videomicroscopy of merozoites in the presence of R1 showed that they undergo successful reorientation and attachment to the erythrocyte surface, including forceful pulling, but fail to proceed to invasion.34 Recently, we showed that R1 binds to the PfAMA1 hydrophobic cleft and that binding to this region prevents its interaction with the PfRON complex.13 These properties suggest that R1 might provide a valuable starting point for the development of peptidomimetics that act as malaria antagonists directed at AMA1.
The solution structure of R133 is generally well-ordered over residues 5–10 and 13–17, with a turn-like conformation from residues Pro7-Ser10 and a distorted type I β-turn encompassing Gly13-Met16. In order to determine the structural features and residues on R1 important for binding to AMA1, peptides with point mutations within the turn-like structure were tested for their ability to bind AMA1. NMR spectroscopy was then used to compare the solution structures of strongly and weakly binding peptides. 15N-labelled peptides were also produced as a basis for probing dynamics and interactions with AMA1. Knowledge of the residues involved in ligand binding, together with the orientations of these key functional residues, will facilitate the design of constrained peptides or non-peptidic scaffolds that project side-chain mimetics in an orientation similar to that of the native peptide, and hence bind strongly to the target.
MATERIAL AND METHODS
Peptides and Recombinant AMA1
Peptides were produced recombinantly as fusions to the N-terminal SH2 domain of the intracellular phosphatase SHP2 (SHP2-N).35 Complementary forward and reverse oligonucleotides encoding the peptides were used, incorporating the M16L substitution either alone or together with one of the following mutations: F5A, P7A, L8A, F9A or F5A+F9A. Cleavage of the peptides from the SHP2-N fusion partner was carried out with cyanogen bromide (CNBr) in 70 % (v/v) trifluoroacetic acid instead of 70 % (v/v) formic acid as described by Fairlie et al.,35 because the latter resulted in formylation of the peptides (data not shown). The correct mass of each peptide was verified by mass spectrometry, and the overall purity (> 95 %) confirmed by reversed-phase high performance liquid chromatography (RP-HPLC). The method used for expression of 15N-labelled peptides was similar to that described above except that M9 medium containing 15NH4Cl and glycerol was used in place of rich medium. The expressed 15N-labelled peptides were cleaved and purified as described above. The R1(s) peptide was synthesized by AusPep Pty. Ltd. (Melbourne, Australia) to >76% purity. Recombinant PfAMA1 was produced essentially as described previously.36 This protein corresponded to residues 25–545 of 3D7 AMA1 but contained a glutamate to glycine substitution at position 120 and a glutamate to lysine substitution at position 537.19,37
ELISA Competition Assays
A competitive ELISA was used to test the ability of each peptide to block the binding to recombinant PfAMA1 of R1 displayed on M13 phage particles. Microwell plates (Maxisorp) were coated with 0.2 μg PfAMA1 in 100 μL per well of phosphate buffered saline (PBS) at 4 °C overnight. Wells were blocked for 90 min at room temperature with 300 μL/well of 6 % (w/v) skim milk powder in PBS, then washed three times with PBS containing 0.1 % (v/v) Tween 20 (PBS/Tween). R1 phage particles were added to the wells in the presence of increasing concentrations of the soluble peptides in 100 μL PBS/Tween containing 1 % (w/v) skim milk (PBS/Tween/milk). After 90 min incubation at room temperature with constant shaking, the wells were washed 15 times with PBS/Tween. An HRP-conjugated anti-M13-phage monoclonal antibody (Amersham-Pharmacia), diluted 1:5000 in PBS/Tween/milk, was then added at 100 μL/well and incubated for 30 min at room temperature. The wells were then washed 15 times with PBS/Tween and the substrate o-phenylenediamine/hydrogen peroxidase in citrate-phosphate buffer (50 mM citric acid, 0.1 M Na2HPO4) (Sigma-Aldrich) was added at 100 μL/well. The reaction was allowed to proceed for 10 min at room temperature, after which it was stopped by the addition of 50 μL/well of 1 M HCl. The absorbance at 450 nm was then read using a microtitre plate reader.
To analyse the pH dependence of peptide binding to PfAMA1, an ELISA was performed as described above except that phage particles displaying R1 were serially diluted in either 20 mM sodium phosphate buffer at pH 6.0 or 7.0, or in 50 mM sodium acetate buffer at pH 4.0 or 5.0 together with 0.1 % (v/v) Tween 20 and 1 % (w/v) BSA, and incubated with the PfAMA1 for 3 h at room temperature. R1 phage diluted appropriately in PBS/Tween, pH 7.4, with 1 % (w/v) BSA was included as a control.
NMR Spectroscopy
Two NMR samples were prepared for R2 by dissolving lyophilized peptide in 10 mM sodium acetate buffer containing 5% (v/v) 2H2O and 0.03% (w/v) sodium azide at concentrations of 2 mM (pH 4.1) and 1 mM (pH 4.7). Two-dimensional homonuclear spectra including TOCSY (spin-lock time 50 ms), NOESY (mixing times 75 and 300 ms) and DQF-COSY were acquired at 278 K. For the analogue R2(P7A), two samples were prepared using the same buffer conditions and at similar concentrations and pH to those of R2. Two-dimensional TOCSY (50 ms), NOESY (250 ms) and DQF-COSY spectra were recorded at 278 K using the 2 mM sample at pH 4.1. NMR samples for the remaining R2 analogues, R2(F5A), R2(L8A), R2(F9A) and R2(F5A+F9A), were prepared by dissolving lyophilized materials in 10 mM sodium acetate buffer containing 5% (v/v) 2H2O and 0.03 % (w/v) sodium azide at a concentration of 1 mM and pH 4.1. Chemical shift assignments for all of these analogues were made based on standard 2D homonuclear TOCSY (60 ms), NOESY (300 ms) and DQF-COSY spectra recorded at 278 K. All 2D spectra were recorded on a Bruker DRX600 spectrometer.
Uniformly 15N-labelled R2 and R2(P7A) samples were prepared by dissolving lyophilized 15N-labelled peptides in 10 mM sodium acetate buffer containing 5% (v/v) 2H2O and 0.03% (w/v) sodium azide at a concentration of 1 mM and pH 4.1. Two-dimensional 1H-15N-HSQC and three-dimensional 1H-15N- HSQC edited NOESY (300 ms) spectra were recorded at 279 K using a Bruker Avance-500 equipped with a TXI-cryoprobe. Additional HMQC-J spectra with dephasing times of 45, 60, 80, 100, 120, and 140 ms were also acquired for R2 at 279 K.
1H chemical shifts were referenced to the water resonance and 15N chemical shifts were referenced indirectly using the 15N/1H γ-ratios.38 All spectra were processed using XWINNMR (Version 3.5, Bruker Biospin) and analyzed with XEASY (Version 1.3). Translational diffusion coefficients reported were measured using a pulsed-field-gradient longitudinal eddy-current delay pulse sequence as implemented by Yao et al,39 and the data were processed and analyzed using XWINNMR (Version 3.5, Bruker Biospin) as described previously.40
The uniformly 15N-labelled R2 sample described above was also used for backbone 15N relaxation measurements. Backbone 15N relaxation parameters (T1, T2 and the steady-state 15N-{1H} NOE) of R2 were measured at 279 K on a Bruker Avance-500 spectrometer equipped with a TXI-cryoprobe and at 296 K on a Bruker Avance-500 spectrometer equipped with a BBI probe using pulse sequences similar to those described previously.41 Ten relaxation delays ranging from 10 ms to 1.2 s for T1 and 19.2 to 192 ms for T2 were used for measurements at 279 K, and 12 relaxation delays ranging from 10 ms to 2.2 s for T1 and 15.6 ms to 1.6 s for T2 were used for measurements at 296 K. All T1 and T2 measurements included two duplicate delays for the estimation of peak volume uncertainties.
Structure Calculations
Structures for R2 were calculated using distance restraints derived from 2D NOESY (300 ms) and 3D 1H-15N- HSQC edited NOESY (300 ms) spectra. 3JHNHα coupling constants were obtained from the HMQC-J spectra and converted to dihedral restraints as follows: 3JHNHα > 8 Hz, φ=−120±30°; 3JHNHα < 6 Hz, φ= −60±30°. If a positive φ angle could be excluded on the basis of NOE data, φ angles were restricted to the range −180 to 0°.42 Distance restraints for R2(P7A) were obtained from 2D NOESY (250 ms) and 3D 1H-15N- HSQC edited NOESY (300 ms) spectra. 3JHNHα coupling constants were obtained from a DQF-COSY spectrum and converted to φ dihedral restraints as described above for R2. Amide exchange rates for R2 were monitored by dissolving lyophilized peptide in 2H2O, then recording a series of 1D spectra, followed by TOCSY (60 ms) and NOESY (250 ms) spectra at 600 MHz. No hydrogen bonds were included as structural restraints. Intensities of NOE cross-peaks measured in XEASY were converted to distance restraints using the CALIBA macro supplied with CYANA. Distances obtained from CALIBA were then scaled by a factor of 1.2 before use in the structural calculations to allow for the possible effects of spin diffusion. NOE cross-peaks providing no restraints or representing fixed distances were removed.
Initial structures were calculated using torsion angle dynamics and simulated annealing protocols in CYANA.43 Following optimization for a low target function comprising terms for NOE and dihedral angle violations, the final constraint list from CYANA was then used to calculate a new family of 200 structures using the standard simulated annealing script supplied with XPLOR-NIH.44 The lowest energy structures were selected for water refinement. A water box was built around the peptide structure and then energy minimized based on NOE and dihedral restraints and the geometry of the bonds, angles and impropers. From this set of structures, a final family of 20 structures was chosen for further analysis based on stereochemistry and energy considerations using the programs PROCHECK,45 and MOLMOL.46 Structural figures were prepared using MOLMOL and InsightII (Accelrys, San Diego, USA). The final structures had no experimental distance violations > 0.2 Å or dihedral angle violations > 5°.
Analysis of Relaxation Data
15N relaxation times were determined by fitting the measured peak volumes to three- and two-parameter single exponential decay curves for T1 and T2, respectively, using the program SigmaPlot (Systat Software Inc.). Uncertainties for the relaxation times were determined from the nonlinear fitting. Steady-state {1H}-15N-{1H} NOE values were calculated from peak volume ratios obtained from spectra acquired in the presence and absence of proton saturation.
An initial estimate of the rotational correlation times of R2 was obtained from averaged 15N T1/T2 ratio of a group of residues located near the centre of the polypeptide with relatively large steady-state 15N-{1H} NOE values.47,48 Residue-specific backbone motional parameters were obtained through Modelfree analysis using the Modelfree software package (version 4.0, A. G. Palmer, Columbia University) by fitting experimentally measured relaxation parameters to the original form of the spectral density function.49,50
RESULTS
Peptide Expression and Labelling
A recombinant fusion protein expression system was established to provide 15N-labelled peptides for more detailed NMR studies of peptide structure, dynamics and interactions with PfAMA1, as well as for the generation of mutant peptides to probe structure-function relationships; recombinant peptide production was also cheaper than chemical synthesis for these applications. However, as release of the peptide from its fusion partner required CNBr cleavage of methionyl peptide bonds, Met16 in R1 was substituted with Leu to generate a PfAMA1-binding analogue of R1, referred to hereafter as R2.
The solution structure of R1 was found previously to contain two regions of ordered structure, Phe5-Ser10 and Gly13-His17.33 The first region is particularly hydrophobic and contained the amino acid sequence FLPLF, which resembled the L(I)GFGP region of F1, another peptide that binds with high affinity to PfAMA1 and forms a well-defined β-turn involved in binding to PfAMA1.32 As such, the structural features and residues required for peptide binding to PfAMA1 in this region of R1 were investigated following alanine substitution of residues within the region Phe5-Phe9 (Figure 1). These mutant peptides are referred to as R2(F5A), R2(P7A), R2(L8A), R2(F9A), R2(F5A+F9A). Approximately 3 mg of each unlabelled peptide was produced per litre of culture and their identities were confirmed by mass spectrometry. The purified peptides were > 95 % pure as assessed by analytical RP-HPLC (Figure S1, Supplementary Material). In the case of the isotopically-labelled peptides, approximately 1 mg of each was produced per litre of culture, with a similar level of purity achieved.
FIGURE 1.
Amino acid sequences of R2 and its analogues. The parent sequence R133 is shown, as well as the control peptide R1(s), and mutations incorporated into R1 are highlighted in bold.
Peptide Binding to PfAMA1
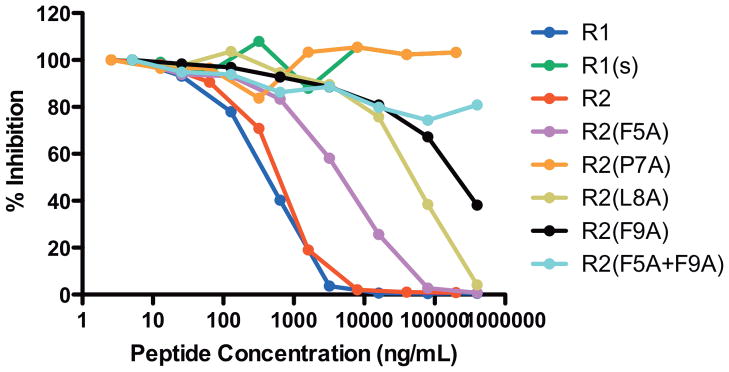
Binding of the R1 and R2 peptides to PfAMA1 was compared by ELISA. A third peptide R1(s) (Figure 1), which is a scrambled and mutated form of the R1 sequence, was included as a negative control. In this assay, R1 and R2 bind to PfAMA1 with very similar affinities, having IC50 values of 177 and 250 nM, respectively (Figure 2). Hence, the Met to Leu substitution at position 16 did not affect peptide binding to PfAMA1, indicating that this residue is not critical for this interaction. As expected, R1(s) did not compete with the R1 phage for binding to PfAMA1.
FIGURE 2.
PfAMA1 binding affinities of peptides. The relative affinities of the peptides were examined by ELISA in which increasing concentrations of peptides were tested for their ability to compete for binding with R1 peptide displayed on M13 phage to plate-bound PfAMA1. The % inhibition indicates the inhibition observed in the presence of peptide compared to in its absence.
We next tested the other peptides containing alanine substitutions within the turn-like structure (Figure 1). Substitution of Phe5 resulted in a 7.5-fold decrease in affinity for PfAMA1, whereas substitution of residues Leu8 and Phe9 both caused more significant decreases (86-fold for L8A and >140-fold for F9A and F5A+F9A) (Figure 2). Mutation of Pro7 alone resulted in the abolition of peptide binding, with no inhibition observed at the highest concentration tested (87 μM), indicating a greater than 350-fold decrease in affinity.
The structures of R2 and R2(P7A) (see below) were determined at slightly acidic pH (pH 4.1–4.7) because they were more soluble in this pH range than around neutral pH, and backbone amide and exchangeable proton resonances give sharper peaks at slightly acidic pH than those at neutral pH. We therefore examined whether R2 would bind to AMA1 at pH values less than pH 7.4, where all previous biological assays had been carried out. The results of ELISA assays of PfAMA1 binding by R1 expressed on phage at different pH values (Figure S2, Supplementary Material) show that R1 phage actually bound with a higher affinity at pH 5.0 and 6.0 than at pH 7.0. Binding at pH 7.0 was similar to that observed under the control conditions in which the phage dilutions were made in PBS at pH 7.4. In contrast, binding of R1 phage to PfAMA1 at pH 4.0 was completely abolished. One-dimensional 1H spectra of R2 over the pH range 4.5–6 showed that its structure, as reflected in 1H chemical shifts was unchanged over this range (Figure S3 and Tables S1–S4, Supplementary Material). This is consistent with the fact that there is only one acidic residue in the sequence, Glu4, which would be expected to have a side-chain pKa ≤ 4.2. Thus, the structures described here are representative of the solution conformation that binds to PfAMA1.
Structure of R2
Chemical shift assignments were made for most backbone and side-chain 1H resonances as well as backbone 15N resonances of R2 (Table S1, Supplementary Material); chemical shifts have been deposited in BioMagResBank (accession number 17253). R2 contains a single proline at residue 7, but there was no evidence of cis-trans isomerization, indicating that this peptide was conformationally constrained in this region. Translational diffusion coefficients measured for R2 were 1.09 × 10−10 m2 s−1 and 2.79 × 10−10 m2 s−1 at 5 and 30 °C, respectively. Comparing these values with those reported previously for peptides of similar length,39 and allowing for viscosity and temperature effects, there was no evidence to suggest that R2 self-associates under these solution conditions.
No long-range NOEs (|i−j| > 4) were observed in the NOESY spectrum and only Glu4, Lys11, and Leu16 had 3JHNHα coupling constants < 6 Hz, (Table S1, Supplementary Material). Changes in the chemical shifts of the amide protons were linear over the temperature range investigated (278–298 K), indicating that there was no conformational change in aqueous solution over this range. The temperature coefficients of backbone amide proton chemical shifts of R2 were determined in order to probe for hydrogen bonding, but no temperature coefficients (Δδ/ΔT) with magnitude smaller than 5.5 ppb K−1 were observed; as a result, no hydrogen bond restraints were included in the structure calculations. However, as for the R1 peptide,33 amide exchange experiments showed that most of the backbone amide protons were still visible after 20 h in a TOCSY spectrum. This extent of protection for so many amides from solvent exchange is unusual for a linear peptide in aqueous solution.
Initial structures for R2 were calculated using distance and dihedral angle restraints in CYANA prior to optimization in XPLOR-NIH. These structures were then further refined and energy minimized in a box of water. No distance violations > 0.2 Å or dihedral violations > 5o were observed in the final 20 structures. Structural restraints used in the calculation and structural statistics for the final 20 structures are summarized in Table I. Plots of backbone and side-chain angular order parameters, S, are shown in Figure S4 (Supplementary Material).
Table I.
Summary of structural restraints used in the structure calculations and structural statistics for the 20 final structures of R2 and R2(P7A)
| R2 | R2(P7A) | |
|---|---|---|
| No of distance restraints | ||
| Total | 217 | 118 |
| Intra (i = j) | 95 | 53 |
| Sequential (|i − j| = 1) | 102 | 58 |
| Short/medium range (1 < |i − j| ≤ 5) | 20 | 7 |
| Dihedral restraints | 18 | 0 |
| Energiesa | ||
| Ecdih (kcal mol−1) | 0.14 ± 0.17 | 0 |
| ENOE (kcal mol−1) | 14.99 ± 1.85 | 0.84 ± 0.47 |
| Deviations from ideal geometryb | ||
| Bonds (Å) | 0.0030 ± 0.0002 | 0.0012 ± 0.0001 |
| Angles (°) | 0.55 ± 0.02 | 0.43 ± 0.01 |
| Impropers (°) | 0.36 ± 0.01 | 0.32 ± 0.01 |
| RMS Deviations of site I (Å)c | ||
| All heavy atoms | 2.22 ± 0.61 | 2.49 ± 0.65 |
| Backbone heavy atoms (N, Cα, C′) | 0.96 ± 0.36 | 1.12 ± 0.38 |
| Ramachandran plot (%)d | ||
| Most favoured | 22.2 | 23.8 |
| Allowed | 68.1 | 55.6 |
| Additional allowed | 8.1 | 16.2 |
| Disallowed | 1.6 | 4.4 |
Energies were calculated from a square well potential with force constants of 50 kcal mol−1 Å2. Ecdih was zero for R2(P7A) as no dihedral restraints were included in the structure calculation.
The values for the bonds, angles, and impropers show the deviations from ideal values based on perfect stereochemistry.
Mean pairwise RMS deviations over the backbone heavy atoms (N, Cα, C′) of residues 5–10 for R2 and 4–7 for R2(P7A).
As determined by the program PROCHECK-NMR for all residues except Gly and Pro.45
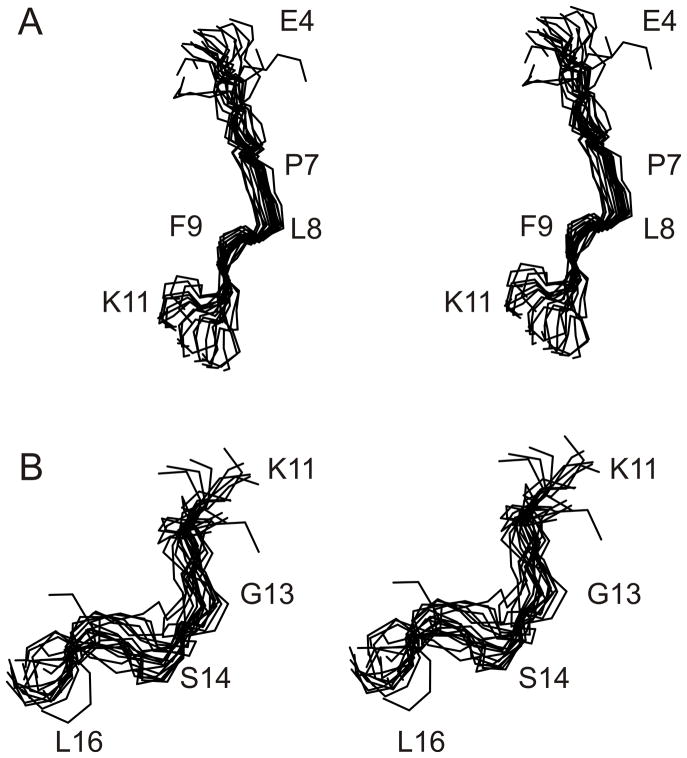
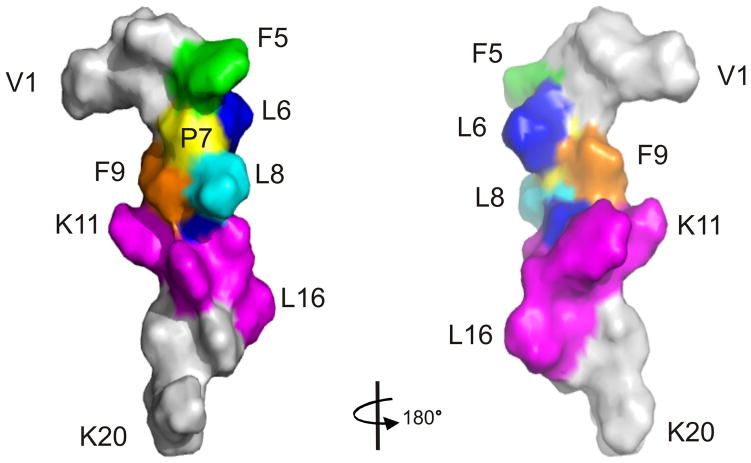
As no long-range NOEs were observed in NOESY spectra and the number of medium-range NOEs was low (Table 1), the ensemble of R2 structures had a global backbone RMSD >2 Å. However, while R2 did not adopt a stable conformation over its entire length in aqueous solution, like the R1 peptide,33 it contained two regions of local structure, referred to herein as Site I (residues 5–10) and Site II (residues 11–16) (Figure 3). When the ensemble of R2 structures was superimposed over residues 5–10 and 11–16, mean pairwise backbone RMSDs of 0.96 ± 0.32 and 1.71 ± 0.53 Å, respectively, were obtained. In the first region of local structure, Leu6-Phe9 adopt a turn-like conformation, with Pro7 and Leu8 occupying the i+1 and i+2 positions. Residues 11–16, which contain the second region of local structure, also form a turn-like conformation encompassing residues Phe12-Arg15. A surface representation of the R2 structure is shown in Figure 4.
FIGURE 3.
Stereo view of the family of 20 lowest energy structures of R2. (A) residues 4–11 and (B) residues 11–16, with the family of 20 structures superimposed over the backbone heavy atoms (N, Cα, C) of residues 5–10 and 11–16, respectively.
FIGURE 4.
Surface representation of R2 with mutated residues highlighted. For Site I (residues 5–10), F5 is coloured in green, L6 in blue, P7 in yellow, L8 in cyan, F9 in orange and S10 in blue, respectively. Residues 11–16 of Site II are coloured in magenta with the rest of the peptide in grey.
Backbone Dynamics of R2
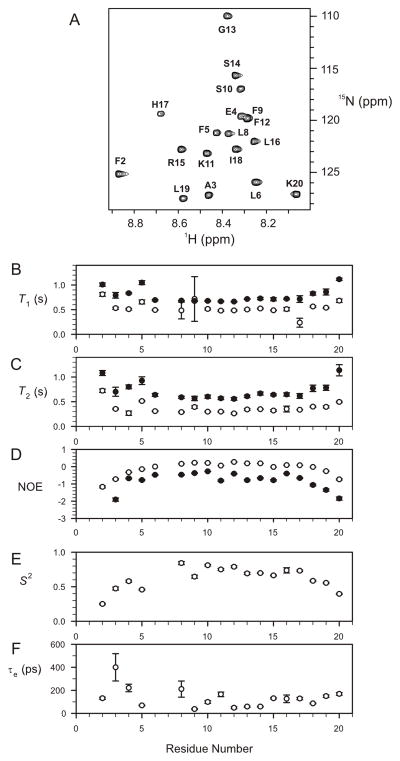
In order to obtain a more detailed understanding of the solution conformation of R2, 15N relaxation measurements were carried out to characterize its backbone dynamics. 15N spin-lattice time, T1, spin-spin relaxation times, T2, and steady-state {1H}-15N NOE values for R2 at 6 and 23 °C are summarized in Figure 5. As expected, R2 tumbled faster at the higher temperature, as reflected in the longer T2 and lower {1H}-15N NOE values. The distribution of T1, T2 and NOE values across the sequence are similar at the two temperatures indicating the relative backbone motion within the molecule may not change much as the temperature increases. At both temperatures, the T1 and T2 relaxation times were longer for residues 1–5 and 18–20 compared with residues 6–17 in the central region of the peptide. A similar pattern was also observed for the {1H}-15N NOE, with lower values at the N- and C-terminal regions than for residues 6–17. This indicates that the backbones atoms in residues 6–17 are less mobile on the picosecond to nanosecond time scale compared with the termini, and further suggests that the two regions of well-defined structure, residues 5–10 and 11–16, could be regarded as a single structured core. Average T1 and T2 values over residues 6–16 (except for Pro7) were 0.51 ± 0.07 and 0.32 ± 0.04 s at 279 K and 0.69 ± 0.02 and 0.61 ± 0.04 s at 296 K, respectively. Correlation times for global reorientation of R2 at both temperatures were estimated to be 2.5 and 0.85 ns, corresponding to T1/T2 ratios of 1.59 and 1.14, respectively. Further analysis was carried out using Modelfree formalism for relaxation data measured at 6 279 K only (although this was not attempted for relaxation data at 296 K because of the relatively fast global reorientation time of 0.85 ns). The results of Modelfree analysis at 279 K are summarised in Figure 5. The average S2 value for all 17 fitted backbone amides is 0.63 ± 0.16 whereas that for residues 6–16 (except for Pro7 and Leu6, the latter was not fitted) is 0.73 ±0.06.
FIGURE 5.
Backbone relaxation data for 15N-labelled R2 peptide. 1H-15N HSQC spectrum of R2 (A), Summary of backbone 15N relaxation parameters T1 (B), T2 (C), and NOE (D) values of R2 measured at 279 K (open circles) and 296 K (filled circles), respectively. Order parameters (E) and internal correlation times (F) for R2 derived from Modelfree analysis using backbone 15N relaxation parameters at 279 K.
Chemical Shift Assignments for R2(F5A), R2(P7A), R2(L8A), R2(F9A) and R2(F5A+F9A)
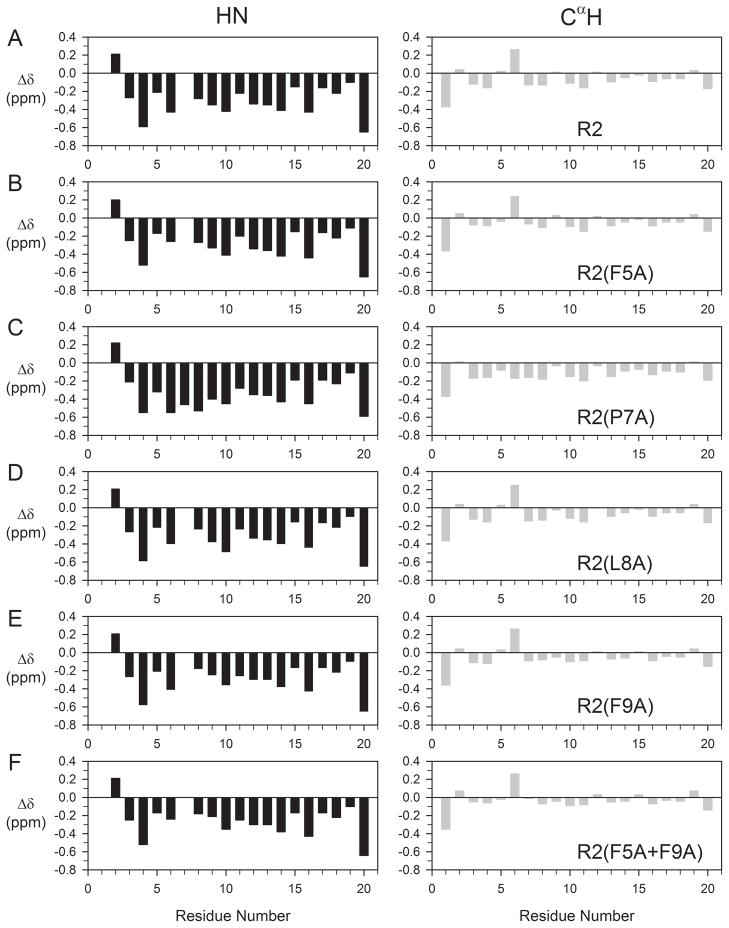
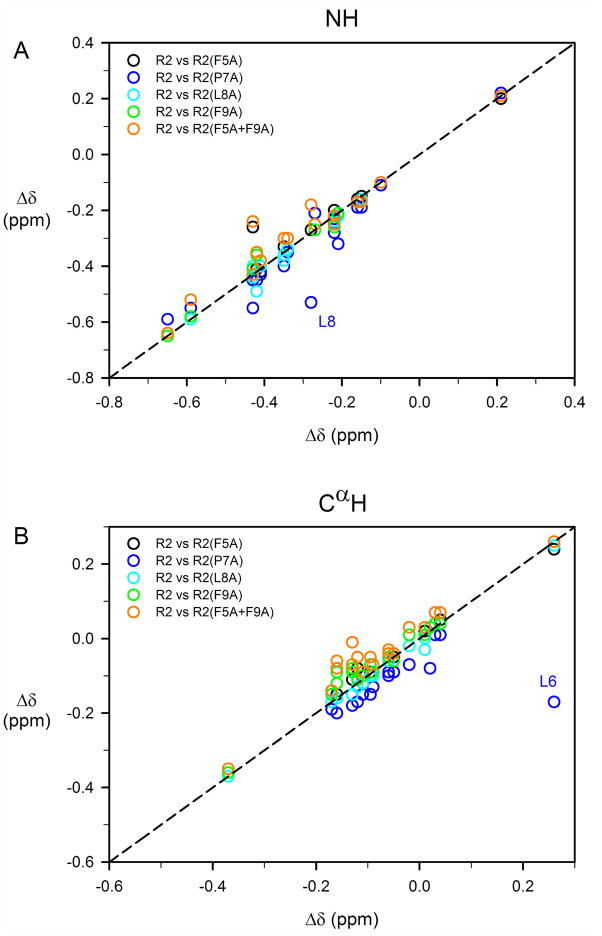
As chemical shifts are a highly sensitive monitor of local structural features in peptides, chemical shift assignments were also made for backbone and most side-chain 1H of all R2 analogues examined here (Tables S5–S9 and Figure S5, Supplementary Material). Chemical shifts for the backbone amide and CαH resonances of these mutant peptides were compared to values of R2; in order to facilitate this comparison, deviations of these chemical shifts from random coil values,51 Δδ, were calculated (Figure 6). Correlation plots of chemical shift deviations from random coil values for amide and CαH resonances between R2 peptide and its analogues, excluding mutated residue(s), are shown in Figure 7. As can be seen from Figures 6 and 7, no significant changes in the secondary chemical shifts (Δδ) were observed for these R2 analogues, with the exception of R2(P7A), where slightly larger differences were observed, particularly for residues flanking position 7. This lack of significant changes in the secondary chemical shifts suggests that these mutations have little effect on the overall solution conformation of R2.
FIGURE 6.
Deviation of 1H chemical shifts (HN, left panel; CαH, right panel) from random coil values for R2 peptide (A) and its analogues R2(F5A), R2(P7A), R2(L8A), R2(F9A), and R2(F5A+F9A), respectively, (B–F). The deviations were calculated using random coil values reported by Merutka et al.51 For Gly13, values of CαH1 and CαH2 were averaged.
FIGURE 7.
Correlation plots of chemical shift deviations from random coil values for amide and CαH resonances between R2 peptide and its analogues, excluding mutated residue(s). Correlation coefficients for amide 1H chemical shifts are 0.98, 0.93, 0.99, 0.99, and 0.96 between R2 and analogues R2(F5A), R2(P7A), R2(L8A), R2(F9A), and R2(F5A+F9A) respectively, (A). Corresponding correlation coefficients for Cα 1H are 0.99, 0.67, 0.99, 0.99, and 0.97 (B). Two heavily perturbed chemical shifts (and amide 1H of L8 and Cα 1H of L6) from R2(P7A) are labeled in (A) and (B), respectively. After excluding the amide 1H of L8, the correlation coefficient for amide 1H between R2 and R2(P7A) is 0.97. Similarly, after excluding L6, the correlation coefficient for CαH between R2 and R2(P7A) is 0.98.
Structure of R2(P7A)
To further investigate whether the loss of binding affinity in R2(P7A) correlated with potential structural changes, we also determined its structure for comparison with that of R2. As with R2, no long-range NOEs (|i−j| > 4) were observed in the NOESY spectrum. However, in contrast to R2, where Glu4, Lys11, and Leu16 had 3JHNHα coupling constants < 6 Hz, all 3JHNHα in R2(P7A) were between 6 and 8 Hz (Table S6, Supplementary Material). Structural restraints used in the calculation of the final family of 20 structures of R2(P7A) and structural statistics are summarized in Table I. Plots of backbone and side-chain angular order parameters, S, are shown in Figure S6 (Supplementary Material).
As for R2, the ensemble of R2(P7A) had a global backbone RMSD value >2 Å, implying that this analogue also did not adopt a stable conformation over its entire length. Superposition of the family of structures revealed that, although residues 11–16 retained local structure similar to that of Site II in R2, the turn-like conformation from residues 5–10 observed in R2 was absent, with a RMSD of >2 Å across those residues. R2(P7A) contained only two small regions of local structure encompassing residues 4–7 and 16–19 (Figure S7, Supplementary Material), with mean pairwise backbone RMSDs of 1.12 ± 0.38 and 1.26 ± 0.49 Å, respectively.
DISCUSSION
The peptide expression system developed by Fairlie et al.35 provided an efficient and cost-effective means of producing both unlabelled and 15N-labelled peptides for this study. A limitation of this system is the use of CNBr cleavage for the separation of the peptide from its fusion partner, SHP2-N. As CNBr cleaves peptide bonds on the C-terminal side of Met residues, it was necessary to modify Met16 of R1 in order to avoid cleavage within the peptide itself. In the present study, Met16 was replaced by Leu to produce the analogue R2, the binding affinity of which for PfAMA1 was essentially identical to that of the parent peptide, R1.
As with R1, the solution structure of R2 did not adopt a stable conformation over its entire length, although the absence of cis-trans isomerization at Pro7 implies a constrained conformation in this region of the peptide. Consideration of RMSD values and angular order parameters suggested that R2 contained two structured regions, encompassing residues 5–10 and 11–16, respectively. In the first of these, Leu6-Phe9 appear to adopt a turn-like conformation, with Pro7 and Leu8 occupying the i+1 and i+2 positions, and in the second residues 11–16 form a turn-like conformation encompassing residues Phe12-Arg15. The advantage of having an efficient peptide expression system is that the solution conformation could be supplemented by residue-specific NMR relaxation measurements on 15N-labelled peptide. Backbone 15N relaxation parameters and subsequent motional parameters resulting from Modelfree analysis showed that the central region of R2, residues 6–17, is much less flexible than both termini. It seems more appropriate, therefore, to regard R2 as containing a structured core centered on turn-like structures involving residues 6–9 and 12–15 than as having two separate and distinct structured regions. The subdivision of this structured region into better defined N- and C-terminal halves on the basis of RMSD values is presumably a reflection of a lack of experimentally-derived structural restraints around residues 10–11 (although there is a slight reduction in the {1H}-15N NOE for Lys11, consistent with slightly decreased order at this position).
The turn-like conformation centered on residues 7–8 appears to be stabilized by interactions between Pro7 and Phe9 and to some extent Phe5, as observed in the closest-to-average structure for R2, and provides an explanation for the lack of cis-trans isomerization, with Pro7 being held preferentially in a trans conformation. This region of R2 is reminiscent of the β-turn forming region (LGFGP) of a previously-identified PfAMA1 binding-peptide, F1, which was shown to bind to PfAMA1 and inhibit erythrocyte invasion by merozoites.31,32 However, R1 was at least 10-fold more effective at inhibiting phage-displayed F1 peptide than synthetic F1 itself.33 It appears that turn-like structures displaying hydrophobic side chains target a common binding region on PfAMA1. These hydrophobic side chains presumably make important interactions with residues in the hydrophobic cleft of PfAMA1.
In considering the degree of structural stabilization in R1 and R2 in solution and the extent to which structural features observed in solution might be retained in the PfAMA1-bound state, the residue-specific backbone relaxation data are again valuable as they afford an independent assessment of structural stability, one that is not influenced by NOEs. A useful comparison can also be made with backbone relaxation data for a recombinant peptide of similar size and structure corresponding to the conserved N-terminal domain of merozoite surface protein 2 (MSP2); this domain contains 25 residues, and the recombinant form includes three additional residues at the N-terminus.52 The overall pattern of backbone relaxation parameters along the polypeptide chain for these two polypeptides is very similar, with longer transverse relaxation times, T2, and more negative {1H}-15N NOE values observed for the termini. The R2 peptide yielded average T1, T2 and {1H}-15N NOE values of 0.54 ± 0.12 s, 0.37 ± 0.11 s, and −0.11 ± 0.40, respectively at 279 K. Corresponding values for MSP21–25 were 0.59 ± 0.04 s, 0.25 ± 0.02 s, and −0.05 ± 0.005 at 278 K.52 The longer T2 and more negative {1H}-15N NOE values for the R2 peptide are consistent with its being eight residues shorter than MSP21–25. The most positive {1H}-15N NOE values in R2 were 0.27, compared with 0.3 in MSP21–25. The N-terminal region of MSP2 also contains some more ordered, turn-like structure centred on the hydrophobic residues Phe11 and Ile12,52–54 although the limited degree of structural stabilization in both R2 and MSP21–25, as reflected in their NMR chemical shifts, 1H-1H NOE values and backbone 15N relaxation parameters, is such that the stabilizing intramolecular interactions are likely to be readily overcome by stronger intermolecular interactions (with PfAMA1 in the case of R2).
The highly hydrophobic pentapeptide segment encompassing residues 5–9 (FLPLF) is essential for PfAMA1binding. Substitution of Pro7 with Ala in R2(P7A) resulted in local changes in conformation, as reflected in the backbone chemical shifts of the flanking Leu residues. Interestingly, the R2(P7A) structure contained two regions of local structure involving residues 4–7 and 16–19 that were not observed in either R1 or R2. Disruption of the hydrophobic turn apparently allows the peptide to sample alternative (non-functional) conformations. Substitution of Pro7 for Ala produces consistent upfield CαH shifts suggestive of a slight tendency toward helical structure in this peptide, but the effect is slight. Substitution of Pro7 with alanine resulted in essentially complete abolition (i.e. >350-fold decrease) of binding, and this loss of affinity probably reflects a combination of local structural changes associated with this mutation, as well as the involvement of Pro7 in direct interactions with the hydrophobic cleft of PfAMA1.
Significant decreases in binding were also observed following substitution of other nearby residues, with mutation of residues Leu8 and Phe9 having significant but comparable effects and mutation of Phe5 the least effect. None of these mutations resulted in any significant disruption of the peptide structure, as monitored by NMR. This indicates that the effects of mutations of Phe5, Leu8 and Phe9 on the interaction with PfAMA1 reflect principally the disruption of hydrophobic interactions between the peptide and PfAMA1, with conformational changes in the peptide playing a minor role.
Our observations complement those of a recent study of the effect of backbone N-methylation of R1 on its affinity for PfAMA1 and ability to inhibit parasite invasion of red blood cells.55 NMR analyses of mono-, di- and tri-N-methylated analogues showed that conformational perturbations were essentially local, with N-methylation neither nucleating local structure in, nor significantly perturbing the conformation of, R1. The most significant enhancement of affinity for PfAMA1 and ability to inhibit parasite invasion of red blood cell was produced by N-methylation at Leu8, consistent with the idea that the hydrophobic nature of this region of R1 is a key factor in mediating binding. N-methylation at Phe5, Leu6 or Phe9 caused slight to moderate enhancements in binding affinity; intriguingly, in the case of Phe5 and Phe9, there were slight reductions in the ability of these modified peptides to inhibit invasion at a fixed concentration, hinting at the possibility of a decoupling of binding affinity and invasion inhibition efficacy for some analogues. N-methylation of Lys11, Phe12, Gly13, and Arg15 resulted in the most significant reductions in PfAMA1binding affinity and invasion inhibition activity, which may be a consequence of local changes in conformation and/or disruption of interactions with PfAMA1 by the introduced methyl groups.
In summary, the results of our studies demonstrate that the cluster of hydrophobic residues in the turn-like structure encompassing residues 5–9 in the R1 and R2 peptides is critical for binding to PfAMA1. This finding has important implications for the design of anti-malarial drugs based on the R1 sequence as such mimetics would be required to incorporate a chemically and structurally similar feature.
Supplementary Material
Acknowledgments
We are grateful to David Keizer, Mark Hinds and Jeff Babon for valuable discussions and to Thao Ngyuen for assistance with production of the R2(P7A) peptide. This work was supported in part by grants to RSN, RFA and MF from the Australian National Health and Medical Research Council of Australia (Project grant 305525 and IRIISS grant 361646) and the National Institutes of Health, Grant NIH RO1AI59229, as well as a Victorian State Government OIS grant.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crewther PE, Culvenor JG, Silva A, Cooper JA, Anders RF. Exp Parasitol. 1990;70:193–206. doi: 10.1016/0014-4894(90)90100-q. [DOI] [PubMed] [Google Scholar]
- 3.Collins WE, Pye D, Crewther PE, Vandenberg KL, Galland GG, Sulzer AJ, Kemp DJ, Edwards SJ, Coppel RL, Sullivan JS, et al. Am J Trop Med Hyg. 1994;51:711–719. doi: 10.4269/ajtmh.1994.51.711. [DOI] [PubMed] [Google Scholar]
- 4.Crewther PE, Matthew ML, Flegg RH, Anders RF. Infect Immun. 1996;64:3310–3317. doi: 10.1128/iai.64.8.3310-3317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anders RF, Crewther PE, Edwards S, Margetts M, Matthew ML, Pollock B, Pye D. Vaccine. 1998;16:240–247. doi: 10.1016/s0264-410x(97)88331-4. [DOI] [PubMed] [Google Scholar]
- 6.Anders RF, Adda CG, Foley M, Norton RS. Hum Vaccine. 2010;6:39–53. doi: 10.4161/hv.6.1.10712. [DOI] [PubMed] [Google Scholar]
- 7.Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, Silva A, Anders RF, Kemp DJ. Mol Cell Biol. 1989;9:3151–3154. doi: 10.1128/mcb.9.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narum DL, Thomas AW. Mol Biochem Parasitol. 1994;67:59–68. doi: 10.1016/0166-6851(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 9.Howell SA, Withers-Martinez C, Kocken CH, Thomas AW, Blackman MJ. J Biol Chem. 2001;276:31311–31320. doi: 10.1074/jbc.M103076200. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. Infect Immun. 2004;72:154–158. doi: 10.1128/IAI.72.1.154-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besteiro S, Michelin A, Poncet J, Dubremetz JF, Lebrun M. PLoS Pathog. 2009;5:e1000309. doi: 10.1371/journal.ppat.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins CR, Withers-Martinez C, Hackett F, Blackman MJ. PLoS Pathog. 2009;5:e1000273. doi: 10.1371/journal.ppat.1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richard D, MacRaild CA, Riglar DT, Chan JA, Foley M, Baum J, Ralph SA, Norton RS, Cowman AF. J Biol Chem. 2010;285:14815–14822. doi: 10.1074/jbc.M109.080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander DL, Mital J, Ward GE, Bradley P, Boothroyd JC. PLoS Pathog. 2005;1:e17. doi: 10.1371/journal.ppat.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander DL, Arastu-Kapur S, Dubremetz JF, Boothroyd JC. Eukaryot Cell. 2006;5:1169–1173. doi: 10.1128/EC.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narum DL, Nguyen V, Zhang Y, Glen J, Shimp RL, Lambert L, Ling IT, Reiter K, Ogun SA, Long C, Holder AA, Herrera R. Infect Immun. 2008;76:4876–4882. doi: 10.1128/IAI.01717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morahan BJ, Sallmann GB, Huestis R, Dubljevic V, Waller KL. Exp Parasitol. 2009;122:280–288. doi: 10.1016/j.exppara.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Triglia T, Healer J, Caruana SR, Hodder AN, Anders RF, Crabb BS, Cowman AF. Mol Microbiol. 2000;38:706–718. doi: 10.1046/j.1365-2958.2000.02175.x. [DOI] [PubMed] [Google Scholar]
- 19.Marshall VM, Peterson MG, Lew AM, Kemp DJ. Mol Biochem Parasitol. 1989;37:281–283. doi: 10.1016/0166-6851(89)90160-6. [DOI] [PubMed] [Google Scholar]
- 20.Peterson MG, Nguyen-Dinh P, Marshall VM, Elliott JF, Collins WE, Anders RF, Kemp DJ. Mol Biochem Parasitol. 1990;39:279–283. doi: 10.1016/0166-6851(90)90067-v. [DOI] [PubMed] [Google Scholar]
- 21.Waters AP, Thomas AW, Deans JA, Mitchell GH, Hudson DE, Miller LH, McCutchan TF, Cohen S. J Biol Chem. 1990;265:17974–17979. [PubMed] [Google Scholar]
- 22.Waters AP, Thomas AW, Mitchell GH, McCutchan TF. Mol Biochem Parasitol. 1991;44:141–144. doi: 10.1016/0166-6851(91)90230-4. [DOI] [PubMed] [Google Scholar]
- 23.Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, Anders RF. J Biol Chem. 1996;271:29446–29452. doi: 10.1074/jbc.271.46.29446. [DOI] [PubMed] [Google Scholar]
- 24.Nair M, Hinds MG, Coley AM, Hodder AN, Foley M, Anders RF, Norton RS. J Mol Biol. 2002;322:741–753. doi: 10.1016/s0022-2836(02)00806-9. [DOI] [PubMed] [Google Scholar]
- 25.Feng ZP, Keizer DW, Stevenson RA, Yao S, Babon JJ, Murphy VJ, Anders RF, Norton RS. J Mol Biol. 2005;350:641–656. doi: 10.1016/j.jmb.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Pizarro JC, Vulliez-Le Normand B, Chesne-Seck ML, Collins CR, Withers-Martinez C, Hackett F, Blackman MJ, Faber BW, Remarque EJ, Kocken CH, Thomas AW, Bentley GA. Science. 2005;308:408–411. doi: 10.1126/science.1107449. [DOI] [PubMed] [Google Scholar]
- 27.Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. Proc Natl Acad Sci U S A. 2005;102:12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, Foley M, Anders RF, Batchelor AH. PLoS Pathog. 2007;3:1308–1319. doi: 10.1371/journal.ppat.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson KA, Streltsov VA, Coley AM, Dolezal O, Hudson PJ, Batchelor AH, Gupta A, Bai T, Murphy VJ, Anders RF, Foley M, Nuttall SD. Structure. 2007;15:1452–1466. doi: 10.1016/j.str.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 30.MacRaild CA, Anders RF, Foley M, Norton RS. Current Topics in Medicinal Chemistry. 2010. in press. [DOI] [PubMed] [Google Scholar]
- 31.Li F, Dluzewski A, Coley AM, Thomas A, Tilley L, Anders RF, Foley M. J Biol Chem. 2002;277:50303–50310. doi: 10.1074/jbc.M207985200. [DOI] [PubMed] [Google Scholar]
- 32.Keizer DW, Miles LA, Li F, Nair M, Anders RF, Coley AM, Foley M, Norton RS. Biochemistry. 2003;42:9915–9923. doi: 10.1021/bi034376b. [DOI] [PubMed] [Google Scholar]
- 33.Harris KS, Casey JL, Coley AM, Masciantonio R, Sabo JK, Keizer DW, Lee EF, McMahon A, Norton RS, Anders RF, Foley M. Infect Immun. 2005;73:6981–6989. doi: 10.1128/IAI.73.10.6981-6989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treeck M, Zacherl S, Herrmann S, Cabrera A, Kono M, Struck NS, Engelberg K, Haase S, Frischknecht F, Miura K, Spielmann T, Gilberger TW. PLoS Pathog. 2009;5:e1000322. doi: 10.1371/journal.ppat.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fairlie WD, Uboldi AD, De Souza DP, Hemmings GJ, Nicola NA, Baca M. Protein Expr Purif. 2002;26:171–178. doi: 10.1016/s1046-5928(02)00521-1. [DOI] [PubMed] [Google Scholar]
- 36.Hodder AN, Crewther PE, Anders RF. Infect Immun. 2001;69:3286–3294. doi: 10.1128/IAI.69.5.3286-3294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saul A, Lawrence G, Allworth A, Elliot S, Anderson K, Rzepczyk C, Martin L, Taylor D, Eisen DP, Irving DO, et al. Vaccine. 2005;23:3076–3082. doi: 10.1016/j.vaccine.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 38.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 39.Yao S, Howlett GJ, Norton RS. J Biomol NMR. 2000;16:109–119. doi: 10.1023/a:1008382624724. [DOI] [PubMed] [Google Scholar]
- 40.Yao S, Babon JJ, Norton RS. Biophys Chem. 2008;136:145–151. doi: 10.1016/j.bpc.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 42.Ludvigsen S, Andersen KV, Poulsen FM. J Mol Biol. 1991;217:731–736. doi: 10.1016/0022-2836(91)90529-f. [DOI] [PubMed] [Google Scholar]
- 43.Herrmann T, Güntert P, Wüthrich K. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 44.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 46.Koradi R, Billeter M, Wüthrich K. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 47.Kay LE, Torchia DA, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 48.Yao S, Hinds MG, Norton RS. J Magn Reson. 1998;131:347–350. doi: 10.1006/jmre.1998.1382. [DOI] [PubMed] [Google Scholar]
- 49.Lipari G, Szabo A. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 50.Lipari G, Szabo A. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 51.Merutka G, Dyson HJ, Wright PE. J Biomol NMR. 1995;5:14–24. doi: 10.1007/BF00227466. [DOI] [PubMed] [Google Scholar]
- 52.Low A, Chandrashekaran IR, Adda CG, Yao S, Sabo JK, Zhang X, Soetopo A, Anders RF, Norton RS. Biopolymers. 2007;87:12–22. doi: 10.1002/bip.20764. [DOI] [PubMed] [Google Scholar]
- 53.Yang X, Adda CG, Keizer DW, Murphy VJ, Rizkalla MM, Perugini MA, Jackson DC, Anders RF, Norton RS. J Pept Sci. 2007;13:839–848. doi: 10.1002/psc.910. [DOI] [PubMed] [Google Scholar]
- 54.Yang X, Adda CG, Macraild CA, Low A, Zhang X, Zeng W, Jackson DC, Anders RF, Norton RS. Biochimie. 2010;92:1287–1295. doi: 10.1016/j.biochi.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris KS, Casey JL, Coley AM, Karas JA, Sabo JK, Tan YY, Dolezal O, Norton RS, Hughes AB, Scanlon D, Foley M. J Biol Chem. 2009;284:9361–9371. doi: 10.1074/jbc.M808762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.