Abstract
Objective
Simulated samples have been widely used in the development of efficient statistical methods identifying genetic variants that predispose to human genetic diseases. Although it is well known that natural selection has a strong influence on the number and diversity of rare genetic variations in human populations, existing simulation methods are limited in their ability to simulate multi-locus selection models with realistic distributions of the random fitness effects of newly arising mutants.
Methods
We developed a computer program to simulate large populations of gene sequences using a forward-time simulation approach. This program is capable of simulating several multi-locus fitness schemes with arbitrary diploid single-locus selection models with random or locus-specific fitness effects. Arbitrary quantitative trait or disease models can be applied to the simulated populations from which individual- or family-based samples can be drawn and analyzed.
Results
Using realistic demographic and natural selection models estimated from empirical sequence data, datasets simulated using our method differ significantly in the number and diversity of rare variants from datasets simulated using existing methods that ignore natural selection. Our program thus provides a useful tool to simulate datasets with realistic distributions of rare genetic variants for the study of genetic diseases caused by such variants.
Key Words: Rare variants, Sequence, Simulation
Introduction
Increasing evidence has suggested that rare and generally deleterious genetic variants might have a strong impact on the risk of not only rare Mendelian diseases, but also many common human diseases and related traits [1,2]. Because genome-wide association studies are inefficient in identifying rare variants that predispose to common diseases, whole-genome and whole-exome sequencing in families or individuals with extreme traits have been proposed to identify the disease-causing variants of these diseases [3].
Simulated datasets have been used to explore the role of genetic variants in human genetic diseases and to evaluate the performance of statistical methods that are designed to detect these variants [4,5]. Under the Common Disease/Many Rare Variants hypothesis [1,4,5,6,7,8], a common disease might be caused by multiple rare variants that are under relatively strong selection pressures. A realistic selection model that reflects the distribution of random fitness effects among newly arising mutations is therefore important for simulating rare variants. Unfortunately, existing coalescent-based simulation programs can only simulate neutral alleles (e.g. ms [9]) or natural selection on a single locus (e.g. SelSim [10]), and existing forward-time simulation programs only focus on a fixed number of mutations that are under selection [11], simulate sequences as independent nucleotides that are under selection [5], or use simple selection models with random fitness effects [12,13,14].
We developed the computer program srv (Simulator of Rare Variants) to simulate large populations of gene sequences using an evolutionary process with realistic demographic, mutation and multi-locus selection models with random fitness effects. The simulated populations can be used to study the impact of demographic and natural selection models on the number and frequencies of mutants (i.e. derived alleles), and, more importantly, generate samples for the development of statistical gene mapping methods for detecting rare variants that predispose to human genetic diseases.
Methods
Our method simulates one or more chromosomal regions that represent genes on the human genome. During a long evolutionary process, single nucleotide mutants are introduced to these genes and cause changes in the fitness of individuals who carry these mutants. The number and frequency of these mutants in the simulated population are affected by mutation, natural selection, and population demography. A gene typically spans from 10,000 to 100,000 base pairs. Although genetic recombination is usually negligible in such short regions, recombination at a constant rate (per base pair per generation) can be used to recombine parental chromosomes before they are transmitted to offspring.
We assume a multi-stage demographic model where a population of size Nt at stage t expands exponentially or reduces instantly to size Nt + 1 in Gt generations (t = 0, …, m − 1, where m is number of stages). More complex demographic models can be approximated with finer stages (e.g. exact population size at each generation). Optionally, the population can be split into several subpopulations with given proportions at a specified generation. An island model is used to migrate individuals between the resulting subpopulations.
We use two diallelic mutation models to mutate alleles at all nucleotides at a fixed mutation rate. A finite-sites model is used by default in which all mutations such as forward, recurrent and back mutations are allowed. A mutation event can happen at a locus with existing mutant, and will mutate a wild-type allele to a mutant allele, and vice versa [12]. Alternatively, a pseudo-infinite-sites model can be used to mimic an infinite-sites mutation model by relocating a mutant if it hits a locus with existing mutants. Although the latter model is less realistic, datasets simulated in this model have the property that all mutants in the simulated population can be traced back to single mutation events.
The fitness effect of a mutant at locus i is modeled by fitness values 1, 1 – hisi, and 1 – si for individuals with 0, 1 and 2 mutants at this locus, respectively. Although the selection coefficients si are usually positive, zero or negative values can be used to simulate neutral loci or loci under positive selection. The dominance coefficient hi is frequently set to 0.5 for an additive model, or 0 for a recessive model. The selection and dominance coefficients at each locus are usually drawn from a random distribution but locus-specific coefficients can be specified using user-provided functions. This program incorporates multiple sets of selection parameters estimated from human genome data using different demographic models [5,15,16,17]. For example, using a mixed gamma distribution, a mutant can have a selection coefficient of zero (neutral alleles) or a random number drawn from a gamma distribution ranging from 0.00001 to 0.1 [5].
Instead of simulating each locus separately [5], we assign an overall fitness value to an individual if he or she carries mutants at more than one locus. Either an exponential (f = expΣ(1 – fi), where fi is the fitness value at locus i, and f is the overall fitness value), a multiplicative (f = Π fi), or an additive (f = max[0, 1 – Σ(1 – fi)]) model can be used. Although these models are different from the models used in SFS_CODE where the fitness effect of multiple mutants (not genotypes) are combined either additively or multiplicatively [12], the differences between these models are small when si is small and hi = 0.5 for all loci.
Our program produces several output files, including a mutant file that contains mutants of all individuals, and a map file that contains the location, frequency, and selection and dominance coefficients of each mutant. Mutation events that happened during the evolutionary process can also be saved and used to trace the age of mutants. The simulated population can be post-processed to generate samples for particular studies. For example, a quantitative trait model can be applied to the simulated populations from which individuals with extreme trait values can be sampled. Pedigree samples can be generated by evolving the simulated population for one or more generations while keeping parental genotype information. Examples on how to generate such samples are provided in the program website.
Results
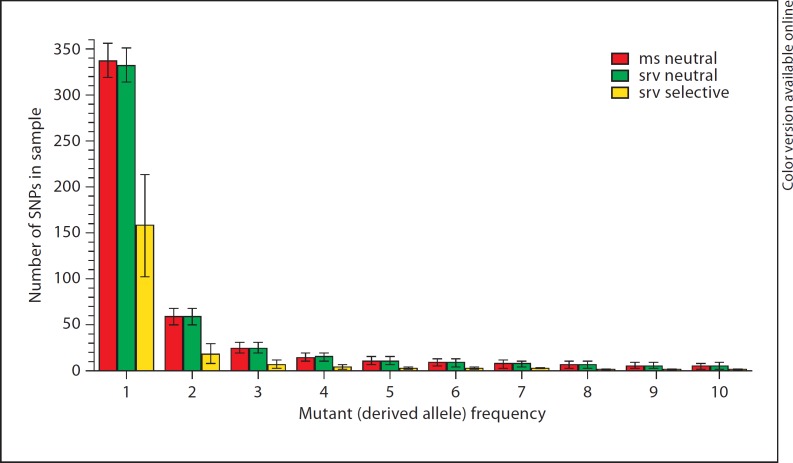
As a demonstration, we evolved initial populations of 8,100 individuals with 63,000 base pairs for 81,000 generations and expanded them to 900,000 individuals in 370 generations after a short bottleneck of 7,900 individuals. This model reflects one of the demographic models of the European population [5]. We used a neutral model and an exponential multi-locus selection model for non-synonymous mutations, where fitness values of new mutants were drawn from a gamma distribution with a shape parameter of 0.184 and a scale parameter of 0.320 [16]. A finite-sites mutation model with a mutation rate of 1.8 × 10−8 per generation per base pair was used. We drew 700 random individuals from the simulated population and obtained the number of SNPs for each mutant frequency class [a class i SNP means there are i copies of mutants (derived allele) on the site, 0 < i < 1,400].
As a comparison, we also simulated sequences with the same neutral model described above using ms [9]. With 10,000 replicates, we compared the average numbers of SNPs for each mutant frequency class obtained from the three simulations. The results obtained with srv and ms under the neutral model were very similar, except the number of singletons of srv was slightly smaller than that of ms (fig. 1). The difference may partly be due to the different models employed by the two methods. srv uses the default finite-sites model for mutations and simulates individuals in discrete time (generation), whereas ms uses an infinite-sites model for mutations and the continuous-time coalescent approximation for simulation. Compared with the neutral models, the mutants under the selection model showed dramatic reductions in the total number of mutants and a proportional concentration of extremely rare variants, as expected (fig. 1).
Fig. 1.
Comparison of three simulation results of the numbers of the first ten mutant (derived allele) frequency classes (1 ≤ i ≤ 10). Means ± SD are shown, which were obtained from the three sets of simulations, each with 10,000 replicates.
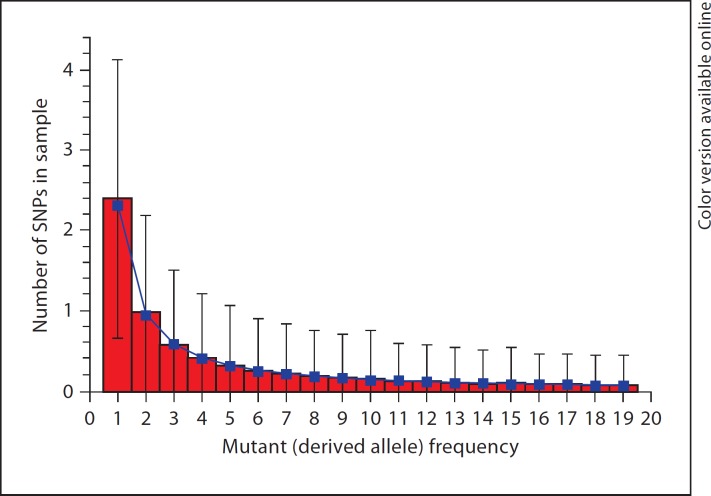
Although the distribution of allele frequencies of sequences with linked loci (nucleotides) is difficult to derive, a computer program (prfreq [16]) has been developed to estimate the distribution of allele frequencies of a nucleotide locus using numerical integration, effectively giving us a distribution of allele frequencies of unlinked loci under certain demographic and genetic assumptions. To validate our simulation method against theoretical estimates, we simulated a constant-size population of 8,100 individuals with 10,000 base pairs for 81,000 generations, using an infinite-sites mutation model with a mutation rate of 1.8 × 10−8 per generation per base pair. We used a multiplicative multi-locus selection model for non-synonymous mutations, with fitness values of new mutants drawn from a gamma distribution with a shape parameter of 0.184 and a scale parameter of 0.320 [16].
We drew 10 random individuals from the simulated population and obtained the number of SNPs for each mutant frequency class. We compared the average number of each SNP class to the expectations obtained from the prfreq program with the same demographic model and selection coefficient distribution (fig. 2). Although prfreq obtains the expectations assuming independent nucleotide sites in contrast to our assumption of a non-recombined DNA strand, the means of the simulation results fit reasonably well to prfreq's expectations, which suggests that the marginal distribution of each nucleotide in our simulation is similar to that of an independent site.
Fig. 2.
Comparison of srv simulation results (columns) with prfreq expectations (squares) [16]. Histograms and error bars show the mean ± SD of the simulation results by srv with 10,000 replicates. prfreq expections = the expected SNP spectrum calculated by prfreq.
Discussion
Due to the stochastic nature of the evolutionary process, the number, location, and frequency of mutants vary from population to population. A random realization of this evolutionary process requires a long evolution time (for example a long period of constant population size). If multiple replicates of the same evolutionary scenario are simulated, a population that is under mutation and selection equilibrium can be simulated in advance and used as the founder population for replicate simulations. Similarities between populations simulated in this manner (e.g. sharing of mutants from the founder population) can be controlled by evolving the founder populations for additional generations.
In order to simulate samples that resemble real-world genetic data, our program provides an option to import alleles from an existing sample (e.g. the HapMap dataset [18]) during evolution. These alleles are usually introduced before rapid population expansion. Because common alleles will most likely remain common during rapid population expansion [19], manually inserted common variants will remain common in the simulated population, but at the same time blend nicely with new mutants that are introduced during the population expansion stage. Such datasets provide an ideal tool to study the power of genome-wide association studies of diseases that are caused by rare and unobserved variants [4].
srv allows the use of a pseudo-infinite-sites mutation model in which mutations only happen at nucleotide loci without existing mutants. Whereas a mutant will be considered new in a coalescent-based simulation if it does not appear in individuals in the coalescent tree, our model requires that no mutant at the same locus exists in the whole population. Because large populations of short sequences may become saturated so that every locus has existing mutants, an infinite-sites model should not be used if the number of segregation sites in the simulated population is close to the length of sequences.
If the underlying evolutionary process can be sufficiently approximated by a Wright-Fisher model, a scaling technique can be used to speed up a forward-time simulation [20,21]. Compared to a regular simulation that evolves a population of size N for t generations, a scaled simulation with a scaling factor λ evolves a smaller population of size N/λ for t/λ generations with magnified (multiplied by λ) mutation, recombination, and selection forces. However, because this scaling technique might not be applicable to all supported selection models, and will result in a final population of size N/λ instead of N, this technique should be used with caution.
A number of computer programs are available to simulate sequences for genetic epidemiological studies. Table 1 lists some of the popular coalescent-based programs and forward-time simulation programs that support natural selection on multiple loci. More complete surveys of such software are provided by Liu et al. [22] and Carvajal-Rodriguez [23]. srv differs from these programs in its ability to simulate diploid and site-specific selection models, to use an infinite-sites in addition to a finite-sites mutation model, and to introduce alleles from empirical data.
Table 1.
P opular applications that simulate genome sequences using a coalescent or forward-time approach
| Program | Method | Selection | Recombination | Mutation models | Disease model | Ref. |
|---|---|---|---|---|---|---|
| MS | coalescent | no | uniform | infinite sites | no | 9 |
| GENOME | coalescent | no | varying1 | infinite sites | no | 25 |
| SelSim | coalescent | single-locus | varying | diallelic for selected site | no | 10 |
| ForSim | forward | based on phenotype | uniform | diallelic | yes | 26 |
| GenomePop | forward | random2 | varying | 2- or 4-allele models | no | 13 |
| FREGENE | forward | random3 | varying | finite sites, diallelic | yes | 14 |
| SFS_CODE | forward | random4 | varying | finite sites, diallelic | no | 12 |
User can define multiple consecutive fragments of the sequence, the recombination only occurs between fragments but not within the fragments.
Multiplicative multi-locus model with selection coefficients drawn from a gamma distribution.
Additive multi-locus selection model with selection and dominance coefficients drawn from a mixture of two Gaussian distributions.
Additive or multiplicative multi-mutant selection model with scaled selection coefficients drawn from a random distribution.
Perhaps the most distinguishing feature of srv is that it is implemented using a general-purpose forward-time population genetics simulation environment (simuPOP [24]) and can take full advantage of the processing power of this software. For example, a single python script can be used to simulate multiple populations using srv, apply different disease or quantitative trait models, draw population- or pedigree-based samples, and use different statistical methods to analyze them. Because of the scripting language design, it is relatively easy to modify srv to reveal details of the evolutionary process or use alternative demographic or genetic models. srv can be executed from a graphical user interface, a command line in batch mode, or called as a function from another script. It takes around 10 min to simulate 90,000 sequences of 63,000 base pairs on a reasonably configured PC using a demographic model with a long constant population size followed by rapid population expansion, and an exponential multi-locus selection model with selection coefficients drawn from a gamma distribution. srv is freely available at the ‘Complete Script's section of the simuPOP online cookbook at http://simupop.sourceforge.net/cookbook. Several examples are provided to demonstrate features of this program.
Acknowledgements
B.P. was supported by grant R01 CA133996 from the National Cancer Institute and by the University of Texas MD Anderson Cancer Center's Support Grant CA16672 from the National Institutes of Health. X.L. was supported by grant 5P50GM065509 from the National Institute of General Medical Sciences. The authors thank Drs. Gregory Kryukov and Shamil Sunyaev for kindly providing details of their simulation method, and Dr. Adam Boyko for providing the prfreq program and the instructions and suggestions for running it.
References
- 1.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 4.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci USA. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price AL, Kryukov GV de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudson RR. Generating samples under a Wright-Fisher neutral model. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 10.Spencer CC, Coop G. SelSim: a program to simulate population genetic data with natural selection and recombination. Bioinformatics. 2004;20:3673–3675. doi: 10.1093/bioinformatics/bth417. [DOI] [PubMed] [Google Scholar]
- 11.Peng B, Amos CI, Kimmel M. Forward-time simulations of human populations with complex diseases. PLoS Genet. 2007;3:e47. doi: 10.1371/journal.pgen.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez RD. A flexible forward simulator for populations subject to selection and demography. Bioinformatics. 2008;24:2786–2787. doi: 10.1093/bioinformatics/btn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvajal-Rodriguez A. GENOMEPOP: a program to simulate genomes in populations. BMC Bioinformatics. 2008;9:223. doi: 10.1186/1471-2105-9-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chadeau-Hyam M, Hoggart CJ, O'Reilly PF, Whittaker JC, De Iorio M, Balding DJ. Fregene: simulation of realistic sequence-level data in populations and ascertained samples. BMC Bioinformatics. 2008;9:364. doi: 10.1186/1471-2105-9-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williamson SH, Hernandez R, Fledel-Alon A, Zhu L, Nielsen R, Bustamante CD. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc Natl Acad Sci USA. 2005;102:7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, White TJ, Nielsen R, Clark AG, Bustamante CD. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyre-Walker A, Woolfit M, Phelps T. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics. 2006;173:891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortia TH. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng B, Kimmel M. Simulations provide support for the common disease-common variant hypothesis. Genetics. 2007;175:763–776. doi: 10.1534/genetics.106.058164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoggart CJ, Chadeau-Hyam M, Clark TG, Lampariello R, Whittaker JC, Iorio MD, Balding DJ. Sequence-level population simulations over large genomic regions. Genetics. 2007;177:1725–1731. doi: 10.1534/genetics.106.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng B, Amos CI. Forward-time simulation of realistic samples for genome-wide association studies. BMC Bioinformatics. 2010;11:442. doi: 10.1186/1471-2105-11-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Athanasiadis G, Weale ME. A survey of genetic simulation software for population and epidemiological studies. Hum Genomics. 2008;3:79–86. doi: 10.1186/1479-7364-3-1-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvajal-Rodriguez A. Simulation of genomes: a review. Curr Genomics. 2008;9:155–159. doi: 10.2174/138920208784340759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng B, Kimmel M. simuPOP: a forward-time population genetics simulation environment. Bioinformatics. 2005;21:3686–3687. doi: 10.1093/bioinformatics/bti584. [DOI] [PubMed] [Google Scholar]
- 25.Liang L, Zollner S, Abecasis GR. Genome: a rapid coalescent-based whole genome simulator. Bioinformatics. 2007;23:1565–1567. doi: 10.1093/bioinformatics/btm138. [DOI] [PubMed] [Google Scholar]
- 26.Lambert BW, Terwilliger JD, Weiss KM. ForSim: a tool for exploring the genetic architecture of complex traits with controlled truth. Bioinformatics. 2008;24:1821–1822. doi: 10.1093/bioinformatics/btn317. [DOI] [PMC free article] [PubMed] [Google Scholar]