Abstract
SATB1 is expressed primarily in thymocytes and can act as a transcriptional repressor. SATB1 binds in vivo to the matrix attachment regions (MARs) of DNA, which are implicated in the loop domain organization of chromatin. The role of MAR-binding proteins in specific cell lineages is unknown. We generated SATB1-null mice to determine how SATB1 functions in the T-cell lineage. SATB1-null mice are small in size, have disproportionately small thymi and spleens, and die at 3 weeks of age. At the cellular level, multiple defects in T-cell development were observed. Immature CD3−CD4−CD8− triple negative (TN) thymocytes were greatly reduced in number, and thymocyte development was blocked mainly at the DP stage. The few peripheral CD4+ single positive (SP) cells underwent apoptosis and failed to proliferate in response to activating stimuli. At the molecular level, among 589 genes examined, at least 2% of genes including a proto-oncogene, cytokine receptor genes, and apoptosis-related genes were derepressed at inappropriate stages of T-cell development in SATB1-null mice. For example, IL-2Rα and IL-7Rα genes were ectopically transcribed in CD4+CD8+ double positive (DP) thymocytes. SATB1 appears to orchestrate the temporal and spatial expression of genes during T-cell development, thereby ensuring the proper development of this lineage. Our data provide the first evidence that MAR-binding proteins can act as global regulators of cell function in specific cell lineages.
Keywords: SATB1, MARs, chromatin structure, gene expression, T-cell development
SATB1 is a nuclear protein that was originally cloned by virtue of its ability to bind to a core unwinding element within the matrix attachment DNA region (MAR) located 3′ of the immunoglobulin μ heavy chain (IgH) gene enhancer (Dickinson et al. 1992). MARs, including those surrounding the IgH enhancer core (Cockerill et al. 1987), have been postulated to contribute to higher order chromatin structure by mediating the attachment of chromatin to the nuclear matrix, thereby folding chromatin into topologically independent loop domains (Cockerill and Garrard 1986; Gasser and Laemmli 1987). Results obtained from transgenic mouse studies indicate that MAR sequences are important in vivo for cell-type-specific gene regulation and chromatin structure. The MAR sequences flanking the IgH enhancer are essential for the transcription of a rearranged μ gene in transgenic B lymphocytes (Forrester et al. 1994) and the generation of an extended domain of chromatin that is accessible to transcription factors (Jenuwien et al. 1997). The IgH MARs together with the core enhancer induce extensive demethylation across the chromatin domain (Jenuwien et al. 1997). Similar to IgH MARs, MARs flanking the immunoglobulin κ enhancer are necessary for B cell-specific demethylation of the Igκ locus (Kirillov et al. 1996).
SATB1 was the first cell-type-restricted MAR-binding protein to be identified and is expressed predominantly in thymocytes. Its expression is virtually undetectable in other tissues except for low levels in testis, fetal brain, and osteoblasts (Dickinson et al. 1992; T. Kohwi-Shigematsu, unpubl.). SATB1 binds to the minor groove of DNA specifically recognizing a unique group of AT-rich DNA sequences. There is no primary consensus sequence for these AT-rich regions; rather, they are represented by a specialized DNA context that contains a cluster of sequences where one strand consists exclusively of well-mixed As, Ts, and Cs, excluding Gs (ATC sequences; Dickinson et al. 1992). Under negative superhelical strain, a region containing an ATC sequence cluster has the intrinsic propensity to unwind by base unpairing. Such a region is referred to as a base unpairing region (BUR) and is typically found in MARs. BURs are usually 100–150 bp in length or less, and can be identified by unpaired DNA-specific chemical probes (for review, see Leonard 1984; Kohwi-Shigematsu and Kohwi 1992). BURs are a potential hallmark of MARs because unwinding propensity is important for MAR activity (Kohwi-Shigematsu and Kohwi 1990; Bode et al. 1992). SATB1 has a homeodomain and a BUR-binding domain, both of which are necessary for recognition of the core unwinding element within a BUR. If the core unwinding element of a BUR is mutated to abolish the unwinding property of the BUR, SATB1 binding is eliminated (Dickinson et al. 1992, 1997; Nakagomi et al. 1994; Wang et al. 1995). Besides SATB1, a B cell-specific protein, Bright (Herrscher et al. 1995) as well as certain ubiquitous proteins have been identified to specifically recognize and bind BURs (Dickinson and Kohwi-Shigematsu 1995; Galande and Kohwi-Shigematsu 1999; Liu et al. 1999a).
Recently, a group of genomic DNA sequences to which SATB1 binds in vivo was cloned and individually characterized. Proof that SATB1 in fact binds to MARs in vivo was obtained by performing fluorescent in situ hybridization (FISH) with each of these sequences and showing that that these sequences hybridize to the genomic DNA remaining attached to the nucleoskeleton (or the nuclear matrix; deBelle et al. 1998). Recent reports suggest that, in at least two different experimental systems, SATB1 can act as a transcriptional repressor mediated by MARs (Kohwi-Shigematsu et al. 1997; Liu et al. 1997). SATB1, a cell-type restricted BUR/MAR-binding protein, may play a critical role(s) during development by regulating multiple genes in specific cell lineages. Unlike classic transcription factors, which bind individual target genes to regulate transcription, SATB1, which binds to multiple sites where chromatin is fastened to form loop domains, may dictate the organization and structure of chromatin domains, thereby orchestrating the transcription potential of multiple genes.
We examined the function of SATB1 by generating SATB1-null mice by gene targeting in ES cells. Because SATB1 is expressed predominantly in thymocytes (Dickinson et al. 1992) and is induced in peripheral T cells as an immediate early gene product (Beadling et al. 1993), we chose to examine the T-cell lineage in SATB1-null mice to understand how a MAR-binding protein might regulate development. Here we show that SATB1 ablation leads to major dysregulation (mostly de-repression) of multiple genes, including cytokine receptor genes. A discrete set of genes was expressed in a disordered manner that was incompatible with specific T-cell developmental stages. SATB1 was found to be necessary at multiple stages in T-cell development and T-cell activation. Our results suggest that SATB1 orchestrates T-cell development by ensuring ordered spatial and temporal gene expression.
Results
Generation of SATB1-null mice
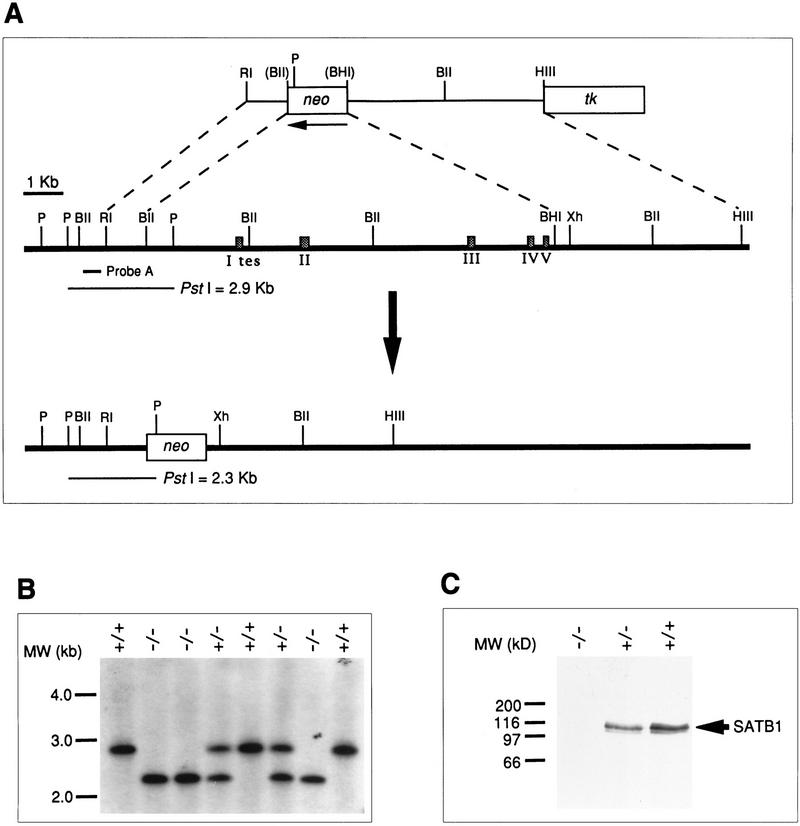
The genomic structure of SATB1 was partially mapped, including eight exons. A targeting construct was designed to replace a 10.9-kb BglII–BamHI fragment, containing the first five exons, which encode 213 amino acids including the translation start codon, with a neomycin resistance gene driven by the PGK promoter (Fig. 1A). The targeting construct, pSB1Neo3.2KO, also contained a PGK–thymidine kinase (tk) gene. The targeting construct was linearized, electroporated into ES cells, and transfectants were grown in selection media containing G418 and gancyclovir. G418r, gancr clones were screened for homologous recombination by the PCR and verified by Southern blot analysis (Fig. 1B). Six homologous recombinant clones were used to generate chimeric mice by injection into 3-day-old C57BL/6 (B6) blastocysts, and three of them yielded chimeric mice that passed the mutated allele to the germ line. Homozygous SATB1-targeted mice were obtained by mating heterozygous F1 offspring from the chimeras. The absence of both SATB1 transcripts and SATB1 protein was confirmed by RT–PCR and Western blotting, respectively, of thymus extracts from homozygous mutant mice (Fig. 1C and data not shown).
Figure 1.
Targeted disruption of the SATB1 gene. (A) Partial genomic structure of SATB1 and the targeting strategy. This deletion replaced the first five exons or 213 amino acids of the mouse thymus SATB1 (Nakagomi et al. 1994), including the translation start codon, with a neomycin-resistance gene. The recombined allele is shown at the bottom. Exons are indicated by shaded boxes. The nucleotide positions encompassed by coding exons are: exon 2, −34–211; exon 3, 212–388; exon 4, 389–515; exon 5, 516–639; exon 6, 640–752; exon 7, 753–1205; exon 8, 1206–N.D. (not determined). The exon labeled I tes encodes the 5′ UTR found in the human SATB1 cDNA isolated previously from testis (Dickinson et al. 1992). The corresponding 5′ UTR for the thymus cDNA has not yet been located. (P) PstI; (BII) BglII; (RI) EcoRI; (BHI) BamHI; (HIII) HindIII; and (Xh) XhoI. The map for PstI is incomplete. Restriction sites in parentheses were destroyed during construction of the targeting construct. (B) Southern blot of PstI-digested tail DNA from offspring of a heterozygous mating. Probe A (400 bp) was used for Southern blot analysis to detect both the wild-type 2.9-kb and mutant 2.3-kb PstI genomic fragments. (C) Western blot of protein lysates from 13-day-old thymi using anti-SATB1 antibody as described (Nakagomi et al. 1994). Symbols represent wild type (+/+) and heterozygous (+/−) and homozygous mutants (−/−).
Growth retardation of SATB1-null mice
Homozygous mice were indistinguishable from wild-type and heterozygous littermates at birth. As early as 3 days after birth, however, the homozygotes were smaller and thinner than their littermates, and this size difference became more pronounced with age (Fig. 2A). The size of knockout mice was variable and may be due to the mixed genetic background of the mice (C57BL/6 and 129/Ola). However, all homozygous mice died by 4 weeks after birth, with the majority dying between 21 and 26 days of age. Lifespan could be extended to at least 42 days by transferring 16-day old SATB1-null mice to a foster mother to prolong nursing. However, no homozygous mice survived weaning. Interestingly, SATB1-null animals exhibited characteristics in common with mice that have neurological defects, such as incomplete eye opening and the clasping reflex (Fig. 2B) (Crowley et al. 1994; Klein et al. 1994; Urbánek et al. 1994). This finding raises the possibility that SATB1 has a role in neurological function, which is consistent with the detection of SATB1 mRNA in fetal brain (Adams et al. 1993).
Figure 2.
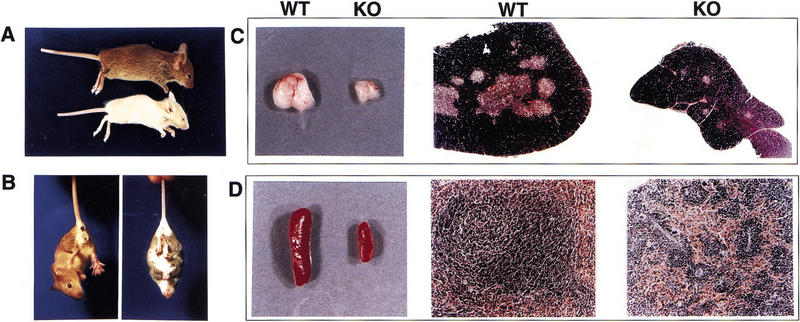
Phenotype of the SATB1-null mice. (A) Shown are 22-day-old wild-type (WT; top) and knockout (KO; bottom) littermates demonstrating a difference in size and ptosis of the eye. (B) A 26-day-old knockout mouse (right) displaying the clasping reflex in comparison with a wild-type littermate (left). The thymus (C) and spleen (D) from the SATB1-null mouse shown in A are smaller than that of the wild-type littermate. These organs were fixed in formalin and embedded in paraffin. Hematoxylin- and eosin-stained sections revealed that the SATB1-null thymus (top right of C) has a higher cortex (purple) to medulla (pink) ratio than the wild-type thymus (top middle of C) at a 40× magnification. The SATB1-null spleen (bottom right of D) has a significantly smaller white pulp (purple) compared with the wild-type spleen (bottom middle of D) visualized at the same magnification (100×).
Despite the marked phenotypic differences between SATB1-null and wild-type mice, gross dissection and histologic examination revealed no obvious defects, except for a striking size difference in the thymus, spleen, and lymph nodes. In the knockout mice, these organs were substantially smaller than in wild-type mice (Fig. 2C,D). The size differences in the thymus, spleen, and lymph nodes were much more pronounced in SATB1-null mice that were older than 2 weeks of age. In SATB1-null thymi, the number of thymocytes was typically reduced 1.5- to 2-fold at 2 weeks of age, and 10- to 100-fold by 3 weeks of age and 10- to 100-fold by 3 weeks of age (data not shown). Profound histologic abnormalities were also found in these organs, suggesting that SATB1 is necessary for T-cell development. Specifically, SATB1-knockout thymi were composed mainly of cortex, with some degenerating cells, and a sparse medulla (Fig. 2C). Also, spleens from these animals had shrunken white pulp, and lymph nodes contained a reduced number of lymphocytes (Fig. 2D; data not shown). Heterozygous and wild-type mice were indistinguishable in this regard.
The CD3−CD4−CD8− triple negative population is markedly reduced in SATB1-null thymi
Immature T cells develop in the thymus by progressing through distinct stages that are characterized by the expression of different sets of cell-surface proteins (for review, see Rothenberg 1992). On the basis of their expression of CD4 and CD8 accessory molecules, developing thymocytes can be subdivided into four populations that correspond to discrete developmental stages.CD4−CD8− cells that lack T-cell receptor (TCR) expression (CD3−CD4−CD8− cells) are designated as triple negative (TN) cells, and represent the most immature thymocytes. TN cells give rise to CD4+CD8+ double positive (DP) cells, which then mature into either CD4+ or CD8+ single-positive (CD4 or CD8 SP) cells.
Initial investigation into the cause of the reduced size of the SATB1-null thymus and spleen focused on the early TN thymocyte population by excluding cells that stain with anti-CD3, anti-CD4, and anti-CD8 antibodies. This trend reflected the fact that TN thymocytes undergo rapid cell proliferation and account for the overall number of thymocytes (for review, see Fehling and von Boehmer 1997). Total thymocytes were isolated from wild-type and SATB1-null mice and mixed with FITC-conjugated anti-CD25 and biotin-conjugated anti-CD3, anti-CD4, and anti-CD8 antibodies. The cells bound by the biotin-conjugated antibodies were detected by streptavidin–Tri-Color. We found that SATB1-null mice had a markedly reduced TN subpopulation (Tri-Color-negative cells). In 14-day-old SATB1-null mice, the TN subpopulation represented only 0.5% of total thymocytes, as opposed to 6% of wild-type thymocytes (Fig. 3A). Thus, a decreased number of TN cells accounts, at least in part, for the small size of the thymus in SATB1-null mice. Additionally, SATB1-null thymi contained a Tri-Color-positive population that expressed an intermediate level of CD25. In contrast, virtually all Tri-Color-positive cells were CD25 negative in wild-type thymi (Fig. 3A). Further FACS analysis showed that the CD25-expressing population in the SATB1-null mice was CD4 and CD8 positive (DP thymocytes; data not shown).
Figure 3.
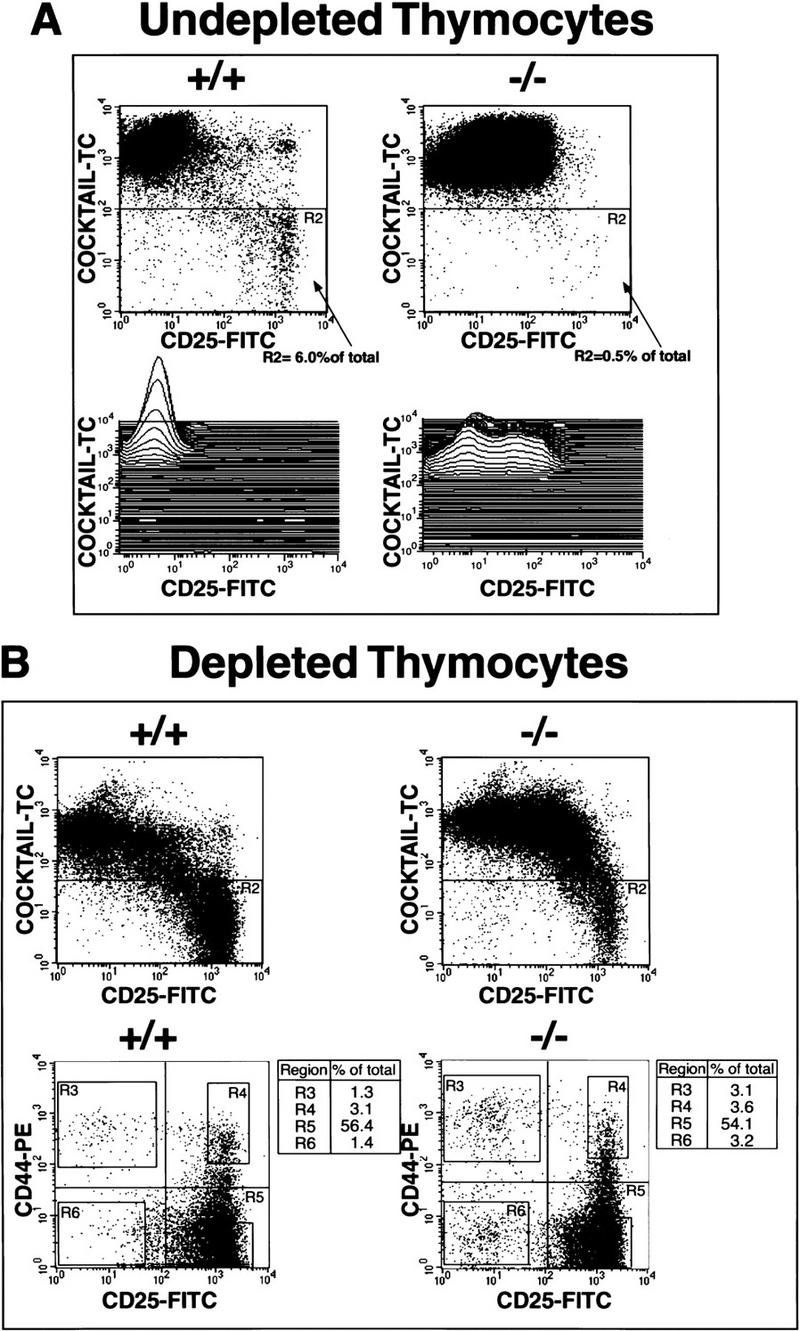
A great reduction in TN thymocytes in SATB1-null thymi without changes in the ratio of specific TN subsets. (A) CD3−CD4−CD8− TN cells were quantified by incubation of total thymocytes with a combination of biotin-conjugated anti-CD3, anti-CD4, and anti-CD8 antibodies plus Tri-Color–streptavidin and FITC-conjugated anti-CD25 antibody and subjecting them to FACS analysis. SATB1-null thymus (−/−) (right) has only 1/10 the number of TN cells detected in a wild-type thymus (+/+) (left). A total of 200,000 events were collected. In each dot plot, 4 × 105 thymocytes are depicted. In SATB1-null thymocytes, there exists a Tri-Color-positive cell population that expresses an intermediate level of CD25 as shown by three-dimensional plot. In contrast, wild-type thymocytes have a well-defined population that is CD25 negative. (B) TN thymocytes were enriched by depletion of CD4- and CD8-expressing cells from total thymocytes. TN cell subsets from depleted thymocytes were purified further by gating out Tri-Color-stained CD3-, CD4-, and CD8-positive T cells, B cells, granulocytes, dendritic cells, NK cells, and monocytes from thymocytes. The TN cell subsets in region R2 in the upper dot plots were subsequently subjected to FACScan analysis with PE-conjugated anti-CD44-antibody and FITC-conjugated anti-CD25 antibody. Again, the SATB1-null thymus contains significantly fewer CD25+ cells than wild-type thymus (cf. top right with top left dot plot); however, the subsets of CD25+ and CD44+ TN cells (bottom right) are represented in their normal proportions (bottom left dot plot). In the top dot plots, 2.5 × 104 events are depicted, in the bottom dot plots, 1 × 104 events are depicted. TN subsets in the order of development are R3 (CD25−CD44+), R4 (CD25+CD44+), R5 (CD25+CD44−), and R6 (CD25−CD44−). The percentage of each subset of the total cell population in the R2 region is shown in the table to the right of each dot plot.
TN cells are divided into four subsets on the basis of the expression of CD25 (interleukin-2 receptor α chain; IL-2Rα) and CD44 (phagocyte glycoprotein-1/Pgp-1). These subsets are, in order of development: CD25−CD44+, CD25+CD44+, CD25+CD44−, and CD25−CD44− (Godfrey et al. 1993). Among the CD25+CD44− subset, those cells that have rearranged one of their TCRβ alleles in frame, the process called β-selection, are selected to rapidly proliferate and progress to the CD25−CD44− stage (Hoffman et al. 1996). To determine whether any of these four TN subsets are deficient in the SATB1-null thymus, the TN population was enriched by depleting most of the CD4- and CD8-expressing cells (Suda and Zlotnik 1993). After depletion, the triple-negative cells in region R2 representing TN thymocytes were examined further for expression of CD25 and CD44 (Fig. 3B, bottom two panels). Wild-type and SATB1-null TN cells exhibited a very similar distribution of the four distinct TN subsets, except that a modest twofold increase in CD25−CD44+(R3) and CD25−CD44− (R6) cells was found in the SATB1-null TN population. This result indicates that no particular developmental stage was skipped in SATB1-null thymocytes (Fig. 3B). Also, the levels of CD25 and CD44 expression were similar in both the wild-type and SATB1-null TN cell subpopulations. We confirmed by Southern analysis that there was no defect in TCRβ-chain gene rearrangement in the SATB1-null thymocytes (data not shown).
T-cell development is blocked at the CD4+CD8+ DP stage in SATB1-null mice
We performed FACS analyses (represented as 1 × 104 events per sample) during the later phase of thymocyte development and found a striking block in thymocyte maturation in SATB1-null mice. During normal development, CD25−CD44− thymocytes spontaneously up-regulate CD4 and CD8 accessory molecules and start to rearrange their TCRα loci resulting in a CD4highCD8highαβTCRlow DP stage (Rothenberg 1992). From this DP stage, full maturation into either CD4highCD8−αβTCRhigh single positive (SP) helper T cells or CD4−CD8highαβTCRhigh SP cytotoxic T cells occurs. SATB1-null thymocytes, however, were arrested mainly at the DP stage, indicating that SATB1 is necessary for maturation into SP cells (Fig. 4A).
Figure 4.
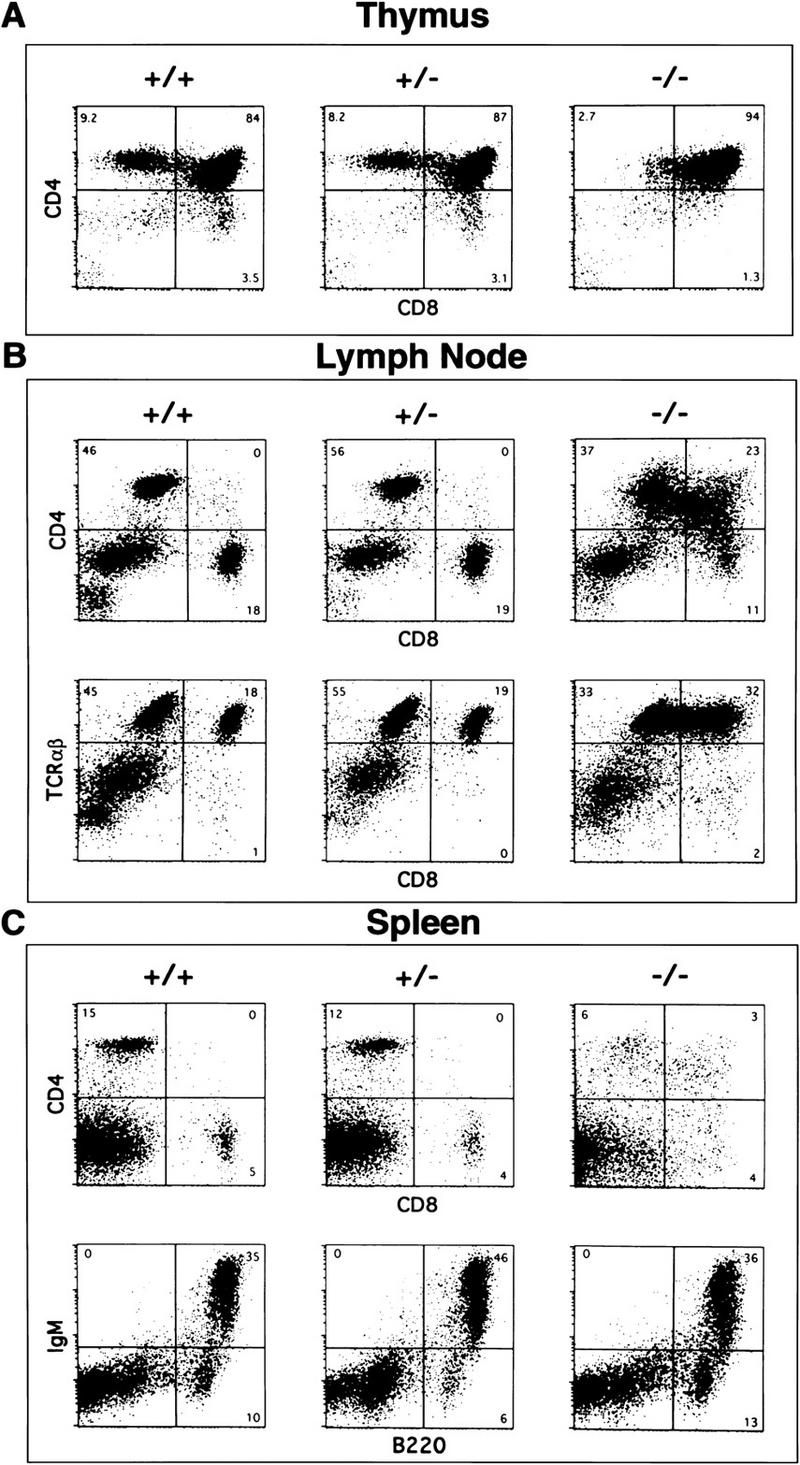
Flow cytometric analysis of lymphocytes from wild-type, heterozygous, and knockout mice at 2 weeks of age. Data is representative of staining from at least four separate mice. Total number of cells collected for thymus in A is 2.7 × 108 +/−, 3.2 × 108 +/−, and 2.8 × 107 −/−, from lymph node in B is 1.4 × 106 +/+, 1.3 × 106 +/−, and 2.8 × 105 −/−, and for spleen in C is 9.1 × 107 +/+, 9.0 × 107 +/−, and 2.6 × 107 −/−. For each FACS analysis, 1 × 104 events are represented. In all cases, the FACS analysis of heterozygotes showed the same pattern as wild-type. (A) SATB1-null (−/−) thymus (right) contains few CD4+ or CD8+ SP cells compared with wild-type (+/+) and heterozygous (+/−) thymi (left and middle) (B) SATB1-null T cells in lymph nodes are greatly reduced in number and contain CD4+CD8+ DP as well as some CD4 SP cells (upper right). These cells express levels of TCRαβ (lower right) comparable to that of SP cells within wild-type and heterozygous lymph node cells (lower left and middle). (C) SATB1-null spleens have fewer T cells and lack distinct CD4+ and CD8+ populations (top right) compared with control wild-type and heterozygous spleens (top left and middle). B cell populations appear to be unaffected in SATB1-null spleen as assessed by IgM and B220 staining (bottom).
The maturational block in thymocytes was apparently not complete as a very small population of CD4highCD8low to CD4highCD8med thymocytes was consistently observed in SATB1-null mice (Fig. 4A). This population presumably represents a stage of development intermediate between DP and CD4 SP stages. The well-defined population of CD8 SP cells, as seen in wild-type and heterozygous thymocytes, was much reduced or virtually undetectable in SATB1-null thymocytes. The FACS profile for SATB1-null thymocytes in Figure 4A, which was obtained from 14-day-old mice, was typical of 9- to 23-day SATB1-null thymi (data not shown). Additionally, spleen and lymph nodes from SATB1-null mice contained a reduced population of T cells that consisted of αβTCRhigh cells. In lymph nodes of SATB1-null mice, most T cells consist of DP (CD4+ cells with varying levels of CD8) and CD4 SP cells (Fig. 4B); the total CD4 SP population was reduced by about sevenfold compared with that in wild-type mice. For SATB1-null mice, it was difficult to quantitate the CD8 SP cells in lymph nodes as well as CD4 SP cells and CD8 SP cells in spleens, because these cells did not fall into well-defined populations as shown by FACS. The total number of cells from each organ was reduced by 3.5- to 5-fold for SATB1-null mice, depending on individual animals (for total number of cells, see Fig. 4 legend). For every 1 × 104 events shown for each FACS analysis, the relative cell number for T and B cells in either lymph nodes or spleens was comparable between wild-type and SATB1-null mice.
Although ablation of SATB1 resulted in a major defect in T-cell development, the FACS profile of B cells in SATB1-null mice was normal with respect to B220, IgM and IgD expression (Fig. 4C and data not shown). The mechanism for the effect of SATB1 ablation in B cell number awaits further study.
SATB1-null peripheral T cells undergo apoptosis in response to activating signals
T-cell activation occurs as a two-step process (Smith 1988). Quiescent T cells are activated at the cell surface by antigen-presenting cells via the T-cell antigen receptor/CD3 complex (TCR/CD3) and co-stimulatory molecules such as CD28 (Fraser et al. 1991), which enable the cells to produce interleukin-2 (IL-2) and its receptor IL-2R. Subsequently, the binding of IL-2 to IL-2R promotes cell cycle progression and DNA replication. We used 12-phorbol 13-myristate acetate (PMA) plus ionomycin that bypass cell-surface signaling by activating protein kinase C and increasing the intracellular Ca2+ concentration to test whether the few SATB1-null T cells present in lymph nodes, expressing high levels of the TCR/CD3 complex, were capable of undergoing T-cell activation. Because lymph nodes from wild-type mice contained CD4 SP and CD8 SP cells whereas those from SATB1-null mice contained mainly CD4 SP and DP cells (Fig. 4), it was necessary to isolate CD4 SP cells from both kinds of mice for a direct comparison. Isolated CD4 SP cells at 98% purification (Fig. 5A, inset) from 11- to 13-day-old mice were placed in a 96-well round-bottom culture plate either in the presence or absence of PMA plus ionomycin, and their cell numbers were monitored at 24, 48, and 72 hr after treatment. At 24 hr after stimulation, SATB1 CD4 SP cells underwent apoptosis as evidenced by cell fragmentation and a reduction in cell number by 50% (Fig. 5A). This loss of cells was due to apoptosis as confirmed by staining with the early apoptotic marker Annexin V followed by FACS analysis (data not shown). Cell death occurred in response to the activating stimuli because at 24 hr, >93% of these cells in media without PMA plus ionomycin were intact. The SATB1-null T cells continued to die rather than proliferate, and cell number was reduced to 38% by 48 hr and 29% by 72 hr (Fig. 5A). There was no evidence of cell proliferation even at 96 hr after stimulation (data not shown). Cell number of wild-type CD4 SP cells was once reduced to 60% after 24 hr stimulation with PMA plus ionomycin. In contrast to SATB1-null CD4 SP cells, however, they began proliferation at about 48 hr and continued to proliferate beyond that time point under the same conditions. The response of the SATB1-null CD4 SP cells to PMA plus ionomycin were not affected by exogenously added IL-2 (data not shown). The amount of secreted IL-2 was quantitated at 24, 48, and 72 hr after activating stimuli for both wild-type and SATB1-null CD4 SP cells, and these values per viable cell are shown (Fig. 5B). Although maximum IL-2 concentration in the media occurred within 24–48 hr for the control cells, SATB1-null T cells produced IL-2 at a very low level at 24 hr and then produced IL-2 at higher levels at later time points. Between 48 and 72 hr, cell number was reduced by an additional 9%, but the amount of secreted IL-2 increased by >13%. The values calculated as IL-2 (picograms) per cell may not represent a totally accurate estimate; nevertheless, the data show that SATB1-null CD4 SP cells can produce IL-2 in response to PMA plus ionomycin. It appears that maximal IL-2 production is significantly delayed for SATB1-null CD4 SP cells compared with that observed for wild-type CD4 SP cells. Clumping of cells, which indicates a normal response of T cells to activating signals, was also observed at a much later time for these SATB1-null cells than for wild-type cells.
Figure 5.
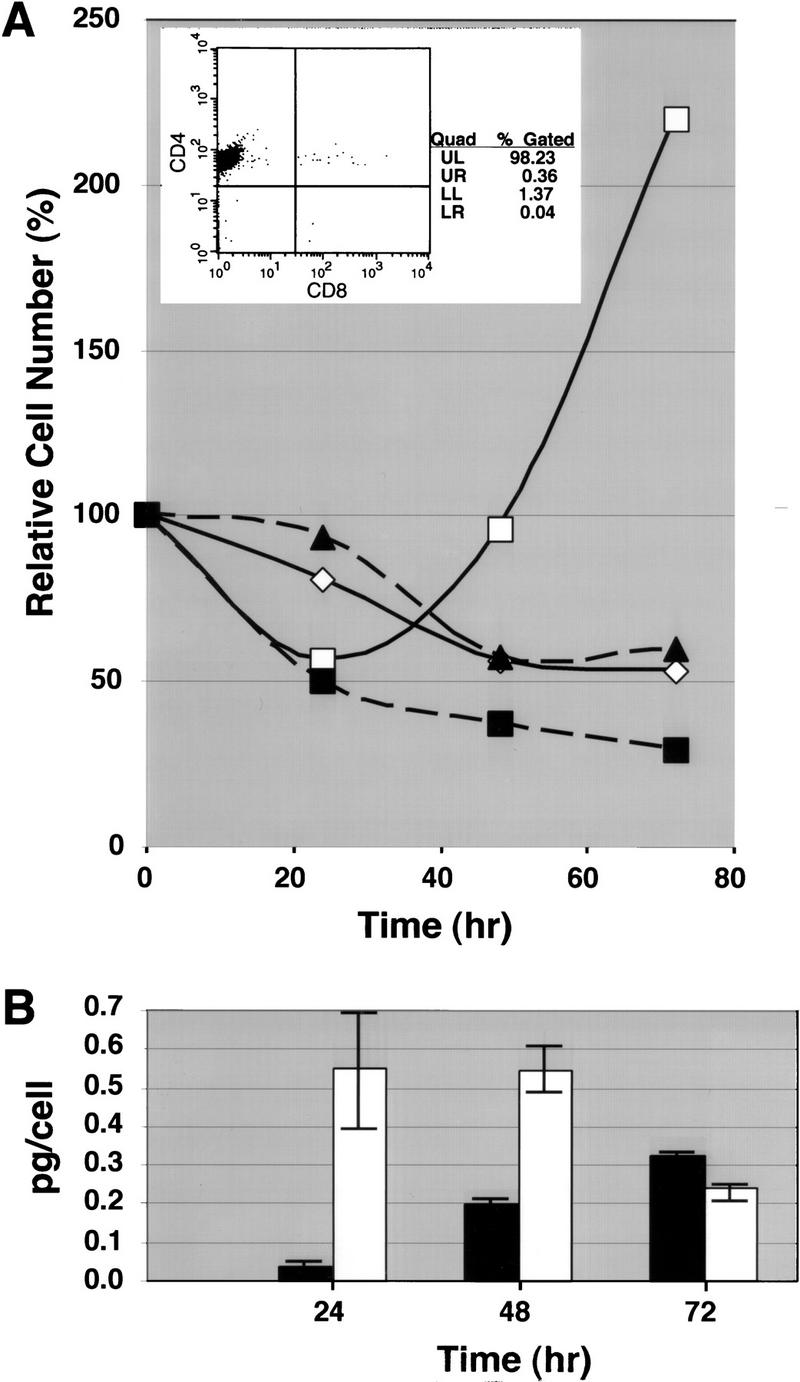
Apoptosis induced by PMA and ionomycin for SATB1-null peripheral CD4 SP cells. CD4 SP T cells were isolated from lymph nodes of control (C, either +/+ or +/−) and SATB1-null (KO) mice as described in Materials and Methods. The +/+ and +/− mice gave identical results. (A) The FACS profile of the purified CD4 SP cells from wild-type lymph nodes is shown in the inset. In each well of a 96-well culture plate, 3 × 104 cells were seeded and cultured in medium with or without PMA plus ionomycin. Cell number was determined at 24-, 48-, and 72-hr time points and the percentages of surviving cells are shown. The cell number did not vary >4% among the three samples. (▴) KO (med.); (█) KO (PMA+I); (⋄) C (med.); (□) C (PMA+I). (B) The IL-2 concentrations in the media were measured by ELISA assay at each time point after stimulation with PMA plus ionomycin. The IL-2 amounts per viable cell (pg/cell) are shown (the average values are shown by an open bar for control and a solid bar for KO CD4 SP T cells). The minimum and maximum pg/cell values obtained are indicated for each bar.
Both IL-2R and IL-7R genes are ectopically transcribed in SATB1-null DP thymocytes
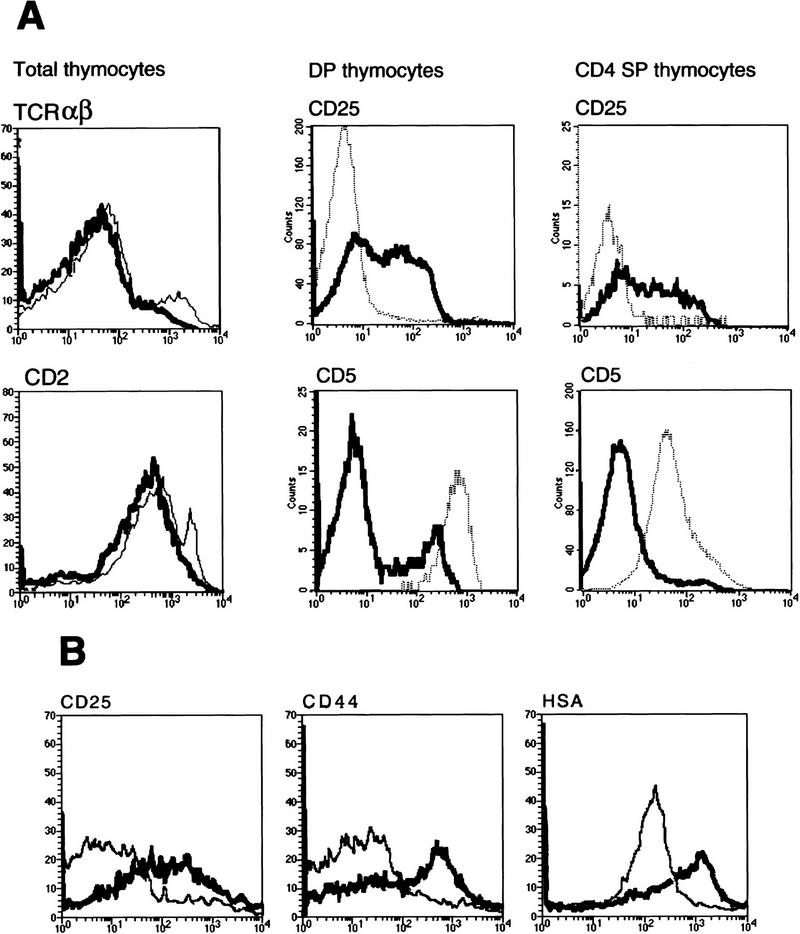
On the basis of the DNA binding specificity of SATB1, its ability to affect transcription, and the various phenotypes observed in SATB1-null mice, we hypothesized that SATB1 ablation resulted in altered regulation of multiple genes. The expression of the developmental markers TCRαβ, CD2, CD44, Thy1.2, and heat-stable antigen (CD24/HSA) was similar in SATB1-null and wild-type thymocytes by FACS analysis (Fig. 6A; data not shown). TCRαβ (high) and CD2 (high) subpopulations were missing in SATB1-null thymocytes presumably because of the lack of mature SP cells. Expression of IL-2Rα (CD25), on the other hand, was significantly higher in isolated SATB1-null DP thymocytes relative to wild-type DP thymocytes (Fig. 6A). This result is consistent with the data shown in Figure 3A. The elevation in CD25 expression persisted even in the very small CD4 SP population in the thymi of SATB1-null mice (Fig. 6A). In wild-type thymocytes, IL-2Rα/CD25 is expressed specifically during the TN stage (Ceredig et al. 1985; Raulet 1985) and is induced upon activation of peripheral T cells (Smith 1988). Therefore, in SATB1-null mice, ectopic expression of IL-2Rα/CD25 was found not only in the DP cells but also in more differentiated CD4 SP cells, in which expression of this protein should be totally repressed. Interestingly, SATB1-null thymocytes were able to up-regulate CD2 (Fig. 6A), which should occur concurrent with down-regulation of CD25 at the TN to DP stage transition (Shinkai and Alt 1994), indicating uncoordinated regulation of the genes in the absence of SATB1.
Figure 6.
Dysregulated expression of certain developmental marker proteins in SATB1-null thymocytes and T cells. (A) Total thymocytes from wild-type and SATB1-null thymi were stained with a biotin-conjugated anti-CD2/Cy-chrome–streptavidin combination or Cy-Chrome-conjugated anti-TCRαβ antibody alone. CD5+ and CD25+ thymocytes in both CD4+ CD8+ DP and CD4+ SP populations were detected by PE-conjugated anti-CD8 antibody and biotin-conjugated anti-CD4 antibody/Tri-Color-streptavidin combined with either FITC-conjugated anti-CD5 antibody or FITC-conjugated anti-CD25 antibody. (B) Peripheral T cells from wild-type and SATB1-null lymph nodes were stained singly with FITC-conjugated anti-CD25 antibody, biotin-conjugated anti-CD44 antibody/Cy-Chrome–streptavidin, and biotin-conjugated anti-HSA antibody/Texas Red–streptavidin and analyzed by FACS. For histogram analysis, CD4−CD8− double negative cells were excluded by gating. In all histograms the bold line represents the distribution of 1 × 104 stained T cells from SATB1-null mice while the staining distribution of T cells from control littermates is depicted by the curve with a narrow line.
Inconsistency in the developmental expression of CD5 is also evident. CD5 is markedly decreased in DP and even in the small CD4 SP population of SATB1-null mice (Fig. 6A). Expression of this protein normally increases during the transition from the TN stage to the DP stage, but SATB1-null thymocytes apparently cannot up-regulate CD5 protein expression.
T-cell-surface markers exhibited an unusual pattern of expression in the small SATB1-null peripheral T-cell population. IL2Rα/CD25 was expressed at a high level (Fig. 6B) even though it is normally absent in resting T cells. High levels of CD5 are normally seen in wild-type T cells, but SATB1-null T cells had an abnormally low level of CD5 (data not shown). Therefore, the T cells in SATB1-null lymph nodes appeared similar in phenotype to those found in the thymus. This result indicates that in SATB1-null mice, the DP thymocytes that normally do not exit the thymus were found in the lymph nodes. Additionally, whereas most activation markers, such as CD3ε, CD28, CD45, and CD122 (IL-2 receptor β-chain) were unchanged in SATB1-null T cells (data not shown), there was elevated expression of CD44 and HSA (Fig. 6B) and CD69 (data not shown). In normal peripheral T cells, these proteins are expressed after antigenic stimulation and activation (Hubbe and Altevogt 1994; Taher et al. 1996). Abnormal expression of developmental markers was not restricted to a specific subpopulation (DP or CD4 SP) of peripheral T cells in SATB1-null mice. Rather, all peripheral T cells, regardless of the level of CD4 or CD8 expression, displayed abnormal protein expression (data not shown).
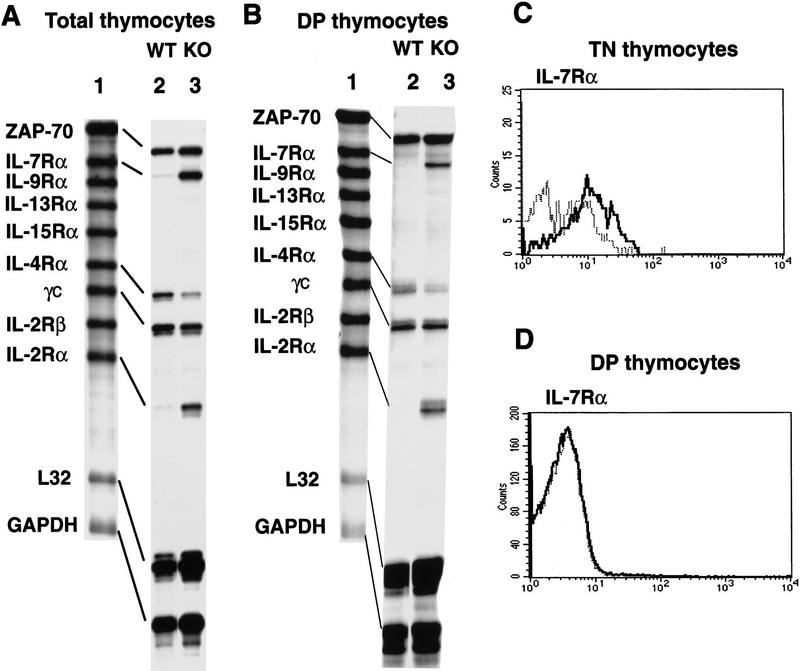
To test whether the transcript levels of cytokine receptors in thymocytes other than CD25/IL-2Rα are altered in SATB1-null thymocytes, an RNase protection assay was employed using a multi-probe template set derived from IL-7Rα, IL-9Rα, IL-13Rα, IL-15Rα, IL-4Rα, the common cytokine receptor γchain (γC), IL-2Rα and IL-2Rα genes. Consistent with the results of our FACS analysis (Figs. 3A and 6A), a striking enhancement in mRNA expression was observed for the IL-2Rα gene in total SATB1-null thymocytes relative to wild-type thymocytes (Fig. 7A, lanes 2,3). In addition, IL-7Rα mRNA was also dramatically increased. The level of IL-4Rα mRNA, on the other hand, was decreased in SATB1-null thymocytes. The abundance of other cytokine receptors mRNAs examined in this assay were unaffected by SATB1 ablation. As a loading control, a ZAP-70 probe was added to the template set. In SATB1-null mice, key players in surface-mediated signaling, including ZAP-70, CD45, Lck and Fyn (for review, see Chan et al. 1994), were found to be unaltered at the RNA and protein levels on the basis of Western blotting and RT–PCR data (data not shown). GAPDH, originally included in the set as a loading control, reproducibly showed modest overexpression in the SATB1-null thymocytes, as determined by RT–PCR (data not shown).
Figure 7.
Derepression of IL-2Rα and IL-7Rα genes in SATB1-null DP thymocytes. (A) RNA isolated from total thymocytes of SATB1-null and wild-type thymi were examined by a multi-probe RNase protection assay system containing a series of cytokine receptor probes. [32P]UTP-labeled antisense RNA probes (lane 1) were hybridized in excess to target total cellular RNA, after which free probe and other single-stranded RNAs were digested with RNase. The remaining RNase-protected probes derived from SATB1-null thymocytes (lane 2) and wild-type thymocytes (lane 3) were purified, and resolved on 6% denaturing polyacrylamide gels. (B) Identical to A except that target RNA was isolated from purified CD4+CD8+ DP cells by cell sorting using a PE-conjugated anti-CD8 antibody and an FITC-conjugated anti-CD4 antibody. (C) The CD3−CD4−CD8− TN cell subset from depleted thymocytes, as described in Fig. 3, were further purified by excluding Tri-Color-stained CD3-, CD4-, and CD8-positive T cells, B cells, granulocytes, dendritic cells, NK cells, and monocytes from thymocytes. The TN cell subset was subsequently subjected to FACS analysis with a PE-conjugated IL-7Rα antibody. (D) IL-7Rα+ cells in a CD4+CD8+ DP population were detected by a PE-conjugated anti-CD8 antibody and an FITC-conjugated anti-CD4 antibody with Cychrome–streptavidin combined with a biotin-conjugated anti-IL7Rα antibody. (C,D) (Bold line) SATB1 null mice; (narrow line) wild-type mice.
Unlike IL-2Rα which is expressed only in TN cells among the total thymocyte population (Ceredig et al. 1985; Raulet 1985), IL-7Rα has been reported to be expressed in TN cells as well as in mature CD4 and CD8 SP cells, but not in DP cells (Sudo et al. 1993). In SATB1-deficient mice, the majority of thymocytes are DP cells (>94%). TN cells, which are expected to express IL-7Rα, comprise <0.5% of the total thymocytes. Therefore, we isolated SATB1-null DP cells and examined their IL-7Rα gene transcription. We found that SATB1-null DP thymocytes actively transcribe the IL-7Rα gene, giving rise to a virtually identical RNase protected band as that obtained from the total thymocytes (Fig. 7A,B). Therefore, at the transcriptional level, the IL-7Rα gene was derepressed, similar to the IL-2Rα gene in SATB1-null DP thymocytes.
Next, we examined the level of surface IL-7Rα in TN, DP, and the small CD4 SP subpopulations of wild-type and SATB1-null thymocytes by FACS analysis. IL-7Rα surface molecules were detected at a similar level in the wild-type and SATB1-null CD4 SP (data not shown), but at a slightly higher level in SATB1-null TN cells compared with wild-type TN cells (Fig. 7C). In contrast to IL-2Rα, however, a very low level of surface IL-7Rα was detected in SATB1-null DP cells, and the FACS histogram profile was identical between SATB1-null and wild-type DP cells (Fig. 7D). From this data, we conclude that the high level IL-7Rα mRNA is untranslated in SATB1-null DP cells (Fig. 7B,D).
Dysregulated transcription of multiple genes occurred during T-cell development in SATB1-null mice
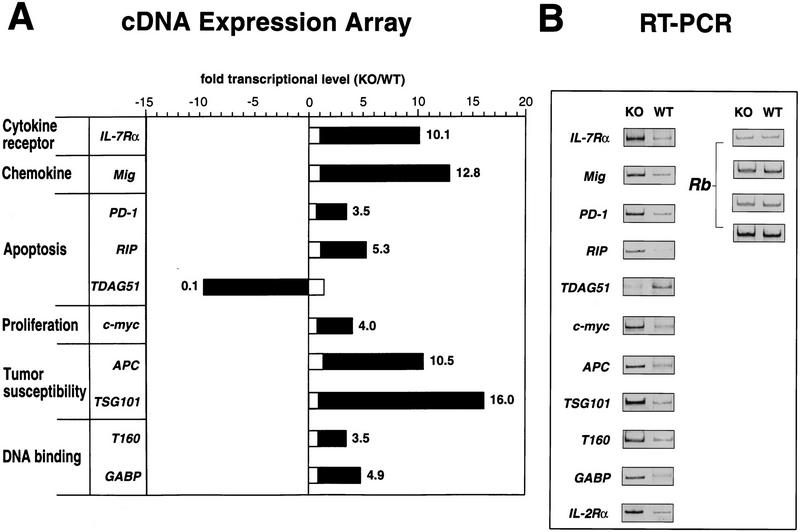
To search for other genes that might be dysregulated as a result of SATB1 ablation, we used the Atlas Mouse cDNA expression Array (Clontech, CA), a nylon membrane containing 588 mouse cDNAs and 9 housekeeping cDNAs. Two identically prepared membranes were each hybridized with cDNA probes prepared from RNA isolated from either wild-type or SATB1-null thymocytes. Two independent analyses, using a new filter set for each experiment, showed >10 genes were significantly up-regulated and one gene was down-regulated in SATB1-null thymocytes. Consistent with our previous data, the IL-7Rα gene was identified as one of these genes (Fig. 8A). Unfortunately, the IL-2Rα gene was not included in this array. On the basis of this initial screening, the genes that exhibited more than a 3.5-fold difference by this expression array were individually assayed by RT–PCR using RNA from SATB1-null and wild-type DP thymocytes. In these experiments, a total of 11 genes, including the IL-2Rα gene, which was absent in the array, were confirmed to be dysregulated in SATB1-null thymocytes (Fig. 8B). This number (11/589) corresponds to ∼2% of the total genes tested. The retinoblastoma (Rb) gene was chosen as a control for RT–PCR. Rb transcripts remained totally unaltered in SATB1-null thymocytes using RT–PCR under all conditions employed (Fig. 8B). In SATB1 thymocytes, all the dysregulated genes except for one were significantly up-regulated, some of them showing an increase as large as that found for the IL-7Rα and IL-2Rα genes. We also examined gene expression in liver, a tissue in which SATB1 is normally totally absent. When liver RNA was examined by array hybridization, no dysregulation in SATB1-null livers in these same 10 genes was detected (Fig. 8A). The other genes displayed on this array, including many highly transcribed genes, were also shown to be unaltered in knockout livers by array hybridization analysis (data not shown). This result indicates that the dysregulated genes are found specifically in those cells that normally express SATB1.
Figure 8.
Dysregulation of multiple genes in SATB1-null thymocytes. (A) Genes that exhibited a >3.5-fold difference in their mRNA levels between SATB1-null and wild-type thymi based on Atlas mouse cDNA Expression Array and confirmed by RT–PCR are shown by solid bars. The ratio of these transcripts between knockout and wild-type liver are shown by open bars. (B) The RT–PCR results for these genes as well as IL-2Rα, which was not included in the expression array, are shown (see Materials and Methods). The Rb gene, which is expressed at a similar level in SATB1-null and wild-type thymocytes, was used as a control under varying RT–PCR conditions.
The microarray data and RT–PCR analyses revealed that gene expression of three apoptosis-related genes, RIP, PD-1, and TDAG51, was highly dysregulated in SATB1-null thymocytes. RIP is a serine/threonine kinase with a death domain and mediates tumor necrosis receptor (TNF)-induced NF-κB activation (Stanger et al. 1995; Hsu et al. 1996; Kelliher et al. 1998). PD-1 is a member of the immunoglobulin superfamily, and is induced by signal transduction through the antigen receptor (Ishida et al. 1992). TDAG51, which is highly expressed in thymocytes, is required to link TCR signaling and Fas expression (Park et al. 1996). The RIP and PD-1 genes were both upregulated, whereas TDAG51 expression was greatly repressed. A significant increase in transcript levels was also detected for the c-myc proto-oncogene in SATB1-null thymocytes compared with wild-type thymocytes. Furthermore, dysregulated genes include two tumor susceptibility genes, the adenomatous polyposis coli (APC) gene, mutation of which leads to multiple colonic adenomas (for review, see Polakis 1995), and the recently identified gene, TSG101 (Li and Cohen 1996). Mig, the monokine induced by interferon-γ, is a CXC chemokine active as a chemoattractant for activated T cells (Farber 1997). The Mig gene was also greatly derepressed in SATB1-null thymocytes. In addition, two DNA-binding proteins (T160 and GABP) were expressed at higher levels in SATB1-null thymocytes. The T160 gene encodes a protein with homology to the non-histone chromosomal protein HMG1 and binds to DNA containing recombination signal sequences (Shirakata et al. 1991). GABP, GA-binding protein, is involved in regulating the ribosomal protein L32 gene (Curcic et al. 1997).
Our results show that SATB1 regulates multiple, but clearly a discrete set of, genes. Whether they are direct or indirect targets of SATB1 regulation, as many as 2% of the total number of genes tested were greatly altered in their temporal and spatial expression during T-cell development in response to SATB1 ablation. Most likely, the net effect of this dysregulation in gene expression is the chaotic defect in T-cell development observed in SATB1-null mice.
Discussion
SATB1 is a BUR/MAR-binding protein expressed predominantly in the nuclei of thymocytes. SATB1 ablation resulted in derepression of multiple genes in thymocytes with impaired T-cell development and function. This study provides the first evidence of the biological function of a MAR-binding protein in vivo. Our data suggest that SATB1 orchestrates the orderly transcription of a series of genes during T-cell development.
SATB1 ablation leads to disordered transcription of multiple genes in thymocytes
Several lines of evidence suggest that genomic DNA is organized into loop domains that vary in size between 5 and 100 kb (for review, see Gasser and Laemmli 1987; Nelson et al. 1986). Such loop domains are presumably formed by the attachment of specific genomic sequences to the nuclear matrix. On the basis of a total DNA content of 6 × 109 bp per diploid nucleus, it is estimated that ∼1 × 105 such loops may exist within a nucleus. A specific group of genomic sequences to which SATB1 binds in vivo are located at the bases of chromatin loops (MARs) that are firmly anchored onto the nuclear matrix (deBelle et al. 1998). The nuclear matrix appears to provide sites at which transcription factors are assembled into factories (Jackson et al. 1993). As SATB1 has been implicated in transcriptional regulation (Banan et al. 1997; Kohwi-Shigematsu et al. 1997; Liu et al. 1997), we reasoned that SATB1 can regulate multiple genes in the loop domains via MAR binding. Therefore, SATB1 ablation might influence transcription of a large number of genes, especially in cells such as thymocytes, which normally express SATB1. On the basis of analyses with mouse cDNA expression arrays and RT–PCR, we confirmed that at least 10 genes were derepressed and that one gene was down-regulated out of 589 genes tested in SATB1-null thymocytes. At this rate, it is likely that hundreds of genes may be dysregulated in thymocytes upon SATB1 ablation, assuming that more than 1 × 104 to 2 × 104 genes are normally expressed in these cells.
The IL-2Rα and IL-7Rα genes were both highly expressed in the SATB1-null DP thymocytes. However, both genes are known to be normally transcriptionally silent at this developmental stage in wild-type thymocytes. Both IL-2Rα and IL-7Rα are necessary for proper T-cell development and function. IL-7Rα-deficient mice suffer from profound lymphopenia (Peschon et al. 1994), and IL-2Rα-deficient mice exhibit autoimmunity, inflammatory bowel disease, and premature death (Willerford et al. 1995). The effect of overexpression of either protein is unknown. In the case of the IL-2Rα gene, two upstream genomic sequences that bind SATB1 in vivo in thymocytes have been identified (data not shown), strongly suggesting that SATB1 directly regulates this gene by acting as a repressor in DP thymocytes. It is likely that the IL-7Rα gene as well as other genes that are similarly derepressed in DP thymocytes are also direct targets of SATB1 regulation. Among these, the PD-1, c-myc, and Mig genes are normally induced upon specific stimulation in thymocytes. The apoptosis-related PD-1 gene is tightly regulated in murine thymocytes and is induced in activation-induced apoptosis by in vivo anti-CD3ε antibody treatment (Ishida et al. 1992; Agata et al. 1996). It is mainly expressed in TN cells and only at a very low level in DP cells (Nishimura et al. 1996). Also, c-myc is known to be induced in thymocytes upon activation (Riegel et al. 1990; Broussard-Diehl et al. 1996). Similarly, a chemokine gene Mig is induced by interferon-γ (Farber 1997). SATB1-null thymocytes that consist largely of DP cells expressed high levels of these genes without any stimulation. These data suggest that SATB1 plays a role in keeping these genes repressed until they are induced for expression.
The ectopic expression of the IL-2Rα and IL-7Rα genes, as well as dysregulation of many other genes including an oncogene, a chemokine gene, apoptosis-related genes, tumor susceptibility genes, DNA-binding protein genes, and developmental surface markers was found in SATB1-null thymocytes and peripheral T cells. These results indicate that the spatial and temporal transcription of multiple genes is disordered in thymocytes and peripheral T cells in the absence of SATB1. In liver, in which SATB1 expression is normally completely absent, no gene was found to be dysregulated in SATB1-null mice.
Multiple defects in T-cell development and function due to SATB1 ablation
SATB1-null mice exhibited multiple defects at almost every developmental stage of T-cell development. This is in contrast to, for example, knockout mice that lack both lymphoid enhancer factor 1 (LEF-1) and T-cell factor 1 (TCF-1; Okamura et al. 1998), which exhibit an arrest at a specific early stage of thymocyte development with marked reduction in TCRα gene transcription. The defects in SATB1-null mice include greatly reduced TN cells, an arrest mainly at the DP stage, inappropriate migration of DP thymocytes to lymph nodes, and apoptosis of peripheral CD4 SP cells in response to activating stimuli. It is clear that SATB1-null thymocytes failed to mature, and there was a dramatic reduction in mature T-cell subsets. Both CD4 and CD8 SP thymocytes and peripheral T cells were very low in number. Failure to up-regulate CD5 is also consistent with the immature phenotype of SATB1-null thymocytes. SATB1 is normally expressed in every thymocyte subpopulation; TN, DP, and SP cells, and in T cells after activation (Beadling et al. 1993; data not shown). Therefore, we interpret the multiple defects as a net effect of disordered transcription of multiple genes due to SATB1 ablation.
We have shown that SATB1-null peripheral T cells undergo apoptosis and fail to proliferate, in response to PMA plus ionomycin. SATB1-null thymocytes, as well as peripheral T cells in lymph nodes, exhibit high incidence of apoptotic cells especially beyond 2 weeks after birth, as evidenced by TUNEL staining of tissue sections (data not shown). The high susceptibility of SATB1-deficient T cells and thymocytes for apoptosis accounts for the greatly reduced size of thymi and lymph nodes in SATB1-null mice compared with wild-type mice, particularly after 14 days of age. Derepressed apoptosis and proliferation-related genes in SATB1-null thymocytes, such as PD-1, RIP, and c-myc, may partially explain the high susceptibility of SATB1-null thymocyes for apoptosis. It is noteworthy that CD44 is not expressed in DP thymocytes (data not shown), whereas it is highly expressed in peripheral T cells of SATB1-null mice. Induction of peripheral SATB1-null CD4 SP T cells apoptosis by activating signals could be due to the activated phenotype of these T cells as evidenced by up-regulation of CD44 and CD25 (Fig. 6B) similar to JAK3-deficient T cells. JAK3-deficient T cells also fail to proliferate in response to mitogenic stimuli and are vulnerable to apoptosis (Thomis et al. 1995, 1997). Further studies are required to understand the role of SATB1 in apoptosis. The effect of apoptosis on the interaction of SATB1 and chromatin will be described in a separate paper (S. Galande, L. Dickinson, G. Salvesen, M. Sikorska, and T. Kohwi-Shigematsu, in prep.).
Although the definitive cause for the death of SATB1-null mice at about 3 weeks of age is unknown, apoptosis of cells other than thymocytes that normally express SATB1 may contribute to this. SATB1 is expressed not only in thymocytes but also in other restricted cell types including absorptive cells of intestinal epithelium, osteoblasts, ameloblasts, and fetal neurons (data not shown). Up to 2 weeks of age, SATB-null mice are healthy and active. At days 19–23, however, they become emaciated as if they are suffering from malnutrition and dehydration. We detected apoptosis of absorptive cells of intestinal epithelium for animals after about 19 days, which would cause problems in absorbing nutrients and water. Together with developmental defects in teeth, SATB1-null mice cannot survive past weaning. Why apoptosis occurs within such a narrow window of time remains a topic of further study.
How might SATB1 regulate multiple genes but a distinct set of genes?
SATB1 is expressed at a very high level in thymocytes and several potential BUR sites containing the ATC sequence cluster are typically found when any given gene locus of 50–100 kb is examined. Yet the percentage of genes affected by SATB1 ablation is estimated to be only about 2% of the total genes examined. How could SATB1 specifically affect certain genes but not others? SATB1 may provide a framework that mediates assembly of specific protein complexes onto a discrete set of BURs. Therefore, SATB1 may act as an architectural protein on the nuclear matrix. The consequence of such protein–protein interactions may be transcriptional repression, but in certain cases it could also be activation. Recently, SATB1 was shown to associate with Cux protein (Liu et al. 1999b). Additional SATB1-binding proteins have been identified (X. Wu and T. Kohwi-Shigematsu, unpubl.). Examining each of these associated proteins may lead to clues about the specificity and activity of SATB1 not only in transcription but also in recombination as well. A DNA segment that is important for recombination of a δ-deleting element (Janowski et al. 1997) binds to SATB1 in vivo (K. Janowski, T. Companion, W.-M. Liu, T. Kohwi-Shigematsu, and R. Hockett, in prep.). SATB1 also binds in vivo to the BCL2 major breakpoint region (Ramakrishnan et al. 2000).
SATB1 may also be able to alter chromatin structure in selected loop domains to change accessibility of these sites to transcription/recombination factors. Future research will examine whether SATB1 recruits enzymes responsible for modification of histone and nonhistone proteins (for review, see Roth and Allis 1996), and/or alters nucleosome positioning at BURs (Fragoso et al. 1995). The precise function of SATB1 and the mechanism for its activity remain to be studied, making the knockout mice an invaluable tool for future studies.
Materials and methods
Isolation and characterization of genomic clones
A probe spanning nucleotides 21–1727 of the human testis SATB1 cDNA (Dickinson et al. 1992) was used to screen a 129Sv mouse genomic library in the Lambda FIX II vector according to manufacturer's instructions (Stratagene, La Jolla, CA). Among 17 clones isolated, clone SB1-3.2 contained the first five exons, encompassing the first 213 amino acids (837 nucleotides of the cDNA). The first exon encoded 5′-untranslated sequence only, and contained the testis-specific 5′-UTR, which is AT rich. The thymus-specific first exon, which is GC rich, may also be contained in SB1-3.2, but its exact location has yet to be determined. Clone SB1-8.1 contained exons 6, 7, and 8 encompassing a 261-amino-acid region of SATB1 containing the 129 out of 150 amino acids of the DNA-binding domain.
Targeting constructs and generation of chimeric mice
Clone SB1-3.2 was used to prepare the targeting vector (pSB1Neo3.2KO). This vector contains a 2.7-kb EcoRI fragment containing the PGK promoter-linked neomycin resistance gene flanked by a 1.1-kb BglII–EcoRII (short arm) and a 5.1-kb BamHI–HindIII (long arm) fragments derived from SB1-3.2, as well as the herpes tk gene.
To generate heterozygous SATB1-targeted ES cell clones, pSB1Neo3.2KO was linearized with SacI, transfected into ES cells (E14 cells supplied by Dr. S. Tonegawa). Drug selection was with 0.3 mg/ml G418 (Gibco BRL, Gaithersburg, MD) and 2.0 μm gancyclovir (Washington University Pharmacy). Drug-resistant clones were screened for homologous recombination of the targeting vector by PCR analysis with primers G7 (5′-GGGTGGGGTGGGATTAGATAAATG-3′) and SB1-PC4 (5′-GCTCATAATTTGTCTTGACCAACAG-3′) yielding a 1.8-kb product. For Southern blotting, a 400-bp BglII–EcoRI fragment derived from the SB1-3.2 clone was hybridized to genomic DNA digested with PstI.
Three independent ES cell clones containing a targeted mutation of SATB1 were injected into C57BL/6 blastocysts. The resulting chimeras were bred to C57BL/6 mice.
Histochemistry
Thymi and spleens were isolated from 11- to 22-day-old mice, fixed in 10% buffered formalin and embedded in paraffin. Sections were cut at a thickness of 5 μm, mounted, and stained with a hematoxylin and eosin.
Flow cytometry of thymocytes and T cells
Thymi and lymph nodes were removed from 12- to 14-day-old wild-type and SATB1-null mice. Mice from the same litter were used in each comparative study. A cell suspension was made in 10 ml of RPMI/ 5% FCS media. The cells were then pelleted by centrifugation and resuspended in 1 ml of staining buffer (1× PBS with 4% FCS and 0.1% sodium azide). Single cell suspensions of lymphocytes (∼2 × 105–5 × 105 cells) were stained with the following antibody conjugates: anti-CD4–FITC (RM4-5), anti-CD8α–phycoerythrin (PE) (53-6.7), anti-αβTCR–Cy-Chrome (H57-597), anti-B220–PE (RA3-6B2), anti-IgM FITC (R6-60.2), anti-CD2–biotin (RM2-5), anti-CD44–biotin (IM7), and anti-HSA–biotin (J11d), and anti-CD69–biotin (H1.2F3). Biotinylated antibodies were detected with streptavidin–Cy-Chrome or streptavidin–Texas Red. All antibodies were purchased from PharMingen (San Diego, CA) unless otherwise specified. Lymphocytes were selected by Forward Scatter (FSC) and Side Scatter (SSC) criteria, and 1 × 104 events were collected per sample. The total number (n) of cells per tissue was counted.
To detect either CD5+ or CD25+ cells in each CD4+CD8+ DP and CD4+ SP subset of thymocytes, total thymocytes were stained with anti-CD4–biotin (RM4-5), anti-CD8–PE (53-6.7) and either anti-CD5–FITC (53-7.3) or anti-CD25–FITC (7D4). The cells were stained further with streptavidin–Tri-Color (Caltag, South San Francisco, CA).
DP thymocytes were purified from SATB1-null and wild-type thymocytes stained with anti-CD4–FITC and anti-CD8–PE by use of a MoFlo MLS flow cytometer (Cytomation, Fort Collins, CO) at a rate of 17,000 cells per second. For RNA analyses, isolated DP thymocytes were sorted directly into TRI Reagent (Sigma Chemical, St. Louis, MO).
Analysis of TN thymocytes
TN thymocytes were isolated by depletion of CD4+ and CD8+ thymocytes as described previously (Suda and Zlotnik 1991). To examine CD25+ and CD44+ TN thymocytes, either depleted thymocytes or total undepleted thymocytes were stained with biotin-conjugated anti-CD3 (145-2C11), anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-B220 (RA3-6B2), anti-Mac-1 (Caltag), and anti-Gr-1 (RB6-8C5). These cells were stained further with anti-CD44–PE (Caltag), anti-CD25–FITC (7D4) and streptavidin–Tri-Color (Caltag). Cells were washed twice between labelings with biotinylated antibodies and streptavidin reagents. A minimum of 200,000 events per sample was collected. For total thymocytes, the percent of the Tri-Color negative population was determined by use of a FACStar-plus (Becton-Dickinson, Milpitas, CA) and analyzed with Cellquest software. For depleted thymocytes with an enriched TN population, 10,000 events of gated, Tri-Color negative cells were analyzed for CD25- and CD44-positive cells.
T-cell stimulation assay
The CD4 SP cell population was isolated by use of the Mouse CD4 Subset Mini Column Kit (R&D Systems, Minneapolis, MN) following the protocol supplied by the manufacturer. Enrichment was routinely 98% as confirmed by FACS analysis. Cells (3 × 104) were seeded into each well of a 96-well round-bottom culture plate (Corning/Costar, Cambridge, MA) and cultured with 10 ng/ml PMA and 500 ng/ml ionomycin. Cell number was determined at each time point by use of a hemacytometer. IL-2 production was measured by use of an anti-mouse IL-2–horseradish peroxidase Biotrak ELISA system (Amersham Pharmacia Inc., Piscataway, NJ). Exogenous IL-2 was added at 100 U/ml (PharMingen, San Diego, CA). The lymph nodes of SATB1-null mice were very small. Typically, 2 × 105 to 4 × 105 cells were isolated from lymph nodes from an 11- to 13-day-old SATB1-null mouse. Among these cells, approximately 0.5 × 105 to 1 × 105 CD4 SP cells were isolated per SATB1-null mouse of this age group.
RNA analysis
Total RNA from flow-sorted DP thymocytes or total thymocytes of day 12 SATB1-null and wild-type thymi was prepared by a one-step acid guanidinium isothiocynate–phenol procedure using the TRI Reagent (Sigma Chemical, St. Louis, MO). The RNA was subsequently incubated at 37°C for 15 min in a solution containing 10 mm MgCl2, 1.0 mm DTT, 0.5 U/μl RNAsin (Promega, Madison, WI) and 0.2 U/ml RNase-free DNase 1 (GIBCO BRL, Gaithersburg, MD), and then extracted with phenol/chloroform and chloroform, precipitated with isopropanol, washed in 75% ethanol, and resuspended in diethylpyrocarbonate-treated water.
RNase protection analysis was performed with a RiboQuant Ribonuclease Protection Assay (RPA) kit and the cytokine receptor template set MCR-1 from PharMingen (San Diego, CA) according to the manufacturer's protocol. A ZAP-70 template was prepared and added to the labeling reaction. This template was prepared by RT–PCR with primers containing KpnI or PstI linkers for amplification of ZAP70 cDNA: 5′-AGTTGGTACCGCTCTGGCAGCTGGTGGAGTACCTGAA-3′ and 5′-TGTTCTGCAGTTCAGCACCTTGATGGCCACGTCA-3′. The resulting ZAP-70 cDNA fragment was cloned into the pBluescript vector (Stratagene). A 600-bp PvuII fragment including the T7 promoter and the ZAP-70 cDNA fragment was gel purified for in vitro transcription. The ZAP70 template (0.5 ng of DNA per reaction) produces a 470-bp transcript, and the final protected product is 410 bp in length.
Analysis by use of the Atlas Mouse cDNA Expression Array (Clontech, Palo Alto, CA) was performed exactly following the manufacturer's protocol with 10 μg of total RNA prepared from either thymus or liver of 12-day-old SATB1-null or wild-type mice.
For RT–PCR, first-strand cDNA was synthesized with a pool of antisense primers for selected genes (listed below) and 4 μg of total RNA isolated from total and flow-sorted DP thymocytes of day-12 SATB1-null and wild-type thymi as described above. Reverse transcription was carried out with the SuperScript RNase H− Reverse Transcriptase (GIBCO BRL, Gaithersburg, MD). Semiquantitative PCR was performed in the linear range of amplification by testing PCR products from different dilutions of first-strand cDNA products. PCR amplification was performed for 26 and 30 cycles with a 1:400 dilution, or 30 and 35 cycles using a 1:800 dilution of the first-strand cDNA synthesized above. Then, PCR products were electrophoresed on a 10% non-denaturing acrylamide gel and detected by staining with SYBR Gold (Molecular Probes, Eugene, OR) fluorescent dye. Subsequently, the intensity of the fluorescent bands was measured on a Storm Imager (Molecular Dynamics, Sunnyvale, CA). Primers used for RT–PCR were obtained from Clontech except for the following: IL-2Rα (sense, 5′-GTGGTGTTCTCTTTCATCTA-3′; antisense, 5′-AGCCCTCTCCTACAAGAA-3′), RIP (sense, 5′-CCACCAGTTCCAAACTGCAT-3′; antisense, 5′-GGCACTTCACCAATGTTG-3′), Mig (sense, 5′-GAGGTCTTTGAGGGATTTGT-3′; antisense, 5′-GACATTCTCGGACTTCAC-3′), c-myc (sense, 5′-GGAAGCAGCTCGAATTTCTT-3′; antisense, 5′-GGGAAACTTTGCCCATTG-3′), T160 (sense, 5′-GACAGGAAGCAGCTTTTACT-3′; antisense, 5′-CCATCTGAGTAACTGCTC-3′), and TSG101 (sense, 5′-GGATAGTGTCCTCACTGATT-3′; antisense, 5′-CCACCAAATACCTCCTAC-3′).
Acknowledgments
We thank Drs. M. Capon and Dr. A. Zlotnik for kind instruction on TN thymocyte experiments and valuable discussion, and Dr. J. Cupp for his kind help on FACSstar analysis. We thank Dr. S. Krajewski for his help with the histological analysis. We thank Drs. K.-I. Nakayama, K. Nakayama, Y. Shinkai, I. Negishi, N. Motoyama, S. Senju, L. Dustin, P. Lucas, I. Chan, and F. Wang for helpful discussion and technical advice, Q. Zhang for blastocyst injection, W.-M. Liu for the ZAP-70 template preparation, C. Oxford for flow cytometry and sorting services and E. Robey for reagents and discussion. This work was supported by National Institute of Health (RO1CA39681 to T.K.-S.), the U.S. Department of Energy Contract DE-AC03-76F00098 (to T.K.-S.), and in part by the Howard Hughes Medical Institute (D.Y.L.), and by the Medical Scientist Training Program (J.D.A.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL terumiks@lbl.gov; FAX (510) 486-4545.
References
- Adams MD, Kerlavage AR, Fields C, Venter JC. 3,400 new expressed sequence tags identify diversity of transcripts in human brain. Nat Genet. 1993;4:256–267. doi: 10.1038/ng0793-256. [DOI] [PubMed] [Google Scholar]
- Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, Honjo T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol. 1996;8:765–772. doi: 10.1093/intimm/8.5.765. [DOI] [PubMed] [Google Scholar]
- Banan M, Rojas IC, Lee W-H, King HL, Harriss JV, Kobayashi R, Webb CF, Gottlieb PD. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J Biol Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- Beadling C, Johnson KW, Smith KA. Isolation of interleukin 2-induced immediate-early genes. Proc Natl Acad Sci. 1993;90:2719–2723. doi: 10.1073/pnas.90.7.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- Broussard-Diehl C, Bauer SR, Scheuermann RH. A role of c-myc in the regulation of thymocyte differentiation and possibly positive selection. J Immunol. 1996;156:3141–3150. [PubMed] [Google Scholar]
- Ceredig R, Lowenthal JW, Nabhol M, MacDonald HR. Expression of interleukin-2 receptors as a differentiation marker on intrathymic stem cells. Nature. 1985;314:98–100. doi: 10.1038/314098a0. [DOI] [PubMed] [Google Scholar]
- Chan AC, Desai DM, Weiss A. The role of protein tyrosine kinases and protein tyrosine phosphatases in T cell antigen receptor signal transduction. Annu Rev Immunol. 1994;12:555–592. doi: 10.1146/annurev.iy.12.040194.003011. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Yuen M-H, Garrard WT. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- Crowley C, Spencer SD, Nishimura MC, Chen KS, Pitts-Meek S, Armanini MP, Ling LH, MacMahon SB, Shelton DL, Levinson AD, et al. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- Curcic D, Glibetic M, Larson DE, Sells BH. GA-binding protein is involved in altered expression of ribosomal protein L32 gene. J Cell Biochem. 1997;65:287–307. [PubMed] [Google Scholar]
- deBelle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to SATB1 in immature T cells in vivo are tightly associated with the nuclear matrix. J Cell Biol. 1998;141:335–338. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson LA, Kohwi-Shigematsu T. Nucleolin is a MAR/SAR DNA-binding protein specifically recognizing a region with high base-unpairing potential. Mol Cell Biol. 1995;15:456–465. doi: 10.1128/mcb.15.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Dickinson LA, Dickinson CD, Kohwi-Shigematsu T. The nuclear matrix attachment region (MAR)-binding protein SATB1 contains a homeodomain that promotes specific recognition of the core unwinding element of a MAR. J Biol Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- Farber JM. Mig and IP-10: CXC chemokines that target lymphocytes. J Leukoc Biol. 1997;61:246–256. [PubMed] [Google Scholar]
- Fehling HJ, von Boehmer H. Early αΒ T-cell development in the thymus of normal and genetically altered mice. Curr Opin Immunol. 1997;9:263–275. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- Forrester WC, van Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglubulin μ gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- Fragoso G, John S, Roberts MS, Hager GL. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes & Dev. 1995;9:1933–1947. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- Fraser JD, Irving BA, Crabtree GR, Weiss A. Regulation of interleukin-2 gene enhancer activity by the T-cell accessory molecule CD28. Science. 1991;251:313–316. doi: 10.1126/science.1846244. [DOI] [PubMed] [Google Scholar]
- Galande S, Kohwi-Shigematsu T. Poly(ADP-ribose)polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- Gasser SM, Laemmli UK. A glimpse at chromosomal order. Trends Genet. 1987;3:16–22. [Google Scholar]
- Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheurmann R, Tucker PW. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: A B cell-specific trans-activator that describes a new DNA-binding protein family. Genes & Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- Hoffman ES, Passoni L, Crompton T, Leu TM, Schatz DG, Koff A, Owen MJ, Hayday AC. Productive T-cell receptor beta-chain gene rearrangement: Coincident regulation of cell cycle and clonality during development in vivo. Genes & Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- Hubbe M, Altevogt P. Heat-stable antigen/CD24 on mouse T lymphocytes: Evidence for a costimulatory function. Eur J Immunol. 1994;24:731–737. doi: 10.1002/eji.1830240336. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski K, Ledbetter S, Mayo MS, Hockett RD. Identification of a DNA segment exhibiting rearrangement modifying effects upon transgenic δ-deleting elements. J Exp Med. 1997;186:91–100. doi: 10.1084/jem.186.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Forrester WC, Fernandez-Herrero LA, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Kirillov A, Kistler B, Mostoslasky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-κB-cell-specific demethylation of the Igκ locus. Nat Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- Klein R, Silos-Santiago I, Smeyne RJ, Lira SA, Brambilla R, Bryant S, Zhang L, Snider WD, Barbacid M. Disruption of the neurotrophin-3 receptor gene trkC eliminates la muscle afferents and results in abnormal movements. Nature. 1994;368:249–251. doi: 10.1038/368249a0. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T, Kohwi Y. Torsional stress stabilizes extended base-unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry. 1990;29:9551–9560. doi: 10.1021/bi00493a009. [DOI] [PubMed] [Google Scholar]
- ————— Detection of non-B DNA structures at specific sites in supercoiled plasmid DNA and chromatin with haloacetaldehyde and diethylpyrocarbonate. Methods Enzymol. 1992;212:155–180. doi: 10.1016/0076-6879(92)12011-e. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T, Maass K, Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated MAR-linked reporter genes. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- Leonard NJ. Etheno-substituted nucleotides and coenzymes: Fluorescence and biological activity. CRC Crit Rev Biochem. 1984;15:125–199. doi: 10.3109/10409238409102299. [DOI] [PubMed] [Google Scholar]
- Li L, Cohen SN. tsg101: A novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross SR, Dudley JP. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997a;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Barnett A, Neufeld EJ, Dudley JP. Homeoproteins CDP and SATB1 interact: Potential for tissue-specific regulation. Mol Cell Biol. 1999b;19:4918–4926. doi: 10.1128/mcb.19.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W-M, Guerra-Vladusic FK, Kurakata S, Lupu R, Kohwi-Shigematsu T. HMG-I(Y) recognizes base-unpairing regions of matrix attachment sequences and its increased expression is directly linked to metastatic breast cancer phenotype. Cancer Res. 1999a;59:5695–5703. [PubMed] [Google Scholar]
- Nakagomi K, Kohwi Y, Dickinson LA, Kohwi-Shigematsu T. A novel DNA binding motif in the MAR-binding protein SATB1. Mol Cell Biol. 1994;14:1852–1860. doi: 10.1128/mcb.14.3.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, Pienta KJ, Barrack ER, Coffey DS. The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Chem. 1986;15:457–475. doi: 10.1146/annurev.bb.15.060186.002325. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Agata Y, Kawasaki A, Sato M, Imamura S, Minato N, Yagita H, Nakano T, Honjo T. Developmentally regulated expression of the PD-1 protein on the surface of double-negative (CD4−CD8−) thymocytes. Int Immunol. 1996;8:773–780. doi: 10.1093/intimm/8.5.773. [DOI] [PubMed] [Google Scholar]
- Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T-cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- Park CG, Lee SY, Kandala G, Lee SY, Choi Y. A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity. 1996;4:583–591. doi: 10.1016/s1074-7613(00)80484-7. [DOI] [PubMed] [Google Scholar]
- Peschon JJ. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. Mutations in the APC gene and their implications for protein structure and function. Curr Opin Genet Dev. 1995;5:66–71. doi: 10.1016/s0959-437x(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan M, Liu W-M, DiCroce PA, Posner A, Zheng J, Kohwi-Shigematsu T, Krontiris TG. Modulated binding of SATB1, a matrix attachment region protein, to the AT-rich sequence flanking the major breakpoint region of BCL2. Mol Cell Biol. 2000;20:868–877. doi: 10.1128/mcb.20.3.868-877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH. Expression and function of interleukin-2 receptors on immature thymocytes. Nature. 1985;314:101–103. doi: 10.1038/314101a0. [DOI] [PubMed] [Google Scholar]
- Riegel JS, Richie ER, Allison JP. Nuclear events after activation of CD4+8+ thymocytes. J Immunol. 1990;144:3611–3618. [PubMed] [Google Scholar]
- Roth SY, Allis CD. Histone acetylation and chromatin assembly: Single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Functionally responsive T-cell development. Adv Immunol. 1992;51:85–214. doi: 10.1016/s0065-2776(08)60487-3. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Alt FW. CD3 epsilon-mediated signals rescue the development of CD4+CD8+ thymocytes in RAG-2−/− mice in the absence of TCR beta chain expression. Int Immunol. 1994;6:995–1001. doi: 10.1093/intimm/6.7.995. [DOI] [PubMed] [Google Scholar]
- Shirakata M, Hüppi K, Usuda S, Okazaki K, Yoshida K, Sakano H. HMB1-related DNA-binding protein isolated with V-(D)-J recombination signal probes. Mol Cell Biol. 1991;11:4528–4536. doi: 10.1128/mcb.11.9.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KA. Interleukin-2: Inception, impact, and implications. Science. 1988;240:1169–1176. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: A novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–512. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- Suda T, Zlotnik A. Origin, differentiation, and repertoire selection of CD3+CD4−CD8− thymocytes bearing either alpha beta or gamma delta T cell receptors. J Immunol. 1993;150:447–455. [PubMed] [Google Scholar]
- Sudo T, Nishikawa S, Ohno N, Akiyama N, Tamakoshi M, Yoshida H, Nishikawa S. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc Natl Acad Sci. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher TE, Smit L, Griffioen AW, Schilder-Tol EJ, Borst J, Pals ST. Signaling through CD44 is mediated by tyrosine kinases. Association with p56lck in T lymphocytes. J Biol Chem. 1996;271:2863–2867. doi: 10.1074/jbc.271.5.2863. [DOI] [PubMed] [Google Scholar]
- Thomis DC, Lee W, Berg LJ. T cells from Jak3-deficient mice have intact TCR signaling, but increased apoptosis. J Immunol. 1997;159:4708–4719. [PubMed] [Google Scholar]
- Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- Urbánek P, Wang Z-Q, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Wang B, Dickinson LA, Koivunen E, Ruoslahti E, Kohwi-Shigematsu T. A novel MAR binding motif identified using a random phage peptide library. J Biol Chem. 1995;270:23239–23242. doi: 10.1074/jbc.270.40.23239. [DOI] [PubMed] [Google Scholar]
- Willerford DM, Chen J, Ferry JA, Davidson L, Ma A, Alt FW. IInterleukin-2 receptor α chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3:521–530. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]