Abstract
Mutations in the PERIANTHIA (PAN) gene of Arabidopsis thaliana specifically transform flowers from tetramerous to largely pentamerous, which is a characteristic of flowers of ancestral plants. We have cloned the PAN gene and here we show that it encodes a member of the basic region/leucine zipper class of transcription factors. Immunohistochemical analysis shows that the encoded protein is present in the apical meristem, the floral meristem, each whorl of organ primordia, and in ovule primordia during wild-type flower development. PAN expression occurs independently of genes affecting floral meristem identity, floral meristem size, or floral organ number. The near absence of a phenotype in transgenic plants overexpressing PAN and the contrast between the broad expression of PAN and the specificity of its mutant phenotype suggest that its activity may be regulated post-translationally or by the presence of partner proteins. Based on these results and on data reported previously, we propose models for the role of PAN in the evolution of flower pattern in the mustard family.
Keywords: Arabidopsis, flower development, PAN, bZIP protein
Arabidopsis flowers arise initially as undifferentiated bulges on the flank of the apical meristem. Soon afterward, whorls of organ primordia arise sequentially. Four sepal primordia develop along the edges of the floral meristem, establishing the first whorl. Then four petal and six stamen primordia initiate in whorls 2 and 3, respectively, followed by development of two carpel primordia in the center of the floral meristem (Smyth et al. 1990).
A proposed mechanism of floral organ primordium initiation and formation must account for how a primordium is formed and also when and where on the floral meristem it develops. The eventual fate of organ primordia is determined by the organ identity genes (Komaki et al. 1988; Bowman et al. 1989, 1991, 1993; Hill and Lord 1989; Kunst et al. 1989; Irish and Sussex 1990; Jack et al. 1992; Weigel and Meyerowitz 1994); however, the position in which they arise (and thus their number) appears to be established independently (Meyerowitz 1997). How proper floral organ number is achieved is not well understood.
One way to understand the molecular mechanisms by which appropriate floral organ number is determined is to find mutations that affect floral organ number but do not affect floral organ identity or floral meristem identity, and then to elucidate the nature and the function of the corresponding wild-type genes. There are a number of reports demonstrating the existence of such mutations, including clavata1 (clv1), clavata2 (clv2), clavata3 (clv3), ettin (ett), fasciata1 (fas1), fasciata2 (fas2), mgoun1 (mgo1), mgoun2 (mgo2), perianthia (pan), revoluta (rev), tousled (tsl), and wiggum (wig) (Leyser and Furner 1992; Clark et al. 1993, 1995, 1997; Roe et al. 1993, 1997; Sessions and Zambryski 1995; Talbert et al. 1995; Running and Meyerowitz 1996; Sessions et al. 1997; Laufs et al. 1998; Kayes and Clark 1998; Running et al. 1998). In clv1, clv3, and wig mutants, the increase in organ number is correlated with increased cell number in the floral meristem (Clark et al. 1993, 1995; Running et al. 1998). This indicates a mechanism by which the positions of floral organs depend on spacing: The organs arise with a fixed distance between them in each whorl, and more organs arise in whorls containing more cells (Meyerowitz 1997).
pan was recognized mainly by its extra organ mutant phenotype and has been well characterized by phenotypic and genetic analysis. Flowers of plants mutant for pan are characterized by an increase in the organ number of the first two whorls, and a decrease in the organ number of the third whorl. Mutant flowers usually have five sepals with the position of the adaxial sepal as in wild type, five petals alternate with the sepals, five stamens alternate with the petals, and two carpels. Unlike the other mutations mentioned above, mutations in PAN affect floral organ number specifically, without detectable defects in floral meristem cell number or in other parts of the plant (Running and Meyerowitz 1996). Thus, PAN seems to affect the spacing mechanism that determines the relative positions of floral organs directly (Meyerowitz 1997). Genetic analysis shows that PAN acts downstream of the floral meristem identity genes LEAFY (LFY), APETALA1 (AP1), and APETALA2 (AP2), but independently of the floral meristem size genes CLV1, CLV3, and WIG and the floral organ identity genes APETALA3 (AP3), PISTILLATA (PI), and AGAMOUS (AG) in specifying early flower patterning (Running and Meyerowitz 1996; Running et al. 1998). In addition, PAN functions redundantly with ETT (Sessions et al. 1997) and TSL (Roe et al. 1997) in both organ primordium initiation and gynoecium development.
To gain insight into the molecular mechanisms underlying the specification of floral organ patterning in Arabidopsis, we have cloned the PAN gene and determined its pattern of expression in developing flowers. PAN encodes a putative basic region/leucine zipper (bZIP) transcription factor. PAN protein is expressed in the apical meristem, the floral meristem, and each of the floral organ primordia. After each whorl of floral organs has been initiated, PAN expression becomes specific to particular regions in developing petals (the base), stamens (the adaxial surface), and carpels (the ovule primordia and mature ovules). Although PAN requires floral meristem identity genes for its activity (Running and Meyerowitz 1996), PAN expression is not dependent on these genes. PAN expression is also independent of the floral meristem size genes and other genes controlling floral organ number.
Results
Molecular cloning of PAN
We described previously the isolation of the pan-1 and pan-2 alleles from a screen of T-DNA insertion-mutagenized lines of ecotype Wassilewskija (Ws) (Fig. 1B; Feldmann 1992; Running and Meyerowitz 1996). The T3 plants of one line (descended from a single transformed seed) segregated both pan-1 and the KanR gene associated with the T-DNA, but tests of T4 plants showed that the mutant phenotype was unlinked to KanR, suggesting that the pan-1 mutation was not caused by a T-DNA insertion. The T3 plants of another line segregated pan-2, but all plants in this family were also kanamycin resistant. Southern blot hybridization of genomic DNA from T4 plants, using the T-DNA left and right border sequences as probes, revealed the presence of multiple T-DNA insertions in the genome of pan-2 plants. Homozygous pan-2 plants were backcrossed to wild-type Landsberg erecta five times, and F2 plants from the fifth backcross showed cosegregation of the pan-2 phenotype with a single KanR marker. We made a λ genomic library from a homozygous pan-2 F3 family derived from the fifth backcross, and screened this library with probes derived from the T-DNA left and right borders. One clone from this library contained the T-DNA left border and plant genomic DNA, and another clone contained the T-DNA right border and plant genomic DNA. Restriction fragments containing both T-DNA and plant DNA were subcloned and used to probe a flower cDNA library (Weigel et al. 1992). We isolated a single cDNA (of 1324 bp in length) out of ∼1 million clones, the sequence of which corresponded to genomic sequences flanking both sides of the T-DNA insert. A genomic clone was isolated by screening a wild-type Arabidopsis genome library, using the cDNA as a probe. The cDNA was mapped to a 7.2-kb XbaI fragment and a 12-kb PvuII fragment of the genomic clone (Fig. 2A).
Figure 1.

(A) Wild-type flower. (B) pan-1 flower. (C) pan-1 flower with a 7.2-kb XbaI genomic fragment containing the PAN gene. (A) Wild-type flowers have four sepals, four petals, six stamens, and two carpels. (B) Most pan mutant flowers have five sepals, five petals, five stamens, and two carpels. (C) pan-1 plants carrying the transgenic wild-type PAN gene produce flowers indistinguishable from wild type.
Figure 2.
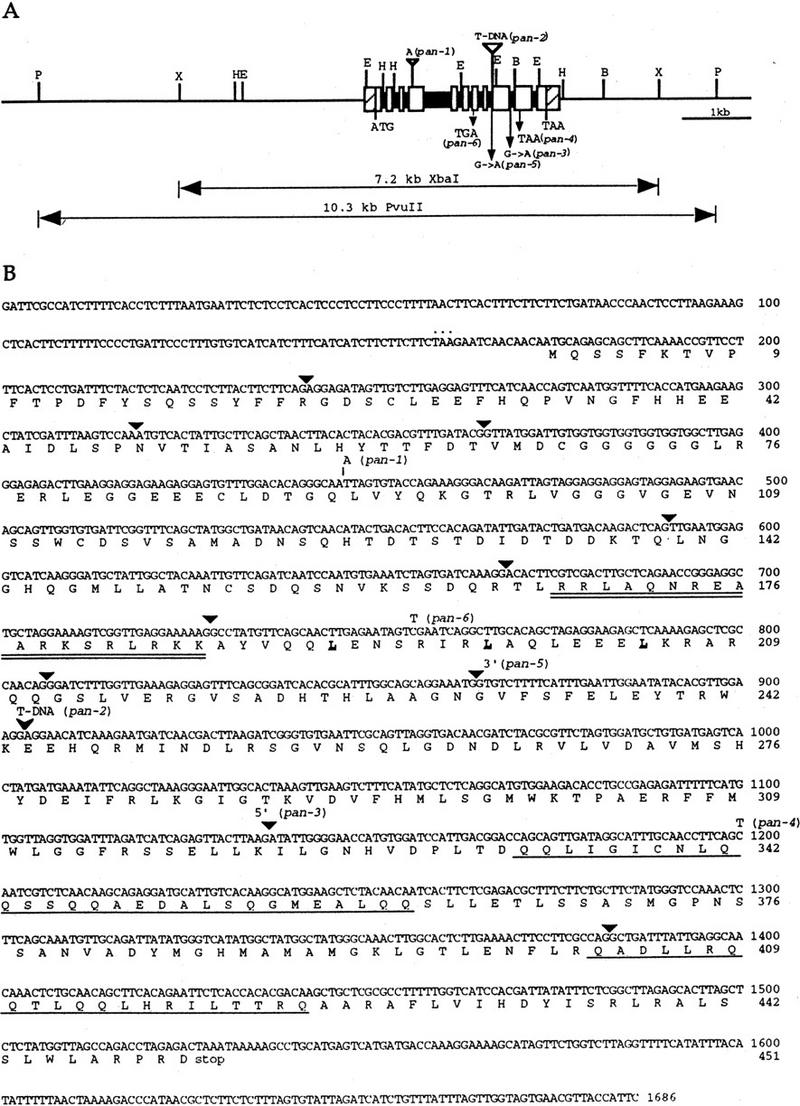
Genomic structure of the PAN gene, and sequence of the PAN cDNA. (A) Structure and restriction map of the PAN genomic region. The open boxes and solid boxes represent exons and introns, respectively. The hatched boxes represent the 5′ and 3′ untranslated regions. The genomic regions used for complementation of pan mutant phenotype are indicated below. ATG and TAA indicate the positions of the putative translational start and stop codons, respectively. Mutations in the six pan alleles are also shown. Restriction sites are indicated as follows: (B) BamHI; (E) EcoRI; (H) HindIII; (P) PvuII; (X) XbaI. (B) The cDNA and deduced amino acid sequence of the PAN gene. The predicted amino acid sequence for the longest open reading frame is shown directly below the nucleotide sequence. Numbers to the right of the sequence refer to the positions of nucleotide and amino acid residues. Triangles indicate the positions of introns. The basic region is doubly underlined and the leucines in the leucine zipper are in bold and underlined. The glutamine-rich regions are singly underlined. The inframe stop codon preceding the first methionine is indicated by dots above the nucleotide sequence. One nucleotide insertion in pan-1 is shown above the nucleotide sequence. The position of the T-DNA insertion in pan-2 allele is indicated by an arrowhead. The pan-3 mutation occurs at the splice donor site of the ninth intron. The pan-5 allele is mutated in the splice acceptor site of the eighth intron. Point mutations resulting in premature translational stop sites in pan-4 and pan-6 are shown above the nucleotide sequence.
To verify that the cloned sequences encode the PAN gene, we transformed these two genomic fragments into pan-1 mutant plants. Flowers from transgenic plants carrying either of the PAN genomic fragments are indistinguishable from those of wild-type plants (Fig. 1A,C). These results (and sequencing results discussed below) demonstrate that the cloned gene corresponds to PAN.
PAN is a bZIP protein with carboxy-terminal glutamine-rich regions
The 1.3-kb cDNA, which contained a 3′ poly(A)+ tail, identified a 1.7-kb mRNA by RNA blot analysis (data not shown), suggesting that the cDNA is not full length and is missing 5′ sequences. The transcriptional start site was identified by rapid amplification of cDNA ends (5′ RACE; Frohman et al. 1988). An in-frame stop codon precedes the first ATG in the longest open reading frame, suggesting that the protein coding sequence is complete (Fig. 2B). The full-length PAN cDNA is 1686 bp and encodes a protein of 451 amino acids. Comparison of these sequences to the GenBank database revealed that the deduced amino acid sequences were similar to members of the bZIP class of transcription factors.
Genomic DNA blot analysis suggests the presence of a single copy of the PAN gene in the Arabidopsis genome (data not shown). Alignment of the cDNA and genomic DNA sequences indicated that the PAN gene is composed of 11 exons (Fig. 2A,B). All exon–intron junctions conform to the GT–AG rule. The intron sizes range from 68 to 96 bp except that the fourth intron is 436 bp, the exon sizes range from 47 to 330 bp. The basic region and leucine zipper are encoded by separate small exons, as in other plant bZIP-encoding genes (Fig. 2B). The PAN protein also contains two glutamine-rich regions (∼30% glutamine residues) near the carboxyl terminus: 9/29 glutamine residues at amino acids 333–361 and 6/21 glutamine residues at amino acids 403–423, which may function as transcriptional activation domains (Figs. 2B, 3; Courey and Tjian 1988).
Figure 3.
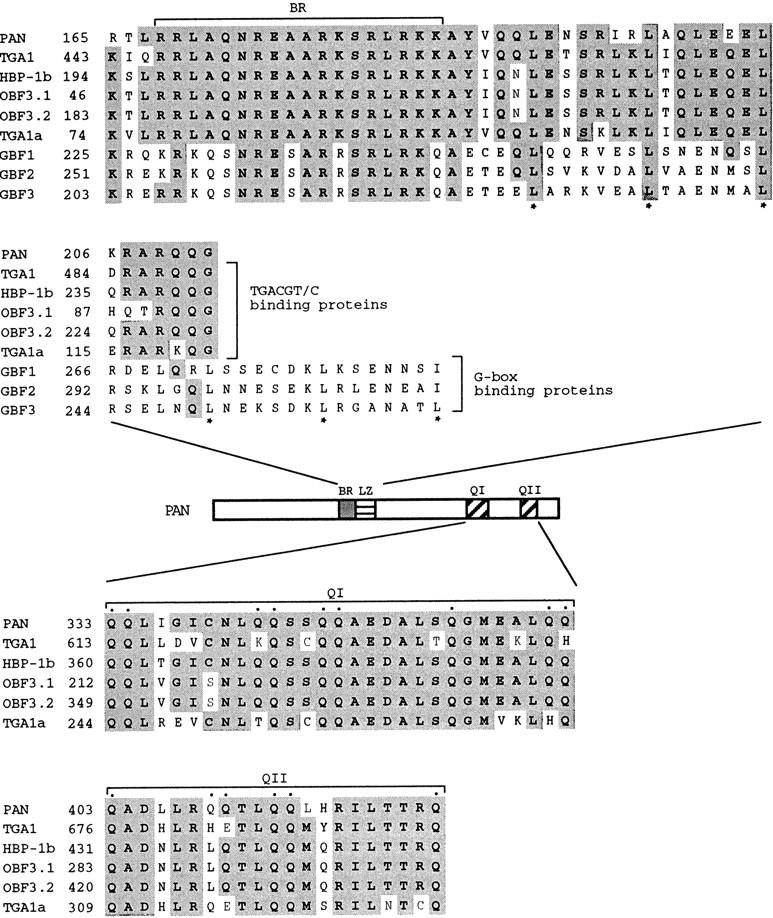
Similarity between PAN and other plant bZIP proteins. (Top) Comparison of the PAN bZIP regions to other plant bZIP proteins: the Arabidopsis protein TGA1 (Schindler et al. 1992a), the wheat protein HBP-1b (Tabata et al. 1991), the maize protein OBF 3.1 and OBF 3.2 (Foley et al. 1993), the tobacco protein TGA1a (Katagiri et al. 1989), and the Arabidopsis proteins GBF1, GBF2, and GBF3 (Schindler et al. 1992b). Asterisks (*) indicate leucine residues within the leucine zipper domain. (Middle) Schematic presentation of PAN, indicating the positions of the basic region (BR), the leucine zipper domain (LZ), and the two glutamine-rich regions (QI and QII). (Bottom) Sequence alignments within the carboxy-terminal glutamine-rich domains of PAN and the TGACGT/C-binding proteins shown in the top. Glutamine residues within the glutamine-rich regions of PAN are indicated by dots. Identical amino acids are shown as shaded boxes. Numbers to the left refer to the amino acid positions within individual proteins.
Sequence of pan mutations
The pan-3 to pan-6 alleles were isolated from a screen for crabs claw (crc) enhancers in ecotype Landsberg erecta (Y. Eshed and J.L. Bowman, unpubl.). To identify mutations in the pan alleles and to confirm that we had identified the PAN gene, we used PCR to amplify genomic DNA or cDNA from each of the six pan alleles and directly sequenced these PCR products. The allele pan-1 contains a single base pair insertion at codon 91, which results in a frameshift at this position (Fig. 2A,B). The pan-2 allele has a T-DNA insertion in the ninth exon (Fig. 2A,B), resulting in the deletion of a 23-bp fragment immediately after the insertion site. The pan-3 allele is a G → A substitution at the splice donor site of the ninth intron (Fig. 2A,B). Sequence analysis of RT–PCR products revealed three improperly spliced forms of pan-3 mRNA, all encoding truncated proteins lacking the glutamine-rich domains. The point mutation found in the pan-4 allele results in a premature stop codon within the glutamine-rich domain I and therefore is expected to eliminate the glutamine-rich domain II. In pan-5, there is a single nucleotide change (G → A) at the splice acceptor site of the eighth intron and the first nucleotide (G) is used as an alternative splice acceptor site, resulting in a frameshift and a premature stop codon after the basic region and leucine zipper. The allele pan-6 has a premature stop codon caused by a nucleotide change in the leucine zipper region. Identification of mutations in these pan alleles, and complementation of the pan-1 mutant phenotype by the cloned gene confirm that we have isolated the PAN gene.
Structural similarity between PAN and other plant bZIP proteins
The basic region/leucine zipper motif and the glutamine-rich regions are highly conserved between PAN and other plant bZIP proteins, including the Arabidopsis protein TGA1 (Schindler et al. 1992a), the wheat protein HBP-1b (Tabata et al. 1991), the maize proteins OBF 3.1 and OBF 3.2 (Foley et al. 1993), and the tobacco protein TGA1a (Katagiri et al. 1989), all of which have been shown to interact with the TGACGT/C motif identified in viral and plant promoters (Fig. 3). These proteins have 100% amino acid identity with PAN within the basic region, which is responsible for binding DNA. In contrast, the DNA-binding domain of PAN is more divergent from the Arabidopsis bZIP proteins GBF1, GBF2, and GBF3 (Schindler et al. 1992b; Fig. 3), all of which interact with the G box (CCACGTGG). These G box-binding proteins contain a leucine zipper consisting of five or six leucines instead of just three leucines as in PAN and the TGACGT/C binding proteins (Fig. 3). In addition, the G-box-binding proteins do not have the glutamine-rich regions.
Expression of PAN RNA and subcellular localization of PAN protein in the apical meristems and organ primordia
To investigate the spatial and temporal expression pattern, as well as the subcellular localization of the PAN protein, immunohistochemical analysis using a PAN-specific polyclonal antibody was performed. During vegetative growth, PAN protein is detected first in the shoot apical meristem of the mature embryo (Fig. 4A) and is localized to the nucleus (Fig. 4B), consistent with the indication that PAN is a potential transcription factor. PAN expression extends from the apical meristem to young leaf primordia in the seedling (Fig. 4C). After the transition from vegetative growth to flowering, PAN protein is expressed at high levels throughout the apical region of the inflorescence meristem (Fig. 4D,E), and PAN remains strongly expressed in the floral meristem in stage 1 and 2 floral buds (Fig. 4D,E; stages as defined in Smyth et al. 1990). At stage 3, when sepals initiate, PAN protein is confined to regions of the floral meristem interior to the sepals where further organ initiation events are taking place (Fig. 4D,E). PAN protein is still detected in the inner three whorls of organ primordia in late stage 4 and 5 flowers (Fig. 4D,E).
Figure 4.
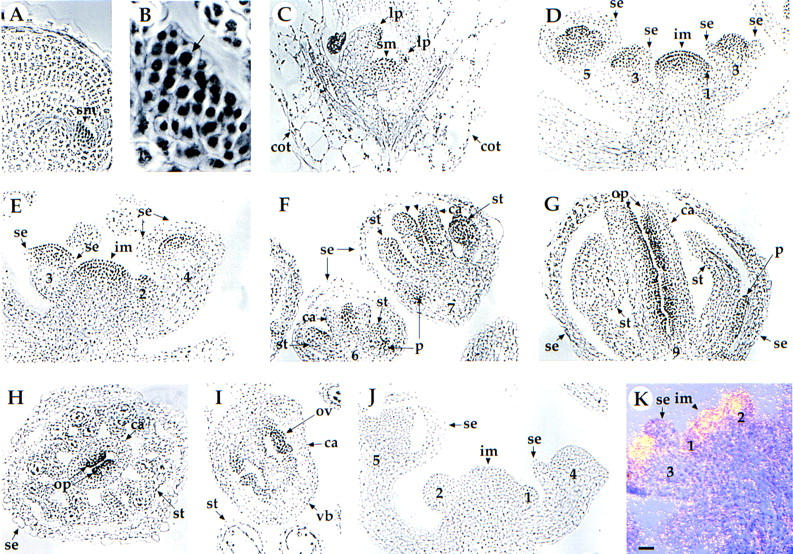
Localization of PAN protein and distribution of PAN RNA in wild-type (A–I, K) and pan mutant (J) plants. (A) Section through the shoot apical meristem (sm) in the mature embryo. Seeds were imbibed at 4°C for 2 days and transferred to constant fluorescent light at 20°C for 1 day. PAN is highly expressed in the shoot apical meristem (arrowhead). (B) High magnification view of the cells in the shoot apical meristem shown in A. The arrow indicates that the PAN protein is localized primarily to the nucleus (arrow). (C) Longitudinal section through a shoot apical meristem of 6-day-old seedling. PAN protein is detected in the shoot apical meristem (sm) and leaf primordia (lp), but not in the cotyledons (cot). (D,E) Longitudinal section of an inflorescence meristem (im) with stage 2–5 flowers. PAN protein is detected at high levels throughout the apical region of the inflorescence meristem (arrowhead), throughout the floral meristem of the stage 2 flower, the central dome of the floral meristem of stage 3–4 flowers, and the inner three whorls of organ primordia in the stage 5 flower. (se) Sepal. (F) Longitudinal section of stage 6–7 flowers. PAN protein becomes restricted in the base of petal (p) primordia, the adaxial side of stamen (st) primordia, and the adaxial side of carpel (c) primordia (arrowhead). Longitudinal section (G) and cross section (H) of a stage 9 flower. PAN protein remains at high levels in the ovule primordia (op). (I) Cross section of a stage 12 flower. Expression level of PAN protein becomes weaker in the ovule (ov). The PAN protein is not expressed at levels above background in sections of inflorescences from pan-1 (J) and pan-2 (data not shown). (K) In situ hybridization of a PAN antisense probe with a longitudinal section of an inflorescence meristem (im) with stage 1–3 flowers. Expression of PAN RNA is detected throughout the apical region of the inflorescence meristem and throughout the floral meristem of stage 1–2 flowers. Expression in the stage 3 flower is limited to regions interior to the sepals (se). In the bright field/dark field double exposures, the silver grains representing PAN expression were made to appear yellow. The number indicated corresponds to the stages of floral development as described by Smyth et al. (1990). All panels shown at the same magnification. Bar, 20 μm.
At stage 6, when each whorl of floral organ primordia has initiated, expression of PAN becomes spatially restricted to specific regions within developing petals, stamens, and carpels. During stages 6–8, PAN is expressed weakly in the bases of the emerging petal primordia, whereas expression is strong in the adaxial half of stamens and the cells lining the central invagination (Fig. 4F). PAN protein is restricted to the ovule primordia and the parietal placental tissue during stages 9–11 (Fig. 4G,H). Late in floral development, at stage 12, PAN expression becomes weaker in ovules and placenta (Fig. 4I). By contrast, only low background is detected in control experiments using the preimmune serum as the primary antibody (data not shown). PAN is not detected in roots, stems, and leaves (data not shown). The PAN protein is not detected above background in pan-1 (Fig. 4J) and pan-2 (data not shown) mutants, suggesting that these are null or severe reduction-of-function alleles, and demonstrating the specificity of the antibody.
In situ hybridization experiments showed that PAN RNA is present in the inflorescence meristem, the floral meristem (Fig. 4K), and in developing petals, stamens, and carpels (data not shown) in the same spatial and temporal pattern as that of PAN protein.
Localization of PAN in lfy, ap1, and ap2 mutants
Double-mutant analyses demonstrate that the floral organ identity mutants lfy, ap1, and ap2 are epistatic to pan, suggesting that PAN acts downstream of these genes (Running and Meyerowitz 1996). To determine whether PAN expression is regulated by these upstream genes, we analyzed the expression of PAN protein in lfy, ap1, and ap2 mutant flowers.
Mutations in the LFY gene cause partial transformation of flowers into inflorescence shoots (Weigel et al. 1992). Secondary flowers occasionally arise from the axils of the outer organs in strong lfy flowers. In lfy-6 mutants, PAN protein localization is largely normal in the inflorescence meristem and the floral primordia of primary (Fig. 5A) and secondary flowers (Fig. 5B).
Figure 5.
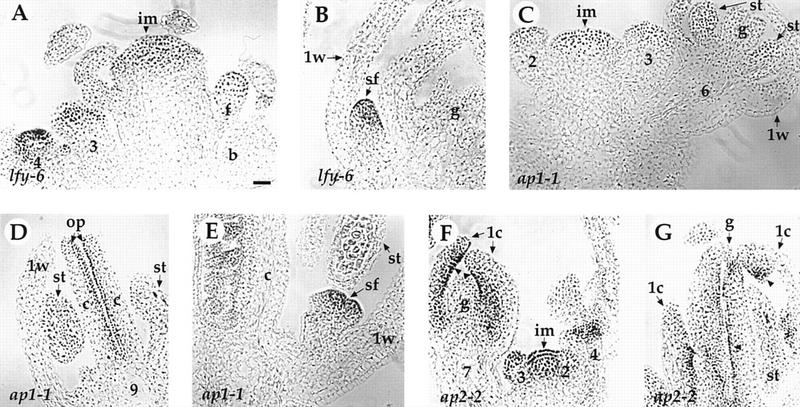
Expression of the PAN protein in lfy, ap1, and ap2 mutants. (A–B) lfy-6. (A) PAN protein is expressed in the inflorescence meristem (im) and the center of abnormal young flowers. The stages of flowers (3 and 4) and the floral bud (f) in the axil of the bract (b) are indicated. (B) PAN protein is detected in the secondary floral buds (sf) in the axils of the first-whorl organs (1w) of an old lfy-6 flower. (g) Gynoecium. (C–E) ap1-1. (C) Early expression in the inflorescence meristem (im) and in the stage 2, 3, and 6 floral buds is normal (compared with Figs. 4D–F). (st) Stamen. (D) PAN protein is expressed normally in the ovule primordia (op) at stage 9 flowers (cf. with Fig. 4G). (c) Carpel. (E) Secondary floral buds (sf) in the axil of the first-whorl organs (1w) show PAN protein expression at levels comparable to the early flower primordia arising on the inflorescence apex (Figs. 4D,E). (F–G) ap2-2. PAN protein expression is largely normal in the inflorescence meristem (im) and young floral buds (stage 2–4). At later stages, PAN protein is detected in the ovule primordia (arrowhead) in both of the first-whorl carpels (1c) and the central gyneocium (g). The number indicated corresponds to the developmental stage of each flower (Smyth et al. 1990). Bar, 20 μm.
Similarly to lfy, mutations in AP1 result in the development of secondary flowers in the axils of the first whorl organs, indicating a partial conversion of a floral meristem into an inflorescence meristem (Irish and Sussex 1990). In ap1-1 mutants, PAN expression is largely normal in the mutant inflorescence meristem, abnormal floral primordia (Fig. 5C), and ovule primordia (Fig. 5D). In addition, PAN is also expressed in the secondary floral buds (Fig. 5E).
Mutations in AP2 usually cause homeotic transformation of floral organs in the first and second whorls. In strong ap2 mutants, sepals are transformed into carpels, petal development is suppressed, and stamens are reduced in number (Bowman et al. 1991). In ap2-2 mutants, PAN expression is largely normal in the inflorescence meristem, young floral primordia (Fig. 5F), and ovule primordia in the fourth whorl carpels (Fig. 5F,G). PAN is also expressed in the ovule-bearing carpels in the first whorl (Fig. 5F,G), indicating that PAN expression in the carpels is cell-type specific, and not whorl specific.
We conclude from these results that the spatial and temporal expression pattern, as well as the expression level of PAN protein are largely normal in lfy, ap1, and ap2 mutants, suggesting that LFY, AP1, and AP2 protein functions are not required for PAN expression during flower development.
Expression of PAN in clv1-4, clv3-2, ett-3, wig-1, and tsl-1
To determine whether PAN expression patterns are altered in other mutants with abnormal floral organ number but with normal floral organ identity, we examined the PAN protein level in mutant flowers. Mutations in CLV1, CLV3, ETT, and WIG increase organ number within the flower, whereas mutations in TSL decrease organ number (Clark et al. 1993, 1995, 1997; Roe et al. 1993, 1997; Sessions and Zambryski 1995; Sessions et al. 1997; Running et al. 1998). Plants carrying a mutation in either of the CLV1 or CLV3 genes also produce flowers with additional whorls of carpels in the center of flowers. PAN protein is expressed normally in the inflorescence meristem and the floral meristem in these mutants (Fig. 6A,C–F). In addition, PAN is also expressed in the fifth whorl of clv1-4 (Fig. 6B) and clv3-2 flowers (data not shown).
Figure 6.
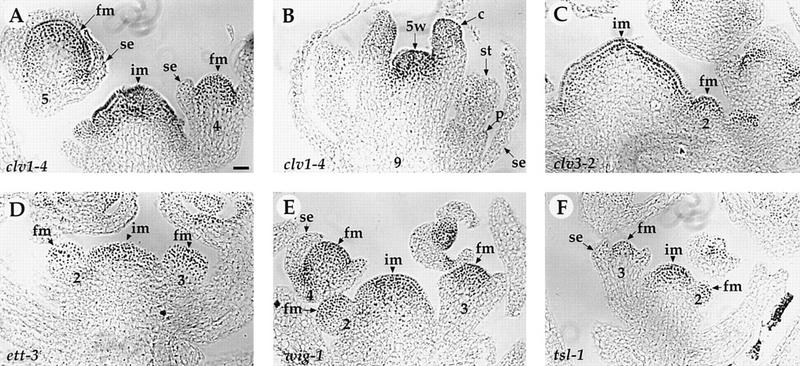
Expression of PAN protein in clv1-4, clv3-2, ett-3, wig-1, and tsl-1 mutants. (A–B) clv1-4. PAN is expressed in the inflorescence meristem (im), the floral meristem (fm, A), and the fifth-whorl (5w) of clv1-4 flowers (B). PAN protein expression is normal in the inflorescence meristem and the floral meristem in clv3-2 (C), ett-3 (D), wig-1 (E), and tsl-1 (F) flowers. (se) Sepal; (p) petal; (st) stamen; (c) carpel. The number indicated corresponds to the developmental stage of each flower (Smyth et al. 1990). Bar, 20 μm.
Ectopic expression of PAN
To examine the effect of ectopic PAN expression on flower development, we constructed transgenic lines that constitutively express PAN. To select a functional transgene, the PAN cDNA driven by the constitutive 35S promoter from cauliflower mosaic virus was transformed into pan-1 plants and the rescued plants were crossed to wild-type plants. The resultant F2 plants that contained the 35S::PAN transgene, but did not contain the pan-1 allele, were analyzed.
Western blot analysis of plant proteins from flower tissues showed that the expression of PAN protein was increased in 35::PAN flowers compared to wild-type (data not shown). Immunolocalization assays on sections of flower tissues further demonstrated that the PAN protein was expressed uniformly throughout the flower in the 35S::PAN transgenic plants (data not shown). The 35S::PAN transgene, which can rescue the mutant phenotype in pan-1 plants, does not cause any noticeable phenotype with respect to floral organ number in wild-type plants (data not shown).
Discussion
Arabidopsis flowers consist of four concentric whorls of organs. Most wild-type flowers have four sepals, four petals, six stamens, and two fused carpels, from the outermost first whorl to the innermost fourth whorl. Whereas much is known about the establishment of floral organ identity, less is known about how the proper number of floral organs is achieved. Recently we identified one mutant, pan, specifically affecting floral organ number (Running and Meyerowitz 1996). Here we describe the isolation of the PAN gene. Two lines of evidence demonstrate that we have cloned the PAN gene: (1) Mutations were found in the six pan alleles. (2) Two different genome fragments containing the cloned gene can complement the pan mutant phenotype.
PAN encodes a bZIP protein
PAN shares homology with members of the bZIP class of transcription factors. The basic region is implicated in DNA binding and the leucine zipper mediates homo- and heterodimerization. Several plant bZIP proteins have been shown to bind the ACGT cis-acting element identified in the promoters of bacterial, viral, and plant genes (Mikami et al. 1987; Giuliano et al. 1988; Bouchez et al. 1989; Lam et al. 1989; Schindler et al. 1992a, 1992b). Based on their DNA-binding specificity and heterodimerization characteristics, these plant bZIP proteins can be classified into at least two classes: the G box (CCACGTGG)-binding factor (GBF) family and the TGACGT/C-binding proteins (Schindler et al. 1992a; Foster et al. 1994). Proteins from each class are also distinct in their overall protein structure. Members of the GBF family are characterized by an amino-terminal proline-rich region and a bZIP region at the carboxyl terminus (Schindler et al. 1992b). In contrast, proteins of the second class, including the Arabidopsis protein TGA1 (Schindler et al. 1992a), the tobacco protein TGA1a (Katagiri et al. 1989), the wheat protein HBP-1b (Tabata et al. 1991), the maize proteins OBF 3.1 and OBF 3.2 (Foley et al. 1993), contain an amino-terminal bZIP domain and a carboxyl terminus enriched in glutamine and acidic amino acids. Sequence alignment analysis revealed that PAN is more similar to members of the second class of bZIP proteins than it is to members of the GBF family (Fig. 3). However, there is an 118 amino acid domain at the amino terminus of PAN that shows no similarity to previously cloned genes.
PAN expression pattern is correlated with floral organ initiation
Immunohistochemical analysis revealed that PAN protein is localized in both floral tissues and vegetative tissues including the apical meristems during vegetative and reproductive growth, the young leaf primordia, four whorls of floral organ primordia, and developing petals, stamens, and ovules. In situ hybridization showed the same spatial and temporal pattern as that of PAN protein.
Mutations in the PAN gene only affect the floral organ number in the first three whorls. However, expression domains of PAN protein in wild-type plants are not restricted to the tissues which are affected in pan flowers. There are two possibilities to explain why some tissues expressing PAN are not affected in pan plants. PAN may be functionally redundant with other factors in some tissues, or PAN does not participate in vegetative growth and apical meristem development. PAN expression in the gynoecium may explain a delay in carpel fusion in some pan flowers (Running and Meyerowitz 1996), and is consistent with the previous findings that PAN functions redundantly with TSL (Roe et al. 1997), ETT (Sessions et al. 1997), and CRC (Y. Eshed and J. L. Bowman, unpubl.) in gynoecium development.
The PAN protein expression pattern is largely normal in mutants affected in meristem identity (LFY, AP1, and AP2), and in organ number without changes in organ identity (CLV1, CLV3, ETT, WIG, and TSL). We conclude from these results that the activity of these genes is not required for PAN expression.
Implications from PAN ectopic expression
Overexpression of PAN can rescue the phenotype of pan mutant plants but does not confer any phenotype in wild-type plants. This result suggests the existence of spatially and/or quantitatively limited factors that modulate PAN protein activity, or heterodimerize with PAN; thus, overexpression of PAN alone is not enough to cause a dominant phenotype. PAN functions downstream of the meristem identity genes LFY, AP1, and AP2. However, PAN expression is not altered in lfy, ap1, and ap2 mutants and PAN is expressed in a wider domain than LFY and AP1. Therefore, LFY, AP1, and AP2 may activate a spatially limited partner of PAN.
It is possible that the activity of PAN is regulated post-translationally. Much evidence suggests that phosphorylation of transcription factors can affect their DNA-binding properties (Prywes et al. 1988; Yamamoto et al. 1988), or their ability to activate transcription in animal systems (Cherry et al. 1988; Tanaka and Herr 1990; Weiss et al. 1991). In plants, it has been shown that the phosphorylation of an Arabidopsis leucine zipper protein, GBF1, by casein kinase II from broccoli stimulates its binding to the specific DNA site (Klimczak et al. 1992). Western blot analysis revealed the existence of two forms of PAN protein (data not shown). One may represent a phosphorylated form of PAN, supporting this hypothesis. It is also possible that PAN needs to function with other factors in a complex. Protein/DNA-binding studies using cotranslated bZIP templates have shown that Arabidopsis proteins GBF1,2,3 (Schindler et al. 1992b), maize proteins OPH1 and Opaque2 (Pysh et al. 1993), and parsley proteins CPRF1, 2, 3 (Armstrong et al. 1992) were able to form heterodimers.
Possible models for PAN in regulation of floral organ number
By analogy to a morphogen theory of leaf primordium positioning, the position in which the floral organ primordia arise could be controlled by inhibitory and promoting types of mechanisms. In this model, an inhibitor of adjacent organ inception is produced at the position of initiation of each organ. The inhibitor decreases in effectiveness with increasing age of the primordium, meaning that the inhibitor decays with time. New organ primordia will arise at concentration minima in the inhibitory field. However, different floral whorls seem to have a promoting rather than an inhibitory effect on each other’s positioning (Lyndon 1990).
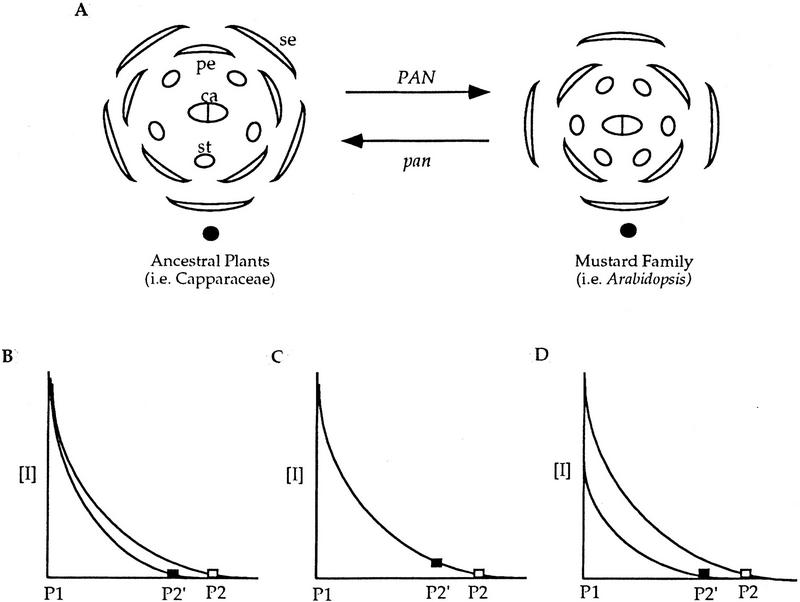
Flowers of the family Brassicaceae (mustards), including Arabidopsis, are tetramerous. Interestingly, flowers of pan mutant plants become pentamerous, which is characteristic of flowers of ancestral plants such as members of the family Capparaceae (Meyerowitz et al. 1997). This suggests that PAN may be involved in the switch from pentamerous to tetramerous flowers during the evolution of flower form in the mustard family (Fig. 7A). We propose that the tetramerous flower of the mustard family was established by introducing the PAN gene into the pre-existing inhibitory pathway mentioned above.
Figure 7.
Possible models for PAN in regulating floral organ number. We postulate that there is an inhibitory mechanism controlling the relative positions of first and second whorl floral organ primordia in ancestral plants (i.e., Capparaceae) of the mustard family (i.e., Arabidopsis) and PAN participates in the pre-existing inhibitory pathway to establish tetramerous flowers in the mustard family. (A) Introduction of PAN into the morphogen pathway leads to more distant spacing of organ primordia and thus fewer organs. When PAN activity is reduced or eliminated (pan), plants produce flowers with a pattern resembling the ancestral flower form. (B–D) The inhibitor concentration [I] is plotted as function of distance. The diffusing inhibitor decays with time and thus distance. P1, the first primordium. (█) pan; (□) PAN. (B) PAN might facilitate inhibitor diffusion or persistence to allow the inhibitor to spread more widely, resulting in the formation of the second primordium (P2) at a more distant position than in pan (P2′). (C) PAN might enhance inhibitor reception, leading to formation of the next primordium at a lower inhibitor concentration than in pan. (D) PAN might increase inhibitor production, indirectly leading to its wider diffusion. (se) Sepal; (pe) petal; (st) stamen; (ca) carpel; (solid ovals) inflorescence meristem; (□) and (█) the lowest concentration in the inhibitory field allowing the new primordium to arise in PAN and pan floral meristems, respectively.
We propose three possible ways by which PAN may participate in the inhibitory pathway to control floral organ patterning. First, PAN might facilitate inhibitor diffusion, thereby allowing the inhibitor to spread more widely (Fig. 7B), thus, the second organ primordium in the first or second whorl can form at the position opposite of the first one (Fig. 7A, right). The inhibitory fields created by the first and the second primordia combine to determine the position of lateral primordia. When PAN activity is reduced or eliminated, the inhibitor reaches a concentration that allows organ formation at a position closer to the first primordium because of reduced concentration and normal decay (Fig. 7B), resulting in two primordia formed in the first inhibitory field (Fig. 7A, left). Similarly, the first three inhibitory fields combine to determine the positioning of the remaining organs (Fig. 7A, left). Second, PAN could enhance reception of the inhibitor. In this scenario, loss of PAN activity decreases the sensitivity of cells to the inhibitor, allowing primordia to form at a higher inhibitor concentration and thus narrower spacing of organ primordia (Fig. 7A,C). Third, PAN might activate inhibitor production, indirectly leading to its wider diffusion (Fig. 7A,D).
Expression of PAN throughout the floral meristem supports the first and second models in which PAN functions in the whole meristem to regulate its response to the inhibitor. However, we cannot rule out the possibility that PAN could interact with spatially limited factors to regulate localized expression of the diffusible inhibitor. In any case, introduction of PAN activity to the morphogen model leads to a wider spacing of organ primordia and thus fewer organs in the first two whorls. The increase in organ number in the third whorl is perhaps explained by duplication of the medial stamens, resulting in six instead of four third-whorl organs (Bowman 1994). Loss of PAN function would cause a flower form similar to that of ancestral plants. Future experiments expressing PAN in relatives of ancestral plants should help to test these models.
Materials and methods
Cloning of PAN
Genomic DNA was isolated from the five times-backcrossed pan-2 line, partially digested with Sau3AI, then partially filled in and ligated to XhoI half-site arms of λ GEM11 (Promega), and packaged in vitro. The library was then screened using the T-DNA left and right borders as probes.
We screened ∼1 × 106 clones of an Arabidopsis flower cDNA library in λZAPII made from wild-type Landsberg erecta ecotype with mixed probes containing left and right border T-DNA and the flanking plant genomic DNA. We isolated a single clone, which was converted to a plasmid by in vivo excision according to manufacturer’s instructions (Stratagene). Remaining sequences (residues 1–556) of the full-length PAN cDNA were determined by 5′ RACE System (BRL) using total RNA from floral buds of Wassilewskija ecotype. A λ phage genomic library made from wild-type Columbia ecotype of Arabidopsis (Hua et al. 1995) was screened at high stringency (65°C hybridization and a final wash with 0.1× SSPE) with a 1353 bp PCR fragment (residues 174–1526) of PAN cDNA as a probe.
DNA sequencing
The cDNA and genomic DNA fragments were subcloned into pBluescript SK(+) (Stratagene), pGEM3Zf(+) (Promega) and pUC19 vectors for sequencing. Sequencing was done using the Sequenase Kit (U.S. Biochemical) or the dye terminator cycle sequencing kit (Perkin-Elmer Co.) according to the protocols provided. Both strands were sequenced.
Growth of plants
Plants were grown in a 4:3:2 or 1:1:1 mixture of soil:vermiculite:perlite in 3 × 5-in pots placed in plastic trays, imbibed at 4°C for 4 days, then maintained at 20°C beneath 600 foot candles of constant cool-white fluorescent light. Plants were fertilized at regular intervals.
Complementation constructs and Agrobacterium- mediated transformation
The 7.2-kb XbaI and 12-kb PvuII genomic fragments, which contain the PAN gene, were subcloned into the BamHI site of the T-DNA expression vector pDHB321.1. These T-DNA constructs were transformed into the Agrobacterium strain C58C1(pMP90). Agrobacterium carrying either of the PAN genomic fragments was then used to transform plants by vacuum infiltration (Bechtold et al. 1993). Transformants were selected by sowing seeds on sand subirrigated with water containing 7.5 mg/l of Basta herbicide. Basta-resistant plants were then transferred to soil.
Production of PAN-specific polyclonal antiserum
The PAN protein was translationally fused to a series of six histidine residues, which allows simple purification of recombinant proteins by immobilized metal affinity chromatography (Hochuli et al. 1987). To make this construct, PAN cDNA encoding the full-length PAN protein was cloned into the expression vector pRSETA (Invitrogen) using BamHI and KpnI restriction sites. This plasmid was transformed into Escherichia coli strain BLR(DE3), and upon induction with 0.8 mm IPTG for 3 hr at 37°C, significant levels of PAN fusion protein were produced. The protein was partially purified using the Ni-NTA resin (QIAGEN), and used to immunize rabbits. The titer of antiserum was determined by Western blot analysis. The specificity of the antibody was demonstrated by control experiments that showed no immunolocalization signal in pan-1 and pan-2 mutants.
Immunolocalization
Tissue was fixed in 1× PBS containing 4% paraformaldehyde, 0.1% Triton-X-100, and 0.1% Tween-20 at 4°C overnight. Fixed tissue was dehydrated with ethanol, cleared with histoclear (National Diagnotics), embedded in paraffin (Paraplast Plus), and sectioned at 8 μm. Antibody reactions and staining were done using the Blocking kit, the VECTASTAIN Elite ABC kit and the DAB substrate kit (Vector Laboratories) according to the protocols provided.
In situ hybridization
A 306-bp fragment of the PAN cDNA (bases 190–495), which contained sequences not closely homologous to previously reported sequences on database searches, was amplified by PCR and cloned into the TA cloning vector (Invitrogen). In vitro transcription and 35S labeling of probes was performed using the Promega Riboprobe kit, with template linearized by BamHI and transcribed by T7 polymerase (sense probe), or linearized by EcoRV and transcribed by SP6 polymerase (antisense probe). In situ hybridization was performed as described by Drews et al. (1991), with modifications by Sakai et al. (1995). Tissue was sectioned at 5.5 μm thickness. Exposure time was 5–7 weeks.
Acknowledgments
We thank Yuval Eshed and John Bowman for the isolation and gift of the pan-3 to pan-6 alleles, Patricia Zambryski for the ett and tsl seeds. We also thank Catherine Baker, Xuemei Chen, Jennifer Fletcher, Neil Hopper, Toshiro Ito, Carolyn Ohno, Robert Sablowski, Doris Wagner, and Eva Ziegelhoffer for critical comments on the manuscript, and L. Medrano for technical assistance. This work was supported by USDA-National Research Initiative Competitive Grants Program (NRICGP) grant 9602558 to E.M.M. M.P.R. was a Howard Hughes Predoctoral Fellow. R.W.W. was supported by National Institutes of Health predoctoral training grant GM07616 and the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The nucleotide sequence data of PAN reported in this paper will appear in the EMBL, GenBank, and DDBJ databases under accession number AF111711.
Footnotes
E-MAIL meyerow@cco.caltech.edu; FAX (626) 449-0756.
References
- Armstrong GA, Weisshaar B, Hahlbrock K. Homodimeric and heterodimeric leucine zipper proteins and nuclear factors from parsley recognize diverse promoter elements with ACGT cores. Plant Cell. 1992;4:525–537. doi: 10.1105/tpc.4.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci. 1993;316:1194–1199. [Google Scholar]
- Bouchez D, Tokuhisa JG, Llewellyn DJ, Dennis ES, Ellis JG. The ocs-element is a component of the promoters of several T-DNA and plant viral genes. EMBO J. 1989;8:4197–4204. doi: 10.1002/j.1460-2075.1989.tb08605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, editor. Arabidopsis: An atlas of morphology and development. New York, NY: Springer-Verlag; 1994. Flowers: Introduction; pp. 135–145. [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- Cherry JR, Johnson TR, Dollard C, Shuster JR, Denis CL. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell. 1988;56:409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–2067. [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: Seed infection/transformation. In: Koncz C, Chua N-H, Schell J, editors. Methods in Arabidopsis Research. Singapore: World Scientific; 1992. pp. 274–289. [Google Scholar]
- Foley RC, Grossman C, Ellis JG, Llewellyn DJ, Dennis ES, Peacock WJ, Singh KB. Isolation of a maize bZIP protein subfamily: Candidates for the ocs-element transcription factor. Plant J. 1993;3:669–679. [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua N-H. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994;8:192–200. doi: 10.1096/fasebj.8.2.8119490. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts-amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR. An evolutionarily conserved protein binding sequence of a plant light-regulated gene. Proc Natl Acad Sci. 1988;85:7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JP, Lord EM. Floral development in Arabidopsis thaliana: Comparison of the wild type and the homeotic pistillata mutant. Can J Bot. 1989;67:2922–2936. [Google Scholar]
- Hochuli E, Döbeli H, Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J Chromatography. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. Function of the APETALA1 gene during Arabidopsis floral development. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LB, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua N-H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Kayes JM, Clark SE. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- Klimczak LJ, Schlindler U, Cashmore AR. DNA binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell. 1992;4:87–98. doi: 10.1105/tpc.4.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki MK, Okada K, Nishino E, Shimura Y. Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development. 1988;104:195–203. [Google Scholar]
- Kunst L, Klenz JE, Martinez-Zapater J, Haughn GW. AP2 gene determinates the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell. 1989;1:1195–1208. doi: 10.1105/tpc.1.12.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Benfey PN, Gilmarten PM, Fang R-X, Chua N-H. Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci. 1989;86:7890–7894. doi: 10.1073/pnas.86.20.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Dockx J, Kronenberger J, Traas J. MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development. 1998;125:1253–1260. doi: 10.1242/dev.125.7.1253. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Furner IJ. Characterization of three shoot apical meristem mutants of Arabidopsis thaliana. Development. 1992;116:397–403. [Google Scholar]
- Lyndon RF. Plant development: The cellular basis. Winchester, MA: Unwin Hyman; 1990. Chapter 3: Meristem functioning: Formation of branches, leaves, and floral organs. Chapter 4: Shape, growth directions, and surface structure; pp. 39–79. [Google Scholar]
- Meyerowitz EM. Genetic control of cell patterns in developing plants. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM, Running MP, Sakai H, Williams RW. Multiple modes of cell division control in Arabidopsis flower development. In: Greenland AJ, Meyerowitz EM, Steer M, editors. Symposia of the Society for Experimental Biology LI: Control of Plant Development: Genes and signals. Cambridge, U.K: The Company of Biologists Limited; 1997. pp. 19–26. [PubMed] [Google Scholar]
- Mikami K, Tabata T, Kawata T, Nakayama T, Iwabuchi M. Nuclear protein(s) binding to the conserved DNA hexameric sequence postulated to regulate transcription of wheat histone genes. FEBS Lett. 1987;223:273–278. doi: 10.1016/0014-5793(87)80303-4. [DOI] [PubMed] [Google Scholar]
- Prywes R, Dutta A, Cromlish JA, Roeder RG. Phosphorylation of serum response factor, a factor that binds to the serum response element of the c-FOS enhancer. Proc Natl Acad Sci. 1988;85:7206–7210. doi: 10.1073/pnas.85.19.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Aukerman MJ, Schmidt RJ. OPH1: A maize basic domain/leucine zipper protein that interacts with opaque2. Plant Cell. 1993;5:227–236. doi: 10.1105/tpc.5.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JL, Rivin CJ, Sessions RA, Feldmann KA, Zambryski PC. The Tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell. 1993;75:938–950. doi: 10.1016/0092-8674(93)90537-z. [DOI] [PubMed] [Google Scholar]
- Roe JL, Nemhauser JL, Zambryski PC. TOUSLED participates in apical tissue formation during gynoecium development in Arabidopsis. Plant Cell. 1997;9:335–353. doi: 10.1105/tpc.9.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running MP, Meyerowitz EM. Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development. 1996;122:1261–1269. doi: 10.1242/dev.122.4.1261. [DOI] [PubMed] [Google Scholar]
- Running MP, Fletcher JC, Meyerowitz EM. The WIGGUM gene is required for proper regulation of floral meristem size in Arabidopsis. Development. 1998;125:2545–2553. doi: 10.1242/dev.125.14.2545. [DOI] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;378:199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR. TGA1 and G-box binding factors: Two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell. 1992a;4:1309–1319. doi: 10.1105/tpc.4.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992b;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC. Arabidopsis gynoecium structure in the wild type and in ett mutants. Development. 1995;121:1519–1532. doi: 10.1242/dev.121.5.1519. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Nakayama T, Mikami K, Iwabuchi M. HBP-1a and HBP-1b: Leucine zipper-type transcription factors of wheat. EMBO J. 1991;10:1459–1467. doi: 10.1002/j.1460-2075.1991.tb07666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 1995;121:2723–2735. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: Interdependent activation domains induce Oct-2 phosphoylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Batut J, Klose KE, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- Yamamoto KK, Gonzalez GA, Biggs WH, III, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of a nuclear factor CREB. Nature. 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]