Abstract
The human positive regulatory domain I binding factor 1 (PRDI-BF1) and its murine homologue Blimp-1 promote differentiation of mature B cells into antibody secreting plasma cells. In contrast, ectopic expression of PRDI-BF1 in lymphoma cells can lead to inhibition of proliferation or apoptosis. However, little is currently known about the regulation of PRDM1, the gene encoding PRDI-BF1. This report establishes that in lymphoma cells stimulation through the B cell receptor rapidly induces endogenous PRDM1 at the level of transcription with minor changes in mRNA stability. The induced PRDM1-encoded protein localizes to its target genes in vivo and suppresses their expression. In vivo genomic footprinting of the PRDM1 promoter in unstimulated lymphoma and myeloma cells reveals multiple common in vivo occupied elements throughout the promoter. Further functional and structural analysis of the promoter reveals that the promoter is preloaded and poised for activation in the B cell lines. The transcription factor PU.1 is shown to be required for the B cell receptor induced expression of PRDM1 in lymphoma cells and in PU.1 positive myeloma cells. Activation of PRDM1 is associated with loss of the co-repressor TLE4 from the PU.1 complex. These findings indicate PRDM1 is poised for activation in lymphoma cells and therefore may be a potential therapeutic target to inhibit lymphoma cell proliferation and survival.
Introduction
The positive regulatory domain I binding factor 1 (PRDI-BF1), encoded by the PRDM1 gene, was originally found to specifically bind the interferon beta (IFN-β) promoter and suppress IFN-β transcription following viral induction (1). Blimp-1, the murine homologue of PRDI-BF1, was originally described by Turner et al. as a transcription factor that could induce the differentiation of B cells (2). PRDI-BF1/Blimp-1 has since been found to be required for the differentiation of a B cell to a plasma cell (3). During the differentiation of mature B cells to plasma cells, PRDI-BF1 represses multiple genes involved in maintaining the B cell phenotype and in maintaining cellular proliferation, such as CIITA (4, 5), c-myc (6), and BSAP (7). Microarray studies have outlined the PRDI-BF1 repression profile and led to the identification of two additional direct targets, Spi-B and Id3 (8). Additionally, expression of the PRDM1 gene has recently been linked to cellular stress and the unfolded protein response in B cells (9).
Anti-IgM cross-linking of the B cell receptor has been reported in multiple studies to induce apoptosis in lymphoma cells (10–14). This response has been correlated with decreased levels of c-myc (6). Inducing PRDI-BF1/Blimp-1 expression in lymphoma cells with histone deacetylase inhibitors also decreased expression of the downstream targets c-myc and BSAP (15). More specifically, introduction of PRDI-BF1/Blimp-1 into lymphoma cells can induce apoptosis, suggesting PRDI-BF1 may be an important mediator of the anti-IgM-mediated apoptotic response (16, 17). However, no direct link between expression of PRDI-BF1/Blimp-1 and anti-IgM mediated B cell receptor activation has been described.
Recently, PRDM1 expression has been detected in a subset of diffuse large B cell lymphomas (DLBCL) (18–20). However, inactivating mutations in the PRDM1 coding sequence were described, indicating a potential tumor suppressor role for this gene (19, 20). Similarly, proliferating myeloma cells and myeloma cell lines abundantly express the truncated PRDI-BF1 isoform, PRDI-BF1β, which has impaired function (21). Additionally, Borson et al. demonstrated PRDM1 expression in B cells isolated from myeloma patients while normal donors lack expression (22). The mutation status of PRDM1 in these myeloma-derived B cells is as yet unknown. Together, these findings indicate PRDI-BF1/Blimp-1 may be important to the pathology of various hematopoietic malignancies, including lymphoma.
Very little is known as to the regulation of PRDM1 expression. Our data now demonstrate PRDM1 is regulated primarily at the level of transcription in both myeloma cells and in lymphoma cells stimulated by cross-linking of the B cell receptor. B cell receptor stimulation leads to rapid increases in newly transcribed PRDM1 RNA levels, while mRNA stability is unchanged. Using promoter deletion constructs, we demonstrate several regions of activation in the PRDM1 promoter in both lymphoma and myeloma cells. In vivo genomic footprinting demonstrates multiple protein-DNA interactions in both lymphoma and myeloma cells. Further analysis of these interactions reveals PU.1 binding is functionally important for promoter activity in stimulated lymphoma cells. These findings demonstrate the PRDM1 promoter is poised for rapid activation in lymphoma cells, which suggests inducing PRDM1 expression in lymphoma cells may be a viable target to inhibit lymphoma progression.
Materials and Methods
Cell lines and reagents
The CA46 EBV-negative Burkitt’s lymphoma and RPMI8226 multiple myeloma cell lines were maintained in RPMI medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (HyClone), and 1% penicillin/streptomycin (P/S) (Invitrogen). Goat anti-human IgM antibody (Southern Biotechnology) was used at 10 μg/ml. Actinomycin D (Sigma) was used at 10 μg/ml.
B cell isolation
B cells were isolated from healthy human donors. Briefly, peripheral blood mononuclear cells were isolated by Ficoll separation and incubated with anti-CD19 microbeads (Miltenyi Biotec) followed by magnetic separation using MS columns. The purified cells were routinely >90% B cells as confirmed by flow cytometry analysis for CD20. Isolated B cells were activated by co-cultured with irradiated CD40L expressing L-cells (23) in presence of cytokines IL-2 (20U/ml), IL-4 (50ng/ml), IL-10 (50ng/ml), IL-12 (2ng/ml) for 4 days. The cells were then divided into two flasks one stimulated with anti-IgM and the other unstimulated for 24 hours.
Apoptosis assay
Cells were treated with anti-IgM continuously for 24 hours, followed by Annexin V-PE and 7-AAD staining per manufacturer’s protocol (BD Pharmingen). ToPro3 staining was used to detect dead cells. Flow cytometry acquisition was done on a FACSCalibur and analyzed with CellQuest software (Becton Dickinson, Carpinteria, CA).
Quantitative PCR
Nascent RNA was isolated as previously described (24, 25). mRNA was isolated from cells using TriZol reagent (Invitrogen). One μg RNA was DNase-treated using RQ1 DNase (Promega), followed by first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad). 1/20th of the final cDNA reaction volume was used in each PCR reaction. PRDM1 mRNA levels were confirmed using two distinct primer sets: PRDM1-set1-FWD, 5′-TACATACCAAAGGGCACACG -3′, PRDM1-set1-REV, 5′-TGAAGCTCCCCTCTGGAATA -3′; PRDM1-set2 were described previously (20). Additional PRDM1 specific primer pairs used to detect mRNA decay rates were PRDM1-setB-FWD 5′-ATCTCAGGGCATGAACAAGG-3′, PRDM1-setB-REV 5′-ATGGGAAGGCTATGCAAACA-3′, PRDM1-setC-FWD 5′-GGCACCCTTGCCTACTGTAA-3′, PRDM1-setC-REV 5′-CTTCCTCCCTGGTTGTTTTG-3′. Control primer sets used were GAPDH-FWD, 5′-GAAGGTGAAGGTCGGAGT -3′, and GAPDH-REV, 5′-GAAGATGGTGATGGGATTTC -3′; and β Actin (Realtimeprimers.com). Quantitative PCR reactions were performed with iQ SYBR Green Supermix (Bio-Rad) using an iCycler (Bio-Rad).
DNA constructs
The PRDM1 promoter construct p2618, originally referred to as PRDM1α, was cloned as previously described (21). Using p2618 as a cloning template, p521 was sub-cloned using a NheI/SmaI digest to remove the 521 bp fragment, blunt-ended, and cloned into pGL3 basic. p1648 was cloned by digesting p2618 with BglII to yield a 1648 bp fragment, which was then ligated into pGL3 basic. p1921 was cloned by digesting p2618 with Kpn/PacI to remove 703 bp. p2391 was derived from p2618 by digestion with HindIII to remove 2391 bp and cloned into pGL3 basic. p983 and p2041 were cloned by PCR.
Site-directed mutagenesis was done by PCR cloning the mutated sequence into the p1921 or p2618 constructs. Briefly, mutations were introduced using a common p2618 forward or reverse primer, paired with either a reverse or forward primer, respectively, containing the site mutation. Primer sets used are as follows, with the mutated bases indicated in lower case letters: PRD 2618 FWD, 5′-TTCCTATTATGGAGCAAGCTTCC -3′, PRD 2618 REV, 5′-CATTTCTCCTTCGACCTGCA -3′, H3 mut FWD, 5′-ATTCTAAAAGGaAcGTtAAATACTCTTTAAC - 3′, and H3 mut REV, 5′-GTTAAAGAGTATTTaACgTtCCTTTTAGAAT - 3′. PCR products were gel-purified and paired with their respective mutation partner and amplified using p2618 common primers. p1921 mut P.H was derived from the p2618 mut P.H construct by digesting with KpnI/PacI. All mutations were confirmed by sequencing. Cross species homology comparison was done using GenomeVISTA (26).
Transfections and luciferase assays
Cells were transfected by electroporation using the Gene Pulser II (Bio-Rad, Hercules, CA). Cells (1 × 107) were pulsed with 250 V at a capacitance of 1070 μF. Transfections for luciferase assays were done with 10 μg of luciferase reporter construct and 50 ng of the internal control plasmid pRL-TK. Firefly luciferase activity was normalized to Renilla luciferase activity in all experiments. For siRNA knock-down experiments in CA46, cells were transfected with 3 μg control non-targeting siRNA or PU.1 siRNA (Dharmacon) for 24 hours. Live cells were separated using a Ficoll gradient and treated with 10 μg anti-IgM for 24 hours. For siRNA knock-down experiments in RPMI8226, cells were transfected with 2μg control non-targeting siRNA or PU.1 siRNA (Dharmacon) for 48 hours.
Dimethylsulfate in vivo footprinting
In vivo methylation of cells with dimethylsulfate and subsequent isolation of DNA was done as previously described (27, 28). Primers for the ligation-mediated PCR were as follows: proximal promoter, PRDLOW2 primer 1, 5′-GTACCTGGGGATTTGAGC -3′, PRDLOW2 primer 2, 5′-AGGAACGTGCGCCCCCTAATTCTGCCGC -3′, PRDLOW2 primer 3, 5′-CGTGCGCCCCCTAATTCTGCCGCGCCAG -3′; distal promoter, PRD C1, 5′-CAAGAAGCAAACATGTGAAG -3′; PRD C2, 5′-GCAAACATGTGAAGGACT TTACATTCCAAC -3′; PRD C3, 5′-GAAGGACTTTACATTCCAACACTCTGTGTC TG -3′; distal promoter, PRD H1, 5′-GAGTCAGCACCACAATG -3′; PRD H2, 5′-CAATGTAGTCATACTGAAGGCTGGC -3′; PRD H3, 5′-TCATACTGAAGGCTGGCCTGCTGTTC -3′.
Nuclear extract preparation and electrophoretic mobility shift assays
Nuclear extracts were prepared according to Dignam et al. (29) and performed essentially as described previously (26). The P.H oligonucleotide is 5′-GATCCAGAGTTTGATTCTAAAAGGGAAGTGAAATACTCTTTAAC -3′. The mutant P.H oligonucleotide contains the same mutation as in construct p2618-mutP.H. The Sp1 oligonucleotide is 5′-GATCTGGCCACGCCCCCACTTCGCGC-3′ and the Sp1 mutant oligonucleotide is 5′-GATCTGGCCACaaaaaCACTTCGCG-3′. Antibodies to Sp1, Ikaros, ATF1 (Santa Cruz) and PU.1 (BD Pharmingen) were used at 0.2 to 1.0 μg.
Chromatin Immunoprecipitation (ChIP)
After 24 hours treatment with anti-IgM or control (no treatment) cells were crosslinked with 1% formaldehyde for 10 minutes at room temperature and the reaction was stopped by the addition of glycine to a final concentration of 0.125M. Cells were washed twice with ice cold PBS and resuspended in ice cold TX-100/NP40 buffer (10mM Tris pH 8.1, 10mM EDTA, 0.5M EGTA, 0.25% TX-100, 0.5% NP40, 1mM PMSF, 0.5x Protease inhibitors) at a density of 4×106 cells/ml. Cells were resuspended in 10ml ice cold Salt-wash buffer (10mM Tris pH 8.1, 1mM EDTA, 0.5M EGTA, 200mM NaCl,1mM PMSF, 0.5x Protease inhibitors) and incubated for 10 minutes at 4 °C. Cells were lysed by adding sonication buffer (10mM Tris pH8.1, 1mM EDTA, 0.5M EGTA, 1% SDS, 1mM PMSF, 1x Protease inhibitors) at a cell density of 1×106 cells/30μl. Lysate was sonicated using a water bath sonicator (Diagenode).
Chromatin immunoprecipitation was carried out using 2×106 cells and 5μg of antibody; IgG (Upstate), PU.1 (Santa Cruz), TLE4 (Santa Cruz). Immunoprecipitated chromatin was washed sequentially with low salt wash (20mM Tris pH8.1, 2mM EDTA, 150mM NaCl, 0.1% SDS, 1% tritonX 100), high salt wash (20mM Tris pH8.1, 2mM EDTA, 500mM NaCl, 0.1% SDS, 1% tritonX 100) and LiCl wash (10mM Tris pH8.1, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholic acid, 1mM EDTA). DNA was eluted with elution buffer (10mM Tris pH8, 1% SDS, 1mM EDTA) and crosslinks were reversed by incubating with 312 mM NaCl at 65 °C for 4 hours. The immunoprecipitated DNA was treated with RNase (Ambion) at 37 °C and proteinase K (Roche) for 1 hour at 45 °C. The DNA was purified with Qiagen PCR spin columns. Purified DNA was analyzed by quantitative PCR using the following primers PRD_PU.1 binding site FWD 5′-ACTCACCAGCAGTTGCATGA - 3′, PRD_PU.1 binding site REV 5′-CAGTCTCACTTGCAGATGTTAAAGA -3′ and proximal PRDM1 FWD 5′ –AGGACCAGACAGCTCCACTG-3′, proximal PRDM1 REV 5′-GCTCAAATCCCCAGGTACAA-3′. Primers to the HLA-DRA promoter or an exon myoglobin locus were used as negative controls for specificity. ChIP data is calculated as relative occupancy (2^(Ct-IgG – Ct-specific antibody)). All the primer sets were run using an annealing temperature of 60°C.
Results
Induction of PRDM1 expression and activity in lymphoma cells
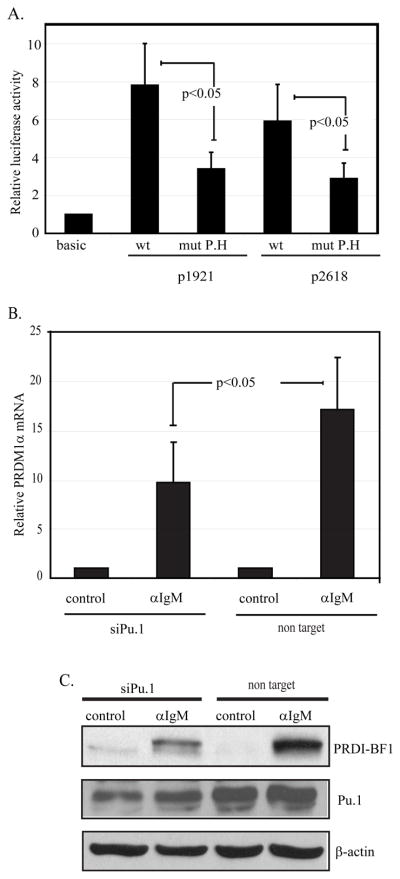
Burkitt’s lymphoma cell lines respond to signaling through the B cell receptor via anti-IgM treatment by undergoing either growth arrest or apoptosis. Similarly, although most Burkitt’s lymphoma cell lines lack detectable levels of PRDM1, ectopic expression of PRDM1 leads to growth arrest or apoptosis (16, 17). In order to dissect the regulation of PRDM1 in B cell lymphoma, we have initially investigated the effects of B cell receptor cross-linking of the EBV-negative lymphoma line, CA46. Twenty-four hour exposure to anti-IgM results in a significant increase in Annexin-V staining, indicative of the early stages of apoptosis (figure 1A) but does not significantly increase the presence of late apoptotic or dead cells (control 3% vs anti-IgM 5%) as detected by ToPro3 staining. This finding is similar to that reported by Kaptein et al in which B cell receptor cross-linking of CA46 cells induced growth arrest and limited apoptosis along with a decrease in c-myc expression (13). This treatment also significantly increases PRDM1 mRNA levels approximately 8-fold above untreated controls (p<0.05) (figure 1B). The induction of PRDM1 is also detectable at the protein expression level as revealed by immunoblot analysis (figure 1C). We have previously reported that a PRDM1-beta isoform can also be expressed from a distinct promoter within intron 3 (21). Neither mRNA nor protein for the PRDM1-beta isoform was detected in the anti-IgM treated cells (data not shown). Whether or not the PRDM1 protein is functional after induction was determined by examining its ability to silence target gene promoters. The steady state levels of BSAP and cmyc mRNA were examined by real time quantitative reverse transcription PCR. Expression of both genes decreased after B cell receptor cross linking consistent with suppression by PRDM1 (figure 1D). The observed 2-fold level of suppression is consistent with previous reports using over-expression of murine PRDM1 (Blimp-1) (7, 30). Given the rapid turn-over rate of c-myc mRNA (31) this may suggest that PRDM1 can attenuate expression of some target genes but does not necessarily silence them.
Figure 1. Anti-IgM treatment induces PRDM1 (PRDI-BF1) and apoptosis in CA46 lymphoma cells.

A) Treatment with anti-IgM (10 μg/ml) for 24 hours induces apoptosis approximately 60% above the control in CA46 lymphoma cells as assessed by Annexin V staining followed by FACS analysis. The data is representative of 3 independent experiments. (Gray histogram, untreated control cells; black outlined histogram, treated cells); B) Treatment of CA46 lymphoma cells for 24 hours with 10 μg/ml anti-IgM induces PRDM1 mRNA levels as assessed by quantitative RT-PCR. The data shown is mean of 3 experiments normalized to GAPDH with SEM shown (p<0.05). C) Immunoblot analysis of PRDI-BF1 protein expression. CA46 lymphoma cells express PRDI-BF1 protein only after treatment with anti-IgM for 24 hours. The RPMI8226 myeloma cell line constitutively expresses PRDI-BF1 and serves as a positive control. D) Quantitative RT-PCR analysis of PRDI-BF1 target genes, BSAP and c-myc. Anti-IgM exposure represses mRNA steady state levels. Data shown is a mean of 3 independent experiments with SEM shown (p<0.05 for c-myc).
PRDM1 regulation occurs at the transcriptional level
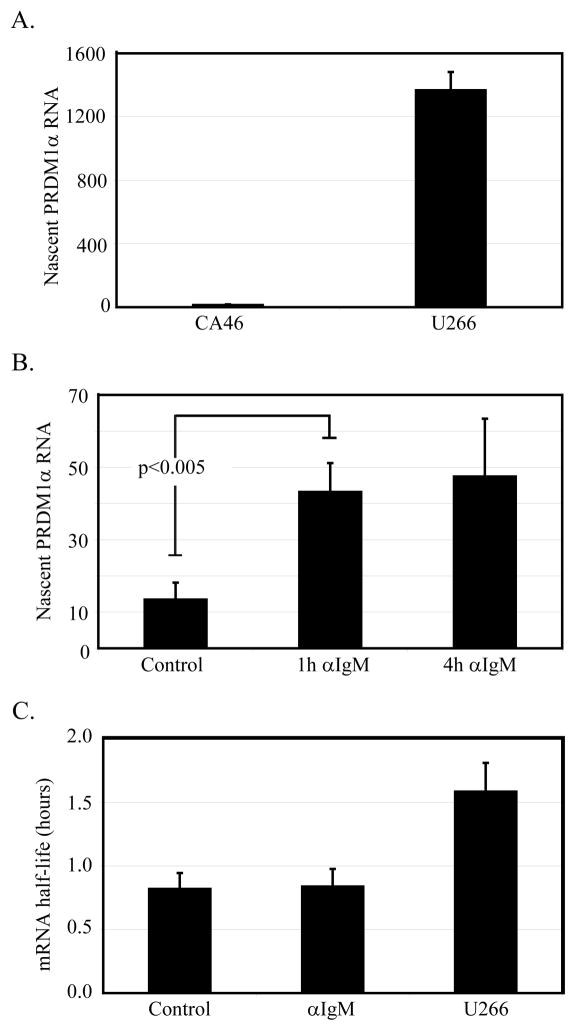
The mechanism by which PRDM1 expression is induced in lymphoma cells is unknown and could occur at multiple levels. We first examined basal levels of nascent RNA production in both lymphoma and myeloma cells. Nascent RNAs, defined as those RNAs still in the process of being transcribed, are an accurate measure of endogenous transcriptional activity (25). The nascent RNA were purified from nuclei after extensive washing to remove the released transcripts and quantified by real-time RT-PCR with specific primers directed to the 5′ end of the RNA transcript. Levels of nascent RNA production in CA46 lymphoma cells are significantly lower than that measured in myeloma cells (figure 2A). This finding is consistent with the high level of PRDM1 protein expression in myeloma cells (figure 1C). Stimulation of the lymphoma cells with anti-IgM increased production of PRDM1 nascent RNA three fold after 1 and 4 hours (figure 2B), indicating a rapid transcriptional activation. Changes in mRNA stability could also contribute to the increase in PRDM1 levels. mRNA stability changes were directly measured by inducing PRDM1 mRNA for one hour and then blocking subsequent transcription initiation with Actinomycin D. The mRNA half-life was indistinguishable before and after anti-IgM treatment of the lymphoma cells (figure 2C). The mRNA half-life was very short (<1 hour) in the lymphoma cells and less than two-fold longer in the myeloma cells. This is consistent with the recent genome wide analysis of mRNA decay rates in mouse embryonic stem cells in which PRDM1 was one of the rare transcripts with a less than 1 hour half-life (32). The PRDM1 mRNA is present is three predominant molecular weights which vary only in the length of the 3′UTR (21, 33). Using real-time PCR probes spanning the 3′UTR we determined the largest mRNA species had a slightly faster decay rate but that this was not affected by anti-IgM treatment (Supplemental Figure S2). Similarly, a proximal 3′UTR probe which detects each of the mRNA species showed a similar decay rate and no change upon anti-IgM exposure. While this does not exclude changes in a minor mRNA species, this indicates that anti-IgM does not have a major role in altering PRDM1 mRNA stability. Together these data indicate regulation of PRDM1 occurs primarily at the level of transcription while the mRNA has a relatively short half-life in both stimulated lymphoma cells and in myeloma cells.
Figure 2. PRDI-BF1 regulation occurs at the level of transcription.
A) Relative levels of active transcription as measured by nascent RNA levels were determined by quantitative RT-PCR. Levels in U266 myeloma cells are significantly higher than that of unstimulated CA46 lymphoma cells. Data represents three independent experiments with SEM shown (p<0.005). B) Anti-IgM induces nascent PRDM1 RNA synthesis as early as one hour. CA46 cells were stimulated with 10μg/ml anti-IgM for 1 or 4 hours before harvest of nascent RNA and analysis by quantitative RT-PCR. Data represents three independent experiments with SEM shown. C) Stability of PRDM1 mRNA is unchanged upon treatment with anti-IgM. CA46 cells were pre-treated for 1 hour with anti-IgM, followed by a inhibition of transcription with Actinomycin D. PRDM1 mRNA levels were determined by quantitative RT-PCR of cells harvested at 15 minute intrvals over 2 hours. Data shown is the mean of at least 3 experiments with SEM shown.
Characterization of PRDM1 promoter activity
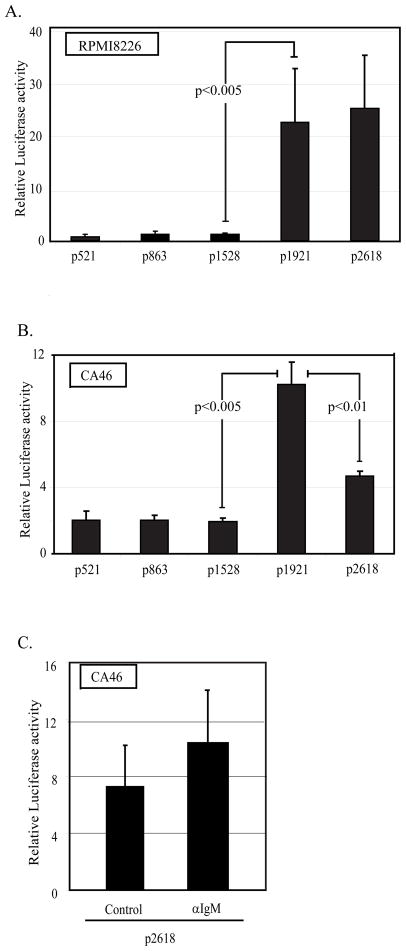
Because PRDM1 expression is primarily regulated at the level of transcription, we cloned the human promoter to assess the regions necessary for activity. Using a series of promoter deletion constructs spanning 2618 base pairs upstream of the transcription start site, potential regulatory regions were identified in lymphoma and myeloma cells (for schematic see Supplemental figure S1). PRDM1 promoter constructs containing 521, 863, or 1528 bp display robust and similar promoter activity in the myeloma cell line, RPMI-8226 (figure 3A). Addition of promoter sequences up to 1921 bp results in a significantly higher level of activity. Similar results were obtained in a second myeloma cell line, U266 (data not shown). PRDM1 promoter activity was also analyzed in the lymphoma cell line CA46 (figure 3B). The pattern of promoter activity in the unstimulated lymphoma cells is similar to that observed in the myeloma cell lines. The 521 bp promoter was sufficient for activity and the activity increased significantly with the addition of the region between −1528 and −1921 bp but larger constructs show a partial but consistent inhibition of activity. The overall level of promoter activity in the lymphoma cell line was lower than that in the myeloma cell lines consistent with the levels of nascent RNAs detected at the endogenous promoter in figure 2A. However comparing activity in two different cell lines requires the assumption that a co-transfected minimal Tymidine Kinase promoter has similar activity in both cell lines. This common assumption has not been proven for these cell lines. The effect of anti-IgM treatment was next examined in the CA46 lymphoma cell line. Constructs containing 2618 bp of the promoter were analyzed 24 hours after stimulation. Stimulation resulted in a small but not statistically significant increase in promoter luciferase activity (Figure 3C). This finding may indicate that a region required for induction lies outside of the 2618 bp promoter. One likely candidate is the intronic regions previously shown to bind the repressor BCL6 (34, 35). It is also possible that the transiently transfected promoter constructs do not fully recapitulate the chromatin structure of the endogenous gene. This may prevent further activation of these promoter constructs by anti-IgM. However, these results reveal that the lymphoma cells have the necessary components to transcribe the PRDM1 gene.
Figure 3. Characterization of PRDM1 promoter activity.
A) RPMI8226 myeloma cells and B) CA46 lymphoma cells were transiently transfected with the indicated PRDM1 promoter deletion constructs fused to a luciferase reporter gene. C) CA46 lymphoma cells were transiently transfected with p2618 construct and stimulated with anti-IgM for 24 hours. Luciferase activity was measured 42 hours after transfection. Data presented is normalized to expression of a co-transfected minimal TK promoter-renilla luciferase construct. Construct names along the x-axis represent the number of PRDM1 promoter base pairs upstream of the transcription start site included in the construct. The region between −1528 to −1921 relative to the transcription start site was required for maximal transcription activity in both cell types. Data shown are the mean of 3 independent experiments with SEM shown. A schematic of the promoter is shown in supplemental figure S1.
In vivo protein/DNA interactions occur across the PRDM1 promoter
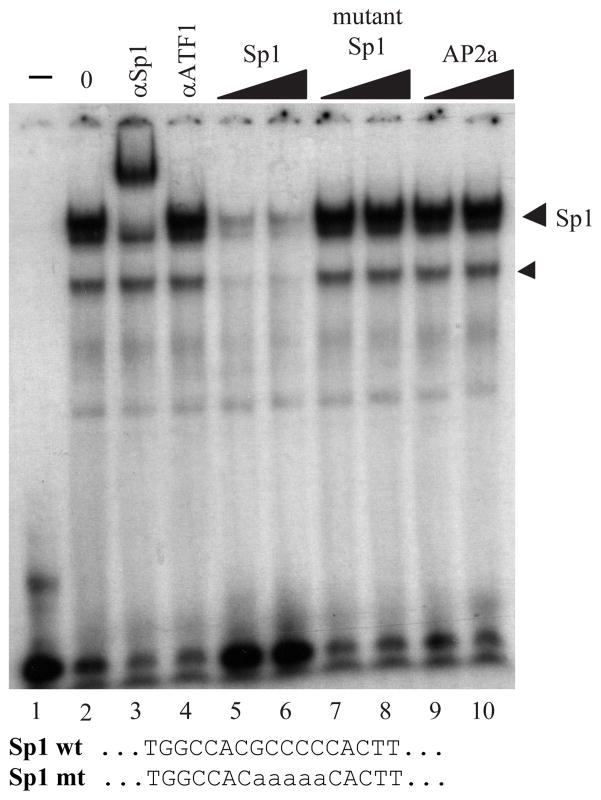
In order to define the important cis-acting DNA elements within the PRDM1 promoter, high resolution mapping of the protein/DNA contact sites was done by in vivo genomic footprinting. Unstimulated CA46 lymphoma cells and RPMI-8226 myeloma cells were treated briefly with dimethylsulfate to induce limited methylation of guanine residues. Close protein/DNA interactions have been demonstrated to inhibit or enhance the methylation activity which can be visualized after chemical cleavage and resolution by sequencing gel electrophoresis. These footprints of altered methylation represent the contact points of transcription factors bound to the promoter. Examination of the first 237 base pair region proximal to the transcription start site demonstrated factor binding in both the lymphoma and myeloma cells (figure 4A). Four clusters of interaction are detected and labeled with brackets on the left side of the sequence. These contacts are indistinguishable between the two cell types. Closest to the transcription start site, nine strongly protected guanine residues map across a sequence with homology to an Sp1 consensus binding element. Sp1 binding to this element was confirmed by in vitro electrophoretic gel mobility shift assay (EMSA) and specific antibody reactivity (Figure 5). This extends the recent findings by Mora-Lopez et al and establishes Sp1 as a regulator of PRDM1 transcription in vivo (36). Three additional occupied sites have been designated P.A, P.B, and P.C. These contact sites do not have obvious homology with known elements.
Figure 4. In vivo genomic footprinting of the PRDM1 promoter reveals multiple protein-DNA interactions.
The RPMI8226 myeloma and CA46 lymphoma cell lines were analyzed with eight different primer sets to reveal interactions across the proximal 2618 base pairs of the PRDM1 promoter. Three regions which revealed contacts are shown: A) +15 to −170 bp, B) −1717 to −1951 bp, and C) −1889 to −2072 bp. In each pane the control lanes show the guanine residue sequence from deproteinized in vitro methylated DNA. The DMS lanes show the in vivo methylated residues. Protections (open circles) and enhancements (filled circles) are shown on the right side of each footprint panel and indicated in the sequence below. Clusters of contacts have been assigned arbitrary names as indicated along the left side and are boxed in the sequence. The bent arrow in panel A indicates the position of the transcription start site. A schematic of the footprint primers is shown in supplemental figure S1.
Figure 5. Sp1 interacts at the PRDM1 proximal promoter.
Electrophoretic mobility shift assay using an oligonucleotide spanning the Sp1 consensus sequence identified at position −52 to −43 in the PRDM1 promoter. Lanes 1 and 2 contain 0 and 2 uL of nuclear extract respectively. The binding reactions in lanes 3 and 4 were incubated with the specific antibody indicated at the top of each lane. Unlabeled competitor oligonucleotides as indicated at the top of lanes 5–10 were added to the binding reaction at 150 or 300-fold molar excess. The sequence of the Sp1 oligonucleotide and the mutant probe are shown at the bottom of the figure. The Sp1 containing complex is indicated by the labeled arrowhead. The smaller arrowhead represents a related specific GC-box binding protein antigenically unrelated to Sp1.
The distal promoter region from −1497 to −2641 base pairs was next examined by genomic footprinting using eight overlapping primer sets. Five additional clusters of contact were detected in the distal promoter (figure 4B, C and data not shown). The region which conferred transcriptional activation (−1528 to −1921) contained three contacts designated P.J, P.H, and P.G. Site-P.J, associated with contacts at −1805 and −1802 and site-P.G associated with contacts at −1648, −1645, −1643, and −1641 overlap with AP1 consensus sequences. Both of these sites were previously predicted due to sequence homology by Vasanwala et al but neither direct assessment of AP1 binding nor functional activity in B cells or unstimulated myeloma cell lines were done (37). These sites show conservation across species (Supplemental Figure S3). Our data demonstrates that both sites are occupied in vivo. Site-P.H associated with strong contacts at −1733, −1732, −1731, −1728 has sequence homology to consensus binding sites for Ets family members known to regulate multiple genes in the B cell lineage. These sites also show conservation across species (Supplemental Figure S3). The region associated with a partial repression of transcription in the lymphoma cell line (−1921 to −2618) contained only two consistent clusters of in vivo contacts, designated sites P.D and P.F. Site P.F was strongly protected in the CA46 lymphoma cell line and weakly protected in the myeloma cell line. Mutation of the P.F site did not effect promoter activity of the 2618 PRDM1-luciferase construct in either lymphoma or myeloma cell lines (data not shown). Site-P.D associated with contacts at −2038 and −2032 only in the lymphoma cell line.
PU.1 binds to the P.H site and regulates PRDM1 promoter activity
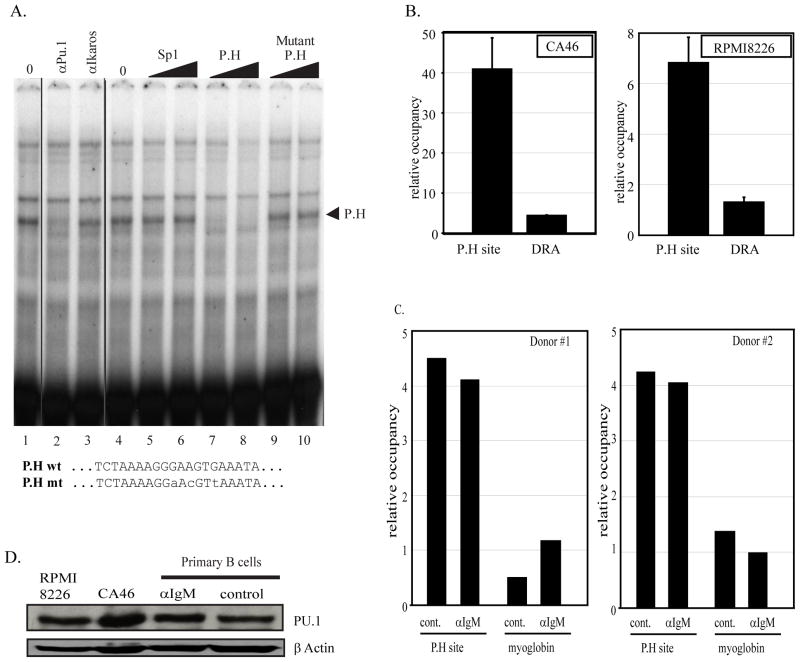
In order to identify the transacting factor bound at site P.H both in vivo and in vitro assays were used. First EMSA binding assays were performed with a 30 base pair probe spanning the P.H site in conjunction with nuclear extracts from CA46 lymphoma cells (figure 6A). A fast migrating protein/DNA complex was detected which was specifically competed by unlabeled P.H probe but not a mutated P.H probe. The P.H site has homology to consensus Ets-family binding sequences. Antibody to PU.1 induced the formation of a super-shifted protein/DNA complex indicating that PU.1 is contained within the complex bound to P.H in vitro (figure 6A). Similar results were obtained with nuclear extracts from the myeloma cell line RPMI8226 (data not shown).
Figure 6. Site P.H is a PU.1 factor binding site.
A) In vitro binding assays were done using an oligonucleotide containing the P.H site sequence identified at position −1745 to −1737 of the PRDM1 promoter and CA46 nuclear extracts. Lanes 1 and 4 contain 2 uL of nuclear extract. Unlabeled competitor oligonucleotides as indicated at the top of lanes 5–10 were added to the binding reaction at 150 or 300-fold molar excess. A single complex indicated by an arrowhead is specifically competed by the wild type but not a mutant or unrelated oligonucleotide. In lanes 2 and 3 antibodies as indicated at the top were added to the binding reaction. The formation of the specific P.H complex is inhibited by addition of PU.1 antibody. The wild type and mutant P.H site sequences are shown at the bottom. B) Chromatin immunoprecipitation assay was performed in CA46 lymphoma cells (left panel) and RPMI8226 myeloma cells (right panel) using PU.1 antibody and quantitative PCR primers spanning the P.H site. PU.1 binding at the P.H. site was significantly higher than on the negative control promoter HLA-DRA. Data shown are mean of 3 independent experiments with SEM shown (p<0.01). C) Chromatin immunoprecipitation assay in activated primary human B cells. The experiment is as described in panel B except a myoglobin B locus is shown as the negative control. Lanes labeled αIgM were treated for 24 hours with anti-IgM while lanes labeled control were mock treated for 24 hours. PU.1 binding is specifically detected at the P.H site and is not altered by anti-IgM treatment. Similar data were obtained in two independent donor samples. D) Immunoblot analysis of PU.1 expression.
PU.1 association at the P.H site was also measured at the endogenous PRDM1 promoter. Chromatin immunoprecipitation (ChIP) was performed with antibodies specific to PU.1 and association with the P.H site was assessed by quantitative PCR (figure 6B). In lymphoma cells PU.1 factor binding at the PRDM1 promoter was approximately 9 fold greater than the negative control promoter HLA-DRA. Similarly, PU.1 binding was also observed in the myeloma cell line. We next examined normal primary B cells for PU.1 binding in vivo. ChIP analysis clearly identified PU.1 binding at the region of the P.H site while essentially no binding was observed at the negative control locus (figure 6C). PU.1 binding was unchanged by anti-IgM treatment. Consistent with this finding the level of PU.1 protein expression in the primary B cells did not change with anti-IgM treatment (figure 6D).
Functional assessment of site P.H. was performed initially by mutating the site in the context of either the p1921 and p2618 PRDM1 promoter-luciferase constructs. Transfection of these mutated constructs into the lymphoma cell line revealed an approximately 50% reduction in transcriptional activity indicating that this site is required for maximal activity of the PRDM1 promoter (figure 7A). PU.1 function in the activation of the endogenous PRDM1 gene was also examined by inhibiting PU.1 protein expression using siRNA. CA46 lymphoma cells were transiently transfected with the PU.1 specific siRNA and incubated with or without anti-IgM stimulation. Anti-IgM mediated induction of endogenous PRDM1 mRNA is significantly abrogated by PU.1 knockdown (figure 7B). Furthermore induction of PRDM1 protein was also inhibited by the reduction in PU.1 expression (figure 7C). The low level of apoptosis as assessed by PARP cleavage was not altered by PU.1 knockdown (data not shown). This indicates that PU.1 is involved in anti-IgM induced transcription of PRDM1 and that additional factors also contribute to the activity.
Figure 7. Site P.H and transcription factor PU.1 are involved in anti-IgM-mediated transcriptional activation of PRDM1.
A) The p1921 and p2618 PRDM1 promoter-luciferase constructs containing a wild type or mutated sequence at the P.H site were transfected into CA46 cells. Mutation of the P.H site in either construct decreased promoter activity. The mutation is the same as shown in figure 6A. The lane marker “basic” represents the activity from the promoterless vector, pGL3-Basic. Promoter activity was normalized as in figure 3 and represents six independent experiments with SEM shown. B) Endogenous PRDM1 expression is inhibited by loss of PU.1. CA46 lymphoma cells transfected with either non targeting control siRNA or an siRNA specific to PU.1 for twenty four hours and then either stimulated with anti-IgM (αIgM) or untreated (control) for an additional twenty four hours. PRDM1 mRNA was assessed by quantitative RT-PCR. Data was normalized to GAPDH and shown as fold induction relative to untreated (control) sample. Data is the mean of 3 independent experiments with SEM shown. C) Immunoblot analysis of PU.1 and PRDI-BF1 expression. PU.1 siRNA decreased PU.1 protein levels and diminished induction of PRDI-BF1 in response to anti-IgM. Experimental conditions are as in panel B.
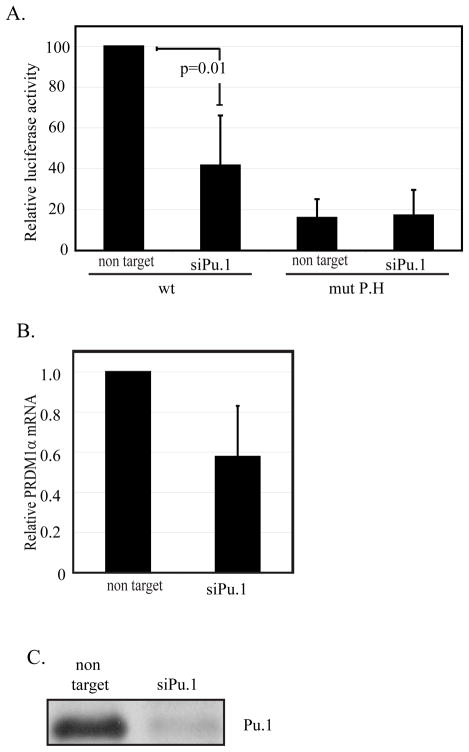
Similarly, to demonstrate the involvement of PU.1 in the expression of PRDM1 in myeloma cells, RPMI8226 were co-transfected with PRDM1 full-length luciferase promoter construct and siRNA specific for PU.1. PU.1 knockdown in these cells decreases the PRDM1 promoter activity by approximately 60% (figure 8A). Mutation of the P.H site results in a greater than 80% loss of PRDM1 promoter activity. Knock-down of PU.1 does not further alter PRDM1 transcription in the context of a mutated P.H site. This confirms that the P.H site is functional in myeloma cells and that PU.1 exerts its effects through the P.H site. Loss of PU.1 also decreases endogenous PRDM1 mRNA levels in the RPMI8226 cell line (figure 8B) further confirming a role for PU.1 in regulating PRDM1 transcription.
Figure 8. Site P.H and transcription factor PU.1 are involved in transcriptional regulation of PRDI-BF1 in myeloma cells.
A) RPMI8226 myeloma cells were transfected with either wild type or mutant Site-P.H p2618 PRDM1 promoter luciferase constructs. In addition the cells received siRNA against PU.1 or the non targeting control as indicated below each graph. siRNA against PU.1 diminished wild type promoter activity. Mutation of the P.H site also significantly lowered transcription but addition of siRNA to PU.1 did not further diminish activity in the context of a mutated P.H site. Data is normalized as in figure 3 and represents four independent experiment with SEM shown. B) Endogenous PRDM1 mRNA levels are decreased by siRNA knockdown of PU.1 in RPMI8226 cells. mRNA was assayed 48 hours after transfection of either a non-targeting siRNA or a PU.1 specific siRNA. Levels were measured by quantitative RT-PCR and shown as the average of 3 independent experiments with SEM shown. C) Immunoblot of PU.1 expression after siRNA knockdown in RPMI8226 cells.
Loss of TLE4 at the P.H site in response to transcriptional stimulation
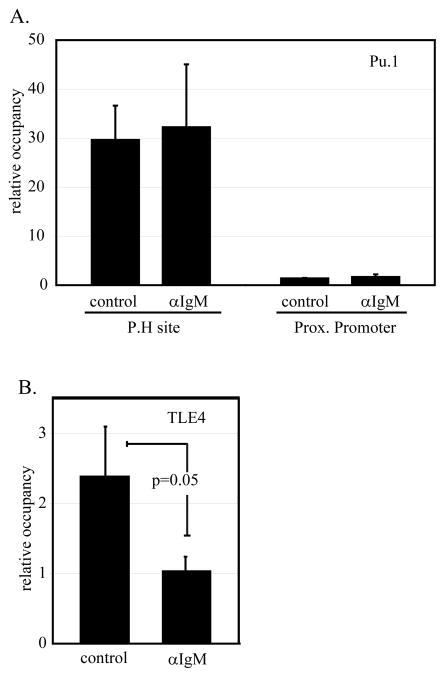
PU.1 has been described to function both as an activator of transcription and as a repressor (38, 39). These divergent activities have been linked to differential PU.1 mediated recruitment of the co-repressor TLE4 and the co-activator CBP. The observed PU.1 dependent activation in response to anti-IgM might be due to changes in PU.1 binding to the PRDM1 promoter or to changes in the co-activator or co-repressors recruited by PU.1. In order to address this question we used chromatin immunoprecipitation to profile binding of the factors in response to anti-IgM. PU.1 binding at the P.H site is robust and unchanged by anti-IgM treatment (figure 9A). Minimal binding was detected near the transcription start site of PRDM1 confirming the localized recruitment of PU.1 to the P.H site. In contrast chromatin immunoprecipitation of the co-repressor TLE4 revealed a significant loss of binding in response to the activation stimulus (figure 9B). This is consistent with a loss of repressive activity and the observed increase in PRDM1 transcription. Similar analyses with antibodies to the co-activator CBP were variable with a general increase in binding observed upon stimulation. However, the results for CBP did not reach statistical significance (data not shown).
Figure 9. Chromatin immunoprecipitation of PU.1 and TLE4 at the distal PRDM1 promoter.
A) ChIP assay using a PU.1 antibody to detect binding at the P.H site before and after anti-IgM stimulation. CA46 cells were left unstimulated (control) or stimulated with anti-IgM (αIgM) for 24 hours prior to harvest. Binding was assessed by quantitative PCR using primers surrounding the P.H site or located at the proximal PRDM1 promoter. PU.1 binding at the P.H site on PRDM1 distal promoter is unaffected by anti-IgM treatment. Data represents the mean of six (P.H site) or four (prox. promoter) independent experiments with SEM. B) ChIP assay using TLE4 antibody to detect binding at P.H site. The same samples assessed in panel A were reassessed for TLE4 binding. TLE4 binding at the P.H site significantly decreases upon treatment with anti-IgM. Data shown in each panel are mean of 4 independent experiments with SEM shown.
Discussion
The transcription factor PRDI-BF1/Blimp-1 is required for the differentiation of a mature B cell to a plasma cell (3). It does this by directly repressing downstream targets, which in turn has a widespread effect on further downstream targets (40). These downstream effector cascades have been well studied, however very little is known as to how PRDI-BF1/Blimp-1 expression is regulated.
This study demonstrates a direct link between B cell receptor cross-linking by anti-IgM and transcriptional activation of PRDI-BF1/Blimp-1. Treatment of CA46 lymphoma cells with anti-IgM significantly up-regulated PRDM1 mRNA and protein levels and induced apoptosis. This is consistent with observations in other B cell lymphoma cell lines (10–14). One previous study using the EBV-negative CA46 cell line reported that this line was unique in responding to anti-IgM with only growth arrest raising the possibility that EBV-negative B cell lymphomas were heterogenous in the apoptosis response (13). However, this report used very late markers of apoptosis (DNA fragmentation) while we measured early markers suggesting that only the kinetics of apoptosis induction may vary. The anti-IgM induced growth arrest and apoptosis has been linked to down-regulation of c-myc (13). This is consistent with the induction of PRDI-BF1/Blimp1 and its known role in directly repressing c-myc transcription (6). Suppression of PRDI-BF1 target genes was approximately 2-fold which is consistent with previous reports using over-expression of murine PRDM1 (Blimp-1) (7, 30). The lower levels of PRDI-BF1 induced by anti-IgM treatment could also be responsible for the attenuation of the suppressive activity of PRDI-BF1/Blimp1. Alternatively, post-translational modifications of PRDI-BF1 after anti-IgM treatment could alter the functional activity of PRDI-BF1, although no such modifications have been described to date. Future investigations of PRDI-BF1/Blimp1 post-translational modifications may reveal additional levels PRDI-BF1/Blimp1 regulation. The increase in PRDM1 expression occurs primarily at the level of transcription as we did not detect any change in mRNA stability but actively transcribing nascent RNA levels were induced within one hour. Unexpectedly in vivo genomic footprinting revealed that the PRDM1 promoter was extensively occupied by transcription factors even in the absence of stimulation or promoter activity. Together these findings indicate that the PRDM1 promoter is in an open and poised state in the lymphoma cells. This provides support that therapeutic approaches to trigger endogenous PRDM1 expression are feasible and could be a viable approach to induce apoptosis in lymphoma cells. Furthermore, PRDI-BF1 has been shown to be an important target in immunotherapy of myeloma by induction of PRDI-BF1-specific cytotoxic T cells, an approach which could also be exploited to kill lymphomas after PRDI-BF1 induction (41).
The transcription factors and cis-acting elements controlling PRDM1 promoter activity have only begun to be investigated. A region of the murine PRDM1 promoter spanning −918 to +207 base pairs was previously shown to have minimal promoter activity but did not confer any cell type specific activity (33). This is consistent with our finding that the sequences between −1528 to −1921 base pairs of the human promoter are required for activation of the promoter in lymphoma and myeloma cells. In vivo genomic footprinting of the proximal promoter region revealed four occupied elements within the first 170 base pairs. These include a bound Sp1 site proximal to the transcription initiation point and is consistent with the absence of a canonical TATA box element and the recent findings by Mora-Lopez et al (36). In vivo genomic footprinting of the upstream activation domain revealed three occupied elements in both lymphoma and myeloma cells. Sites P.J and P.G both have homology to an AP1 consensus binding site sequence. These sites were previously predicted by sequence homology (37). Investigation of the murine PRDM1 promoter has also provided evidence that c-fos can regulate the gene (42). The authors identified an AP1 binding site at a region homologous to the site we designated P.G and demonstrated that c-fos can bind to this site. c-fos was required for maximal activity of the murine PRDM1 promoter. Together these findings strongly support that the PRDM1 gene expression is directly regulated by AP1 through sites P.J and P.G.
The third in vivo occupied element within the PRDM1 promoter required for transcriptional activation is site P.H. In vivo chromatin immunoprecipitation assays and in vitro DNA binding assays established that Ets family member PU.1 specifically binds to site P.H. Basal and anti-IgM stimulated PRDM1 promoter activity was significantly inhibited by mutating the P.H site in lymphoma cells. Additionally, knock-down of PU.1 expression by siRNA decreased PRDM1 transcription after B cell receptor cross-linking by anti-IgM. This indicates that PU.1 and the Ets site is a critical and required component of the PRDM1 promoter which must be present for the promoter to fully respond to anti-IgM stimulation. However, this site is not sufficient for the anti-IgM response. These data do not exclude the possibility that other Ets family members may also function in regulating PRDM1 expression such as Elf-1 which has a DNA recognition sequence similar to that of PU.1. In the myeloma cell line RPMI8226 the P.H site and PU.1 expression were also required for maximal promoter activity. PU.1 is known to have an important role in regulating early B cell development and is continued to be expressed throughout B cell maturation (43). A recent report has shown that PU.1 is also expressed in primary human plasma cells but that expression in myeloma cells and cell lines is variable (44). Our results suggest that PU.1 may contribute to the initial activation of PRDM1 expression in B cells. Furthermore, PRDM1 expression in myeloma cells is significantly enhanced by PU.1. PU.1 is bi-functional and can act to either increase or repress transcription of its target promoters (39, 45). This opposing activity is mediated by differential recruitment of co-regulators by PU.1. The co-activator CBP, a histone acetyltransferase, binds to PU.1 and promotes activation of the promoter (39). In contrast PU.1 can also recruit TLE4, a corepressor which in turn recruits the histone deacetylases HDAC1 and HDAC2 (38). We observed TLE4 recruitment to the PRDM1 promoter at the region of PU.1 binding. This recruitment was significantly diminished upon activation by anti-IgM while CBP binding was largely unaffected. This indicates that the components of the PU.1 complex bound at the PRDM1 promoter are modulated by B cell receptor cross linking to promote gene activation. Conversely knockdown of PU.1 in unstimulated B cells did not induce PRDM1 expression suggesting that the PU.1/TLE4 complex is not acting as a dominant repressor.
BCL6 can repress PRDM1 expression (46). Vasanwala et al showed that AP1 and BCL6 could interact and suggested that the two AP1-like sites, which we have designated as P.J and P.G, may mediate BCL6 repression of PRDM1 (37). Bach2 has also been suggested to repress murine PRDM1 though association with Mafk bound at the site homologous to site P.G (47). Our in vivo data support the hypothesis that the sites are bound by transcription factors but the region conferred activation not repression in the BCL6-positive CA46 cell line. This suggests that BCL6 does not repress through these elements although cell line differences between the reports may have an effect. Recent direct evidence of BCL6 binding to intron 5 of human PRDM1 or introns 3 and 5 of murine PRDM1 are consistent with the absence of a dominant repression domain detected by our promoter studies (34, 35). Our studies revealed that the distal region of the promoter (−1921 to −2686 base pairs) partially decreased overall promoter activity in the B cell line. This region contained only 2 clear in vivo occupied elements and the site P.D was observed only in the B cell line but not in the myeloma line. The factor binding at site P.D remains to be elucidated but the sequence does have partial homology to the transcription repressor ZEB1 (48)
In conclusion, we have demonstrated PRDM1 regulation occurs primarily at the transcriptional level in lymphoma and myeloma cells. We show for the first time that PRDM1 is transcriptionally regulated by PU.1 and the co-repressor TLE4. Furthermore we have shown that two AP1 sites and an Sp1 site within the PRDM1 promoter are occupied in vivo. Importantly, we report that the promoter is poised for activation in lymphoma cells, suggesting that inducing PRDI-BF1 expression in lymphoma cells lacking PRDM1 gene mutations is a viable therapeutic approach to inducing apoptosis in these cells.
Supplementary Material
Acknowledgments
This work is supported by NIH grants CA114504 and CA080990.
We thank the H. Lee Moffitt Cancer Center Flow Cytometry and Molecular Biology Core Facilities.
References
- 1.Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta- interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 2.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 4.Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh N, Gyory I, Wright G, Wood J, Wright KL. Positive regulatory domain I binding factor 1 silences class II transactivator expression in multiple myeloma cells. J Biol Chem. 2001;276:15264–15268. doi: 10.1074/jbc.M100862200. [DOI] [PubMed] [Google Scholar]
- 6.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 7.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 9.Doody GM, Stephenson S, Tooze RM. BLIMP-1 is a target of cellular stress and downstream of the unfolded protein response. Eur J Immunol. 2006;36:1572–1582. doi: 10.1002/eji.200535646. [DOI] [PubMed] [Google Scholar]
- 10.Benhamou LE, Cazenave PA, Sarthou P. Anti-immunoglobulins induce death by apoptosis in WEHI-231 B lymphoma cells. Eur J Immunol. 1990;20:1405–1407. doi: 10.1002/eji.1830200630. [DOI] [PubMed] [Google Scholar]
- 11.Carey GB, Scott DW. Role of phosphatidylinositol 3-kinase in anti-IgM-and anti-IgD-induced apoptosis in B cell lymphomas. J Immunol. 2001;166:1618–1626. doi: 10.4049/jimmunol.166.3.1618. [DOI] [PubMed] [Google Scholar]
- 12.Hasbold J, Klaus GG. Anti-immunoglobulin antibodies induce apoptosis in immature B cell lymphomas. Eur J Immunol. 1990;20:1685–1690. doi: 10.1002/eji.1830200810. [DOI] [PubMed] [Google Scholar]
- 13.Kaptein JS, Lin CK, Wang CL, Nguyen TT, Kalunta CI, Park E, Chen FS, Lad PM. Anti-IgM-mediated regulation of c-myc and its possible relationship to apoptosis. J Biol Chem. 1996;271:18875–18884. doi: 10.1074/jbc.271.31.18875. [DOI] [PubMed] [Google Scholar]
- 14.Zupo S, Isnardi L, Megna M, Massara R, Malavasi F, Dono M, Cosulich E, Ferrarini M. CD38 expression distinguishes two groups of B-cell chronic lymphocytic leukemias with different responses to anti-IgM antibodies and propensity to apoptosis. Blood. 1996;88:1365–1374. [PubMed] [Google Scholar]
- 15.Lee SC, Bottaro A, Insel RA. Activation of terminal B cell differentiation by inhibition of histone deacetylation. Mol Immunol. 2003;39:923–932. doi: 10.1016/s0161-5890(03)00029-4. [DOI] [PubMed] [Google Scholar]
- 16.Knodel M, Kuss AW, Lindemann D, Berberich I, Schimpl A. Reversal of Blimp-1-mediated apoptosis by A1, a member of the Bcl-2 family. Eur J Immunol. 1999;29:2988–2998. doi: 10.1002/(SICI)1521-4141(199909)29:09<2988::AID-IMMU2988>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 17.Messika EJ, Lu PS, Sung YJ, Yao T, Chi JT, Chien YH, Davis MM. Differential effect of B lymphocyte-induced maturation protein (Blimp-1) expression on cell fate during B cell development. J Exp Med. 1998;188:515–525. doi: 10.1084/jem.188.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia JF, Roncador G, Garcia JF, Sanz AI, Maestre L, Lucas E, Montes-Moreno S, Fernandez Victoria R, Martinez-Torrecuadrara JL, Marafioti T, Mason DY, Piris MA. PRDM1/BLIMP-1 expression in multiple B and T-cell lymphoma. Haematologica. 2006;91:467–474. [PubMed] [Google Scholar]
- 19.Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, Aster JC, Murty VV, Shipp MA, Dalla-Favera R. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 21.Gyory I, Fejer G, Ghosh N, Seto E, Wright KL. Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J Immunol. 2003;170:3125–3133. doi: 10.4049/jimmunol.170.6.3125. [DOI] [PubMed] [Google Scholar]
- 22.Borson ND, Lacy MQ, Wettstein PJ. Altered mRNA expression of Pax5 and Blimp-1 in B cells in multiple myeloma. Blood. 2002;100:4629–4639. doi: 10.1182/blood.V100.13.4629. [DOI] [PubMed] [Google Scholar]
- 23.Garrone P, Neidhardt EM, Garcia E, Galibert L, van Kooten C, Banchereau J. Fas ligation induces apoptosis of CD40-activated human B lymphocytes. J Exp Med. 1995;182:1265–1273. doi: 10.1084/jem.182.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchtey J, Pennini M, Pai RK, Harding CV. CpG DNA induces a class II transactivator-independent increase in class II MHC by stabilizing class II MHC mRNA in B lymphocytes. J Immunol. 2003;171:2320–2325. doi: 10.4049/jimmunol.171.5.2320. [DOI] [PubMed] [Google Scholar]
- 25.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and tools for whole-genome alignments. Genome Research. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh N, Piskurich JF, Wright G, Hassani K, Ting JP, Wright KL. A novel element and a TEF-2-like element activate the major histocompatibility complex class II transactivator in B-lymphocytes. J Biol Chem. 1999;274:32342–32350. doi: 10.1074/jbc.274.45.32342. [DOI] [PubMed] [Google Scholar]
- 28.Pfeifer GP, Tanguay RL, Steigerwald SD, Riggs AD. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990;4:1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- 29.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA pol II in a soluble extract from isolated mammalian nuclei. Nucleic Acid Research. 1983;11:1475–1488. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y, Wong Kk, Calame K. Repression of c-myc Transcription by Blimp-1, an Inducer of Terminal B Cell Differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 31.Dani C, Blanchard JM, Piechaczyk M, El Sabouty S, Marty L, Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984;81:7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tunyaplin C, Shapiro MA, Calame KL. Characterization of the B lymphocyte-induced maturation protein-1 (Blimp-1) gene, mRNA isoforms and basal promoter. Nucleic Acids Res. 2000;28:4846–4855. doi: 10.1093/nar/28.24.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parekh S, Polo JM, Shaknovich R, Juszczynski P, Lev P, Ranuncolo SM, Yin Y, Klein U, Cattoretti G, Dalla Favera R, Shipp MA, Melnick A. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 36.Mora-Lopez F, Pedreno-Horrillo N, Delgado-Perez L, Brieva JA, Campos-Caro A. Transcription of PRDM1, the master regulator for plasma cell differentiation, depends on an SP1/SP3/EGR-1 GC-box. Eur J Immunol. 2008;38:2316–2324. doi: 10.1002/eji.200737861. [DOI] [PubMed] [Google Scholar]
- 37.Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 Function: A Mechanism for the Regulation of Blimp-1 Expression and B Lymphocyte Differentiation by the B Cell Lymphoma-6 Protooncogene. J Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- 38.Linderson Y, Eberhard D, Malin S, Johansson A, Busslinger M, Pettersson S. Corecruitment of the Grg4 repressor by PU.1 is critical for Pax5-mediated repression of B-cell-specific genes. EMBO Rep. 2004;5:291–296. doi: 10.1038/sj.embor.7400089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto H, Kihara-Negishi F, Yamada T, Hashimoto Y, Oikawa T. Physical and functional interactions between the transcription factor PU.1 and the coactivator CBP. Oncogene. 1999;18:1495–1501. doi: 10.1038/sj.onc.1202427. [DOI] [PubMed] [Google Scholar]
- 40.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 41.Lotz C, Mutallib SA, Oehlrich N, Liewer U, Ferreira EA, Moos M, Hundemer M, Schneider S, Strand S, Huber C, Goldschmidt H, Theobald M. Targeting positive regulatory domain I-binding factor 1 and X box-binding protein 1 transcription factors by multiple myeloma-reactive CTL. J Immunol. 2005;175:1301–1309. doi: 10.4049/jimmunol.175.2.1301. [DOI] [PubMed] [Google Scholar]
- 42.Ohkubo Y, Arima M, Arguni E, Okada S, Yamashita K, Asari S, Obata S, Sakamoto A, Hatano M, Ebara OWJM, Saisho H, Tokuhisa T. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J Immunol. 2005;174:7703–7710. doi: 10.4049/jimmunol.174.12.7703. [DOI] [PubMed] [Google Scholar]
- 43.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 44.Tatetsu H, Ueno S, Hata H, Yamada Y, Takeya M, Mitsuya H, Tenen DG, Okuno Y. Down-regulation of PU.1 by methylation of distal regulatory elements and the promoter is required for myeloma cell growth. Cancer Res. 2007;67:5328–5336. doi: 10.1158/0008-5472.CAN-06-4265. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Oikawa T. Direct association between PU.1 and MeCP2 that recruits mSin3A-HDAC complex for PU.1-mediated transcriptional repression. Oncogene. 2003;22:8688–8698. doi: 10.1038/sj.onc.1207182. [DOI] [PubMed] [Google Scholar]
- 46.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 47.Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, Tashiro S, Muto A, Igarashi K. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 48.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.