Abstract
Ubiquitin-mediated proteolysis has a central role in controlling the intracellular levels of several important regulatory molecules such as cyclins, CKIs, p53, and IκBα. Many diverse proinflammatory signals lead to the specific phosphorylation and subsequent ubiquitin-mediated destruction of the NF-κB inhibitor protein IκBα. Substrate specificity in ubiquitination reactions is, in large part, mediated by the specific association of the E3–ubiquitin ligases with their substrates. One class of E3 ligases is defined by the recently described SCF complexes, the archetype of which was first described in budding yeast and contains Skp1, Cdc53, and the F-box protein Cdc4. These complexes recognize their substrates through modular F-box proteins in a phosphorylation-dependent manner. Here we describe a biochemical dissection of a novel mammalian SCF complex, SCFβ-TRCP, that specifically recognizes a 19-amino-acid destruction motif in IκBα (residues 21–41) in a phosphorylation-dependent manner. This SCF complex also recognizes a conserved destruction motif in β-catenin, a protein with levels also regulated by phosphorylation-dependent ubiquitination. Endogenous IκBα–ubiquitin ligase activity cofractionates with SCFβ-TRCP. Furthermore, recombinant SCFβ-TRCP assembled in mammalian cells contains phospho-IκBα-specific ubiquitin ligase activity. Our results suggest that an SCFβ-TRCP complex functions in multiple transcriptional programs by activating the NF-κB pathway and inhibiting the β-catenin pathway.
Keywords: Ubiquitin ligase, SCF complex, proteolysis, destruction motifs, NF-κB, β-catenin
The transcription factor NF-κB has a central role in cellular stress and inflammatory responses by controlling cytokine-inducible gene expression and lymphocyte stimulation by antigens (Baeuerle and Baltimore 1996; Gilmore et al. 1996). In addition, NF-κB is required to block cell death in response to tumor necrosis factor α (TNFα) and ionizing radiation, suggesting that it acts to regulate the transcription of survival genes (Beg and Baltimore 1996; Liu et al. 1996; Van Antwerp et al. 1996; Wang et al. 1996). NF-κB is a ubiquitous heterodimeric complex composed of a p65/RelA subunit and a p50 subunit. This complex is normally sequestered in an inactive form in the cytoplasm through interaction with members of a family of inhibitory proteins, the IκBs (Beg et al. 1992; for review, see Baeuerle and Baltimore 1996). These proteins, when associated with NF-κB, obscure the nuclear localization signal in NF-κB and also block the ability of NF-κB to bind DNA. In response to TNFα and other signals, IκBα is rapidly phosphorylated on two serine residues near the amino terminus (Ser-32 and Ser-36 in IκBα) (Beg et al. 1993; Finco et al. 1994; Alkalay et al. 1995; Brown et al. 1995; Chen et al. 1995, 1996; DiDonato et al. 1995; Lin et al. 1995). This modification triggers the rapid destruction of IκBα by ubiquitin-mediated proteolysis, thereby allowing NF-κB nuclear translocation and target gene expression (Chen et al. 1995; Scherer et al. 1995; for review, see Hochstrasser 1996). Recent work has uncovered two IκBα kinases, IκKα and IκKβ, that are responsible for signal-dependent phosphorylation of IκBα (DiDonato et al. 1997; Mercurio et al. 1997; Regnier et al. 1997; Woronicz et al. 1997; Zandi et al. 1997, 1998). These proteins are part of a 700-kD protein complex that is assembled through two structural components IκKγ/NEMO and IKAP (Cohen et al. 1998; Rothwarf et al. 1998; Yamaoka et al. 1998) and are activated by cytokines. In vitro, both IκKα and IκKβ can phosphorylate IκBα specifically on serines 32 and 36, but both kinases are required for efficient IκBα phosphorylation in vivo (Zandi et al. 1997).
Although the pathways leading to IκBα phosphorylation have been described in detail, little is known about the molecules responsible for ubiquitination. Ubiquitin-mediated proteolysis involves a cascade of ubiquitin transfer reactions in which the ubiquitin-activating enzyme E1 uses ATP to form a high-energy thiolester bond with ubiquitin, which is then transferred to members of the E2 ubiquitin-conjugating enzyme family (Hershko et al. 1983; Hochstrasser 1995). Ubiquitin is then transferred from the E2 to lysine residues in the target through an E3–ubiquitin ligase. E3s serve as adaptors that interact with both the target protein and the appropriate E2, thereby providing specificity to the ubiquitin transfer reaction. In some cases, the E3 is also involved in ubiquitin transfer (Scheffner et al. 1995; Rolfe et al. 1995). Multiple rounds of ubiquitination of the initial conjugates lead to polyubiquitination, which targets the protein for proteolysis by the 26S proteasome. Recent studies have elaborated a modular ubiquitin ligase complex, the SCF–ubiquitin ligase, which mediates phosphorylation-dependent ubiquitination of a large number of proteins (for review, see Elledge and Harper 1998; Patton et al. 1998b). The SCF is composed of Skp1, Cdc53/Cul1, and a specificity-conferring F-box protein (Bai et al. 1996; Feldman et al. 1997; Skowyra et al. 1997; Patton et al. 1998a). F-box proteins contain two domains, an F-box motif that binds Skp1 and allows assembly into Skp1/Cdc53 complexes, and a second protein–protein interaction domain that interacts specifically with one or more target proteins (Bai et al. 1996). Cdc53/Cul1, in turn, interacts with both the E2 and the Skp1/F-box protein complex (Skowyra et al. 1997; Patton et al. 1998a). SCF complexes mediate phosphorylation-dependent destruction of a wide array of regulatory proteins in yeast, including the Cdk inhibitors Sic1, Far1, and Rum1, G1 cyclins, the transcription factor Gcn4, and the DNA replication initiator proteins Cdc6 and Cdc18 (for review, see Elledge and Harper 1998; Patton et al. 1998b). In contrast with yeast, targets of vertebrate SCF complexes remain largely unknown. Previously, we identified four vertebrate proteins that contain the F-box motif, linking them to the ubiquitin pathway: mammalian cyclin F, Skp2, MD6, and Xenopus β-TRCP (β-transducin repeat-containing protein; Bai et al. 1996). β-TRCP was originally identified as a suppressor of a temperature-sensitive mutation in the budding yeast CDC15 gene (Spevak et al. 1993), but its mechanism of suppression has not been determined. Recent genetic evidence has implicated Xenopus β-TRCP and its Drosphila homolog, slimb, in control of proteolysis in the Hedgehog and Wingless/Wnt signaling pathways (Jiang and Struhl 1998; Marikawa and Elinson 1998).
We have used biochemical approaches to examine whether IκBα ubiquitination might involve an SCF–ubiquitin ligase. Here we report that mammalian β-TRCP binds to the IκBα destruction motif in a phosphorylation-dependent manner, thereby recruiting IκBα into an SCF–ubiquitin ligase complex. Moreover, SCFβ-TRCP components cofractionate with IκBα–ubiquitin ligase activity from tissue culture cells and SCFβ-TRCP can stimulate ubiquitination of phosphorylated but not unphosphorylated IκBα in an in vitro reconstitution assay. We also demonstrate that the same SCFβ-TRCP complex recognizes a similar destruction motif in β-catenin, a component of the TCF/Lef transcription factor complex that functions downstream of Wingless/Wnt (for review, see Peifer 1997) and whose levels are also controlled by phosphorylation-dependent ubiquitin-mediated proteolysis (Aberle et al. 1997). Our results, together with the effects of loss-of-function mutations in the Drosophila β-TRCP homolog slimb (Jiang and Struhl 1998), suggest that a single SCFβ-TRCP complex functions in diverse signaling pathways that impinge on transcription control mediated by cytokines (NF-κB), Wnt/Wingless (β-catenin), and Hedgehog [Cubitus interruptus (Ci)].
Results
Phosphorylation-dependent association of the IκBα destruction motif with Skp1
IκBα contains two serine residues at positions 32 and 36 that are specifically phosphorylated by the IκK complex in response to TNFα stimulation. Phosphorylation of both of these residues is required for IκBα ubiquitination in vivo. Previous studies have shown that a 21-amino-acid phosphopeptide containing this destruction motif can block IκBα–ubiquitin ligase activity in crude cell extracts and can block NF-κB activation in tissue culture cells (Yaron et al. 1997). In addition, this destruction motif can confer signal-dependent destruction when fused to heterologous proteins (Wulczyn et al. 1998). Given the role for SCF complexes in phosphorylation-dependent ubiquitination of various regulatory proteins, we sought to determine whether SCF complexes might be involved in IκBα ubiquitination. Synthetic 21-residue peptides encompassing the IκBα destruction motif in either the doubly phosphorylated or unphosphorylated forms (Fig. 1a) were immobilized on agarose beads and incubated with HeLa cell lysates. Proteins stably associated with these beads were then examined for the presence of Skp1 by immunoblotting (Fig. 1b). Skp1 was readily detected in proteins bound to the phospho-IκBα peptide but not the unphosphorylated peptide. We estimate that ∼1% of the total Skp1 in these lysates stably associated with the phospho-IκBα peptide under these conditions.
Figure 1.
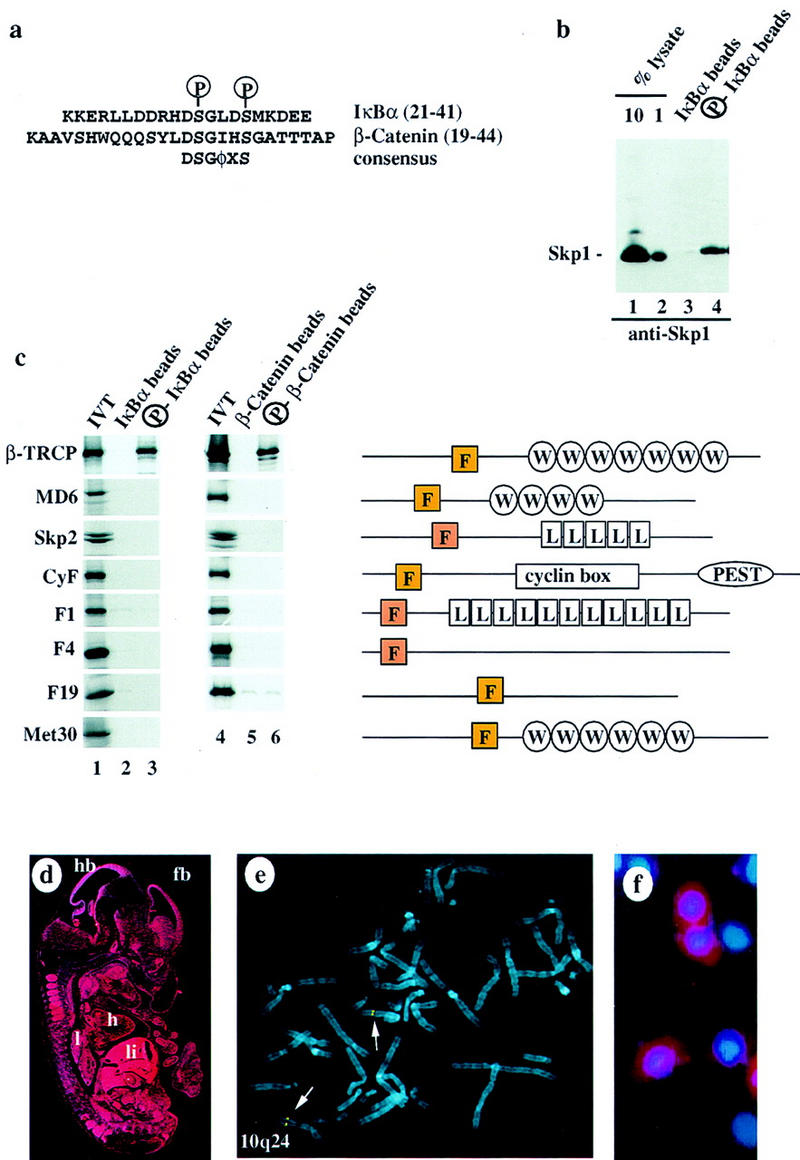
The F-box protein β-TRCP associates with phosphorylated destruction motifs in IκBα and β-catenin. (a) Sequences of the IκBα (p21) and β-catenin peptides used in this study. The positions of phosphorylation in the peptides are shown as is the consensus sequence for the destruction motif. (φ) Hydrophobic amino acid. (b) Phosphorylation-specific association of the IκBα destruction motif with Skp1 in vitro. HeLa cell proteins (600 μg) were incubated with phosphorylated or unphosphorylated IκBα peptides (p21) immobilized on beads. Bound proteins were immunoblotted with anti-Skp1 antibodies. Approximately 1% of the Skp1 contained in these lysates remained bound to the phosphorylated IκBα beads. (c) β-TRCP specifically associated with phosphorylated IκBα and β-catenin destruction motifs. A panel of [35S]methionine-labeled in vitro-translated F-box proteins was used in binding reactions with IκBα (lanes 1–3) or β-catenin (lanes 4–6) peptide beads. Bound proteins were analyzed by SDS-PAGE and autoradiography. (Right) The domain structures of each F-box protein. (d) The pattern of expression of β-TRCP at day 11.5 during mouse development was determined by in situ hybridization. The dark-field signal from the β-TRCP riboprobe is shown in red. (hb) Hindbrain; (fb) forebrain; (h) heart; (l) lung; (li) liver. (e) Chromosomal localization of β-TRCP. A bacmid containing human β-TRCP DNA was hybridized to metaphase chromosomes (blue) and detected using fluorescein. The position of hybridization (yellow) is 10q24 (indicated by arrows). (f) β-TRCP is localized in the cytoplasm. HeLa cells were transiently transfected with pCMV–HA–β-TRCP and subcellular localization determined after 48 hr by indirect immunofluorescence. Anti-HA localization, red; nuclei stained with DAPI, blue.
Recognition of phosphorylated destruction motifs in IκBα and β-catenin by the WD-40 repeat-containing F-box protein β-TRCP
The ability of a phospho-IκBα peptide to associate with Skp1 suggested the existence of an F-box protein capable of recognizing the IκBα destruction motif. Our previous studies identified three vertebrate F-box proteins (Skp2, MD6, and Xenopus β-TCRP) based on homology to the F-box sequence in human cyclin F and the budding yeast protein Cdc4 (Bai et al. 1996). Recently, we have identified cDNAs encoding 20 distinct mouse and/or human F-box proteins, including the WD-40 repeat-containing protein β-TRCP, a leucine-rich repeat containing F-box protein F1, and a number of F-box proteins lacking known protein–protein interaction motifs outside the F-box (Fig. 1c; J. Winston, S.J. Elledge, and J.W. Harper, in prep.). Using in vitro translation products, we asked whether members of a collection of these F-box proteins could associate with IκBα peptides. Only one, β-TRCP, was found to associate with the phospho-IκBα destruction motif, and this interaction was dependent on phosphorylation (Fig. 1c). Mouse and human β-TRCP are 95% identical and both interact equally well with IκBα in this assay (data not shown for human β-TRCP). Our analysis included two other WD-40-containing F-box proteins, human MD6 and Met30, the closest homolog of β-TRCP in budding yeast (31% identity). Importantly, neither of these proteins associated with phospho-IκBα (Fig. 1c), suggesting that the interaction of β-TRCP with phospho-IκBα is highly specific.
Previous studies in Drosophila have demonstrated that mutations in the β-TRCP homolog slimb led to accumulation of Armadillo, the Drosophila homolog of β-catenin (Jiang and Struhl 1998). β-Catenin is known to be ubiquitinated in a glycogen synthase kinase 3β (GSK3β)-dependent manner and contains a motif within a cluster of candidate GSK3β phosphorylation sites that is closely related to the IκBα destruction motif (Fig. 1a; Ikeda et al. 1998). Although the GSK3β phosphorylation sites in β-catenin are not known, we hypothesized based on the sequence similarity between IκBα and β-catenin that Ser-33 and Ser-37 might represent relevant phosphorylation sites. A β-catenin-derived peptide containing phosphoserine residues at these two positions associated with β-TRCP but not other F-box proteins tested, whereas the unphosphorylated peptide failed to associate with β-TRCP (Fig. 1c).
NF-κB is a ubiquitous transcription factor. As assessed by in situ hybridization, β-TRCP is also expressed throughout the developing mouse embryo (day 11.5 postcoitum), with the highest levels in the brain, lung, and liver (Fig. 1d). β-TRCP is largely, if not exclusively, cytoplasmic, as assessed in HeLa cells transiently expressing an HA-tagged β-TRCP protein (Fig. 1f). The gene for human β-TRCP lies on chromosome 10q24, as determined by in situ hybridization of metaphase chromosomes with β-TRCP genomic DNA (Fig. 1e). Cytogenetic data indicate that this region of the genome is altered in a limited number of cancer types (see Discussion).
Association of SCFβ-TRCP with IκBα and β-catenin destruction motifs
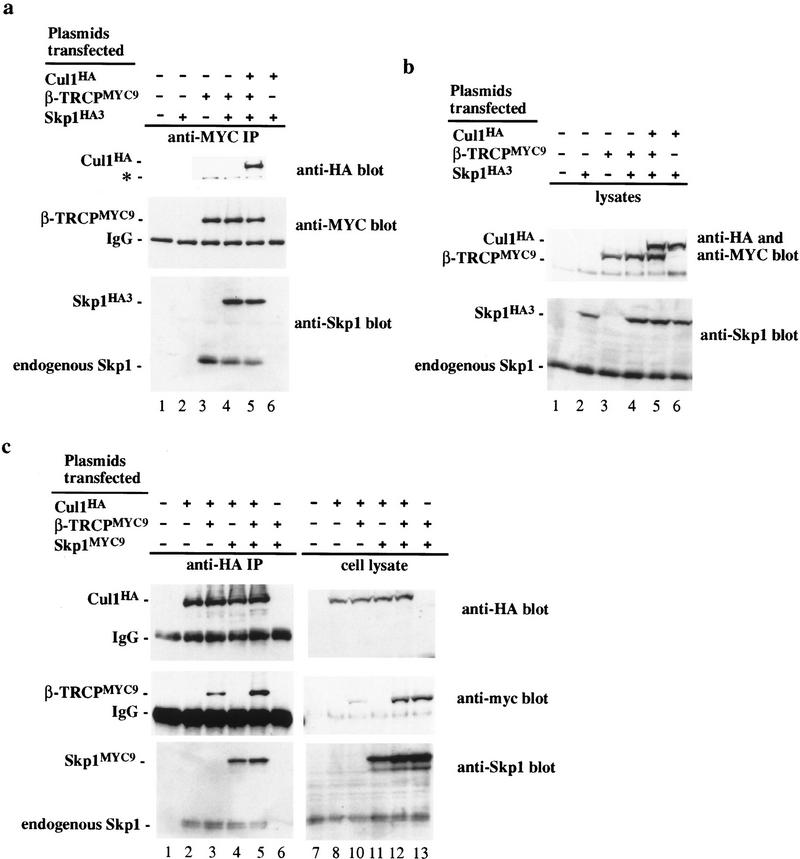
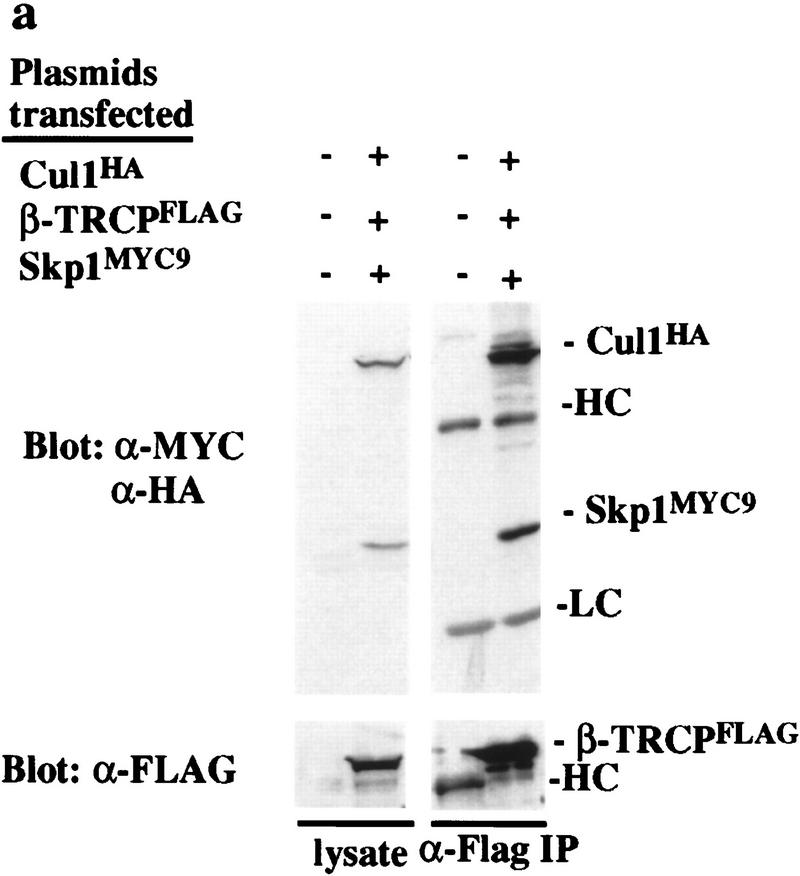
Having identified β-TRCP as a candidate F-box protein for IκBα and β-catenin, we next sought to demonstrate that β-TRCP forms an SCF complex in mammalian cells and that this complex recognizes IκBα and β-catenin destruction motifs. Although there are six mammalian Cullin homologs, the interaction of Skp1 with this family appears to be restricted to Cul1 (Michel and Xiong 1998). 293T cells were transfected with various combinations of plasmids expressing epitope-tagged β-TRCP, Cul1, or Skp1 and anti-Myc immune complexes from cell lysates analyzed by immunoblotting. β-TRCPMyc associated with both transfection-derived Skp1HA (Fig. 2a, lanes 4,5) and Cul1HA (lane 5). In contrast, Cul1HA and Skp1HA were not precipitated from control lysates lacking β-TRCPMyc (lane 6). In the absence of transfection of Skp1 and Cul1, β-TRCPMyc associated with endogenous Skp1 (lane 3) and Cul1 (data not shown). Analogous results were obtained when Cul1HA was immunoprecipitated with anti-HA antibodies from cells expressing Cul1HA, β-TRCPMyc, and Skp1Myc (Fig. 2c). Thus, β-TRCP can form an SCF complex in vivo analogous to that found previously for other F-box proteins (Skowyra et al. 1997; Lisztwan et al. 1998; Lyapina et al. 1998; Michel and Xiong 1998).
Figure 2.
β-TRCP associates with Skp1 and Cul1 in tissue culture cells. 293T cells were transfected with the indicated plasmids and lysates (0.5 μg of protein/250 μl) used for immunoprecipitation as described in Materials and Methods. Immune complexes or crude lysates from each transfection were analyzed for the presence of Skp1, Cul1, and β-TRCP by immunoblotting. (a) Anti-β-TRCPMyc immune complexes. Blots were probed first for β-TRCP, stripped, and probed for Skp1 and Cul1. The bands indicated by the asterisk indicate the position of β-TRCP whose antibody was not efficiently stripped from the blot. (b) Crude cell lysates (50 μg) corresponding to extracts used in a. (c) Anti-Cul1HA immune complexes (lanes 1–6) and corresponding cell lysates (50 μg) (lanes 7–13). The positions of both epitope-tagged and endogenous Skp1 are shown.
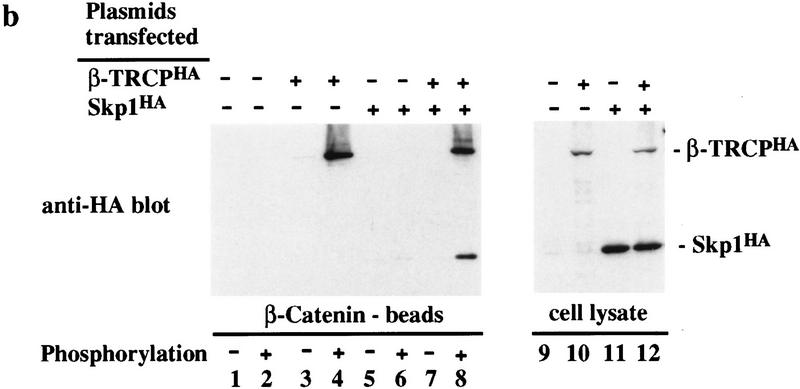
Next, we asked whether the SCFβ-TRCP complex could associate with the IκBα destruction motif peptide. As shown in Figure 3a, the SCFβ-TRCP complex readily associated with phosphorylated IκBα peptide beads (lanes 6,8,10) but was not retained on unphosphorylated IκBα beads (lanes 5,7,9). Although Cul1 associates at low levels with agarose beads containing IκBα peptides in the absence of β-TRCPMyc expression (lanes 11,12) and with agarose beads alone (data not shown), the association with the phospho-IκBα peptide was greatly enhanced by expression of β-TRCPMyc (lane 10). Consistent with the results in Figure 1b, endogenous Skp1 was observed in association with phospho-IκBα peptide beads in a phosphorylation-dependent manner in the absence of transfection of β-TRCP (lanes 1–4), but when the levels of β-TRCP were increased by transfection, the quantity of endogenous Skp1 associated with β-TRCP increased substantially (Fig. 3a, lanes 4,6). Analogous results were obtained in a more limited series of binding reactions employing β-catenin-derived peptides (Fig. 3b).
Figure 3.
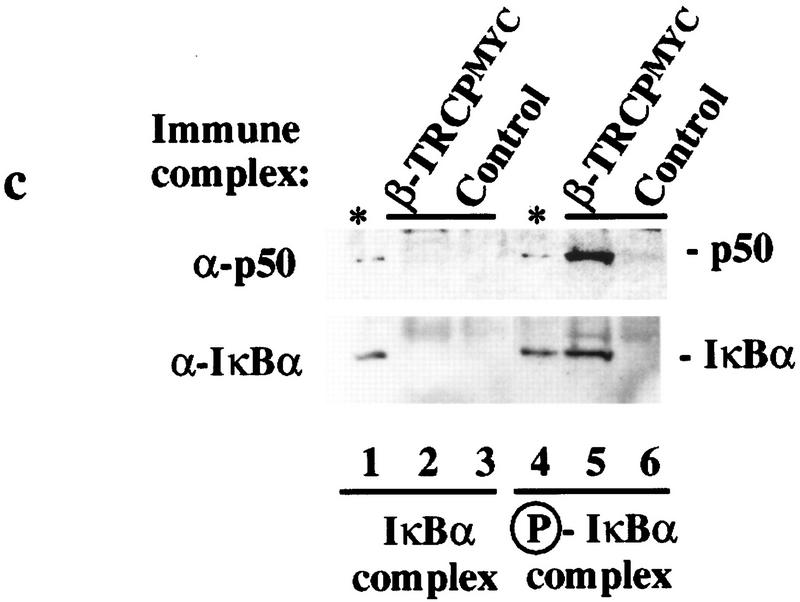
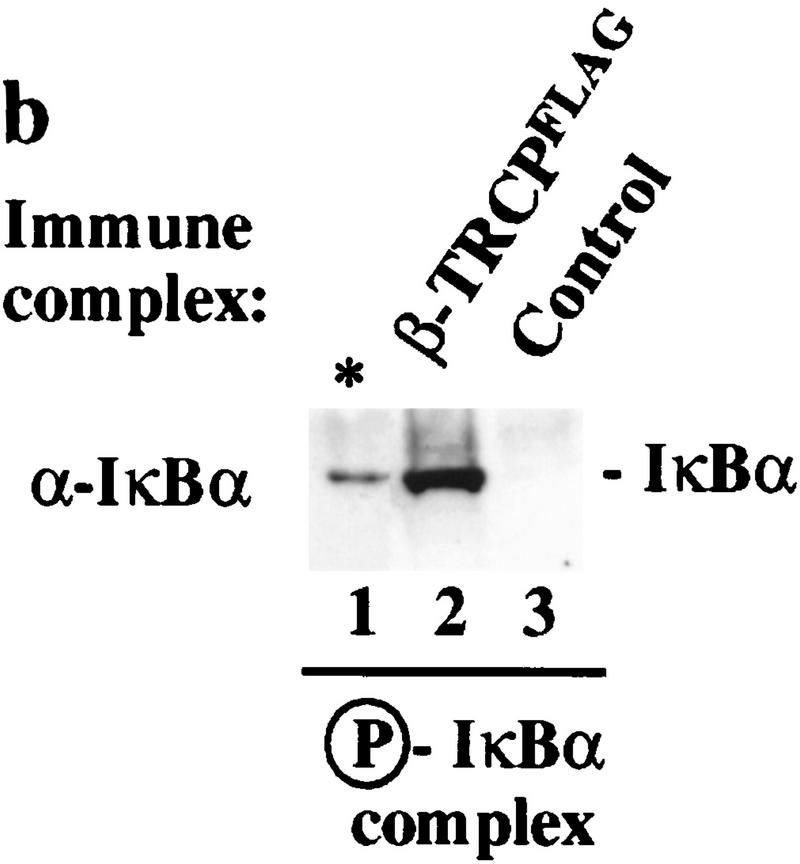
Association of SCFβ-TRCP with IκBα and β-catenin destruction motifs and with the IκBα/NF-κB complex. (a,b) Cell lysates (0.3 μg of protein/150 μl) from Fig. 2 were used in IκBα (a) and β-catenin (b) peptide bead binding reactions as described in Materials and Methods. Bound proteins were analyzed by immunoblotting with the indicated antibodies. (c) Phosphorylation-dependent association of β-TRCPMyc with the IκBα/p50/p65 complex in vitro. β-TRCPMyc immune complexes (lanes 2,5) corresponding to those in Fig. 2a (lane 3) or control complexes (lanes 3,6) corresponding to those in Fig. 2a (lane 1) were used in binding reactions with either IκBα/p50/p65 or IκK-β phosphorylated IκBα/p50/p65 complexes (see Materials and Methods). Bound proteins were separated by SDS-PAGE and immunoblotted using anti-p50 or anti-IκBα antibodies. The asterisk (lanes 1,4) indicates the positions of 15% of the input IκBα complexes used in the binding reaction.
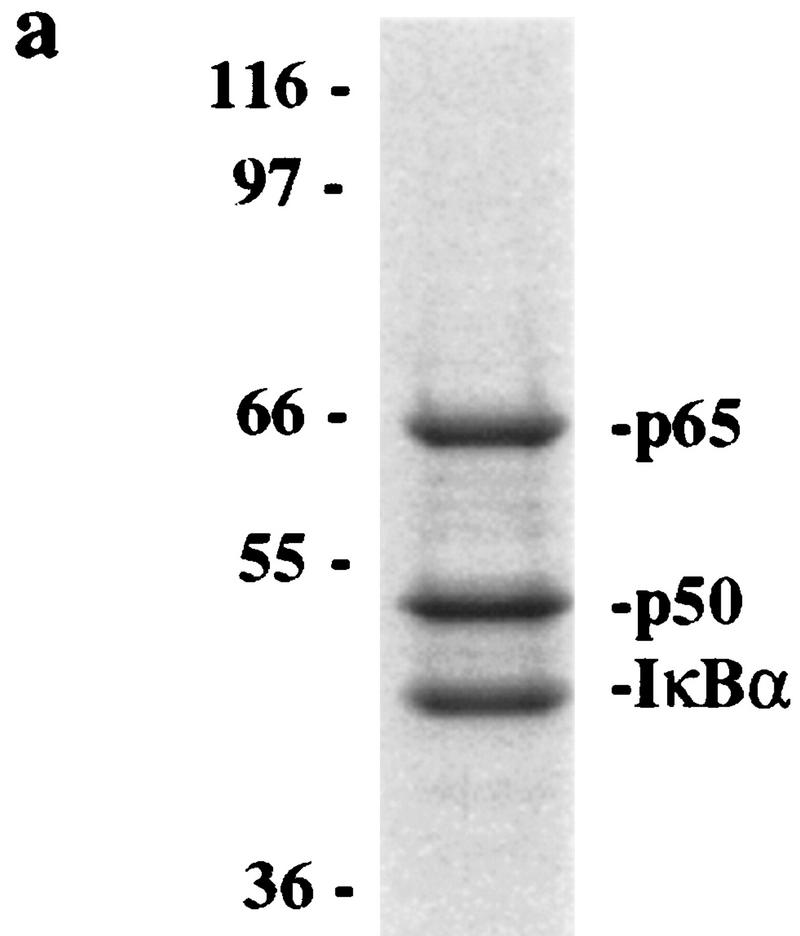
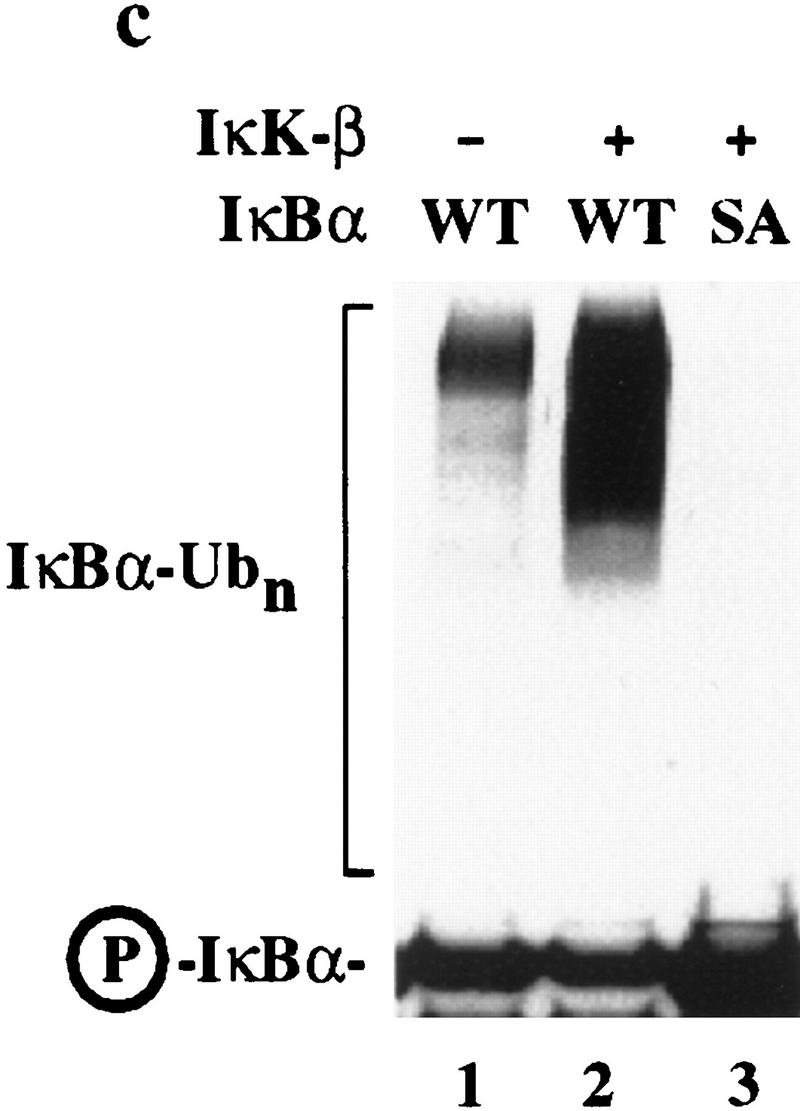
Although it was clear that destruction motif peptides can bind the SCFβ-TRCP complex, it was necessary to demonstrate that this complex also recognized the endogenous ubiquitination substrate, the IκK-phosphorylated IκBα/NF-κB complex. To generate this substrate, IκBα/p50/p65 complexes were produced in insect cells and purified to near homogeneity (Fig. 4a). When incubated with ATP and purified IκK-β, the IκBα protein underwent a mobility shift reminiscent of that observed upon phosphorylation in vivo, and this phosphorylated IκBα protein was recognized by phosphospecific antibodies directed at Ser-32 of IκBα (Fig. 4b, lane 2). In addition, microsequencing of IκK-treated IκBα confirmed that both Ser-32 and Ser-36 were phosphorylated (data not shown). To examine whether SCFβ-TRCP could recognize this complex, binding reactions were performed using immobilized SCFβ-TRCP complexes isolated from 293T cells transiently expressing Myc-tagged β-TRCP or mock transfected cells as a control and either phosphorylated or unphosphorylated IκBα/NF-κB complexes. The β-TRCPMyc immune complexes contain endogenous Skp1 (Fig. 2, lane 3) and Cul1 (data not shown) as determined by immunoblotting. Both IκBα and p50 were found to associate with the SCFβ-TRCP complex but not control immune complex in a phosphorylation-dependent manner (Fig. 3c). Similar results were obtained with GST–β-TRCP complexes purified from insect cells (data not shown).
Figure 4.
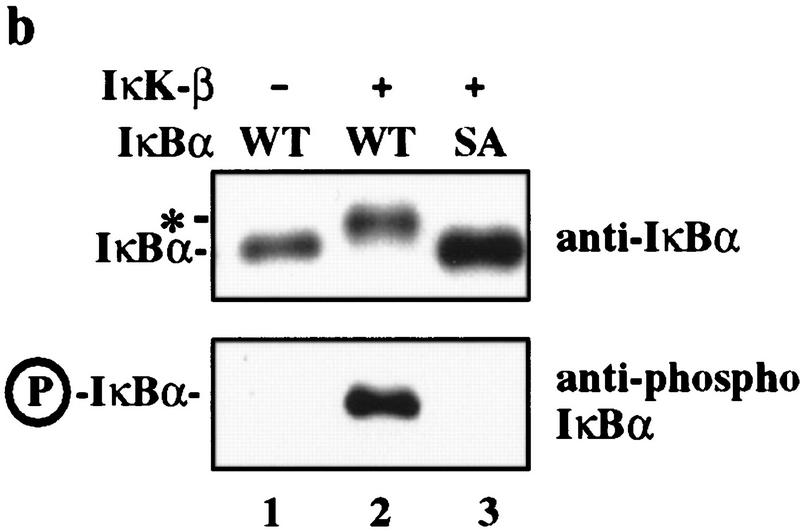
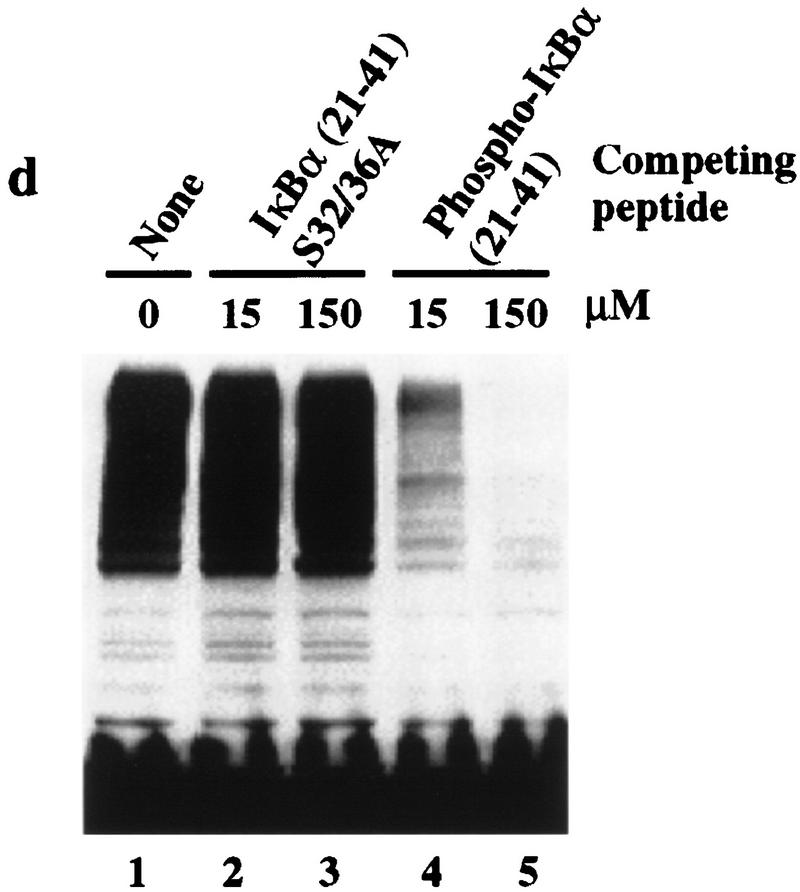
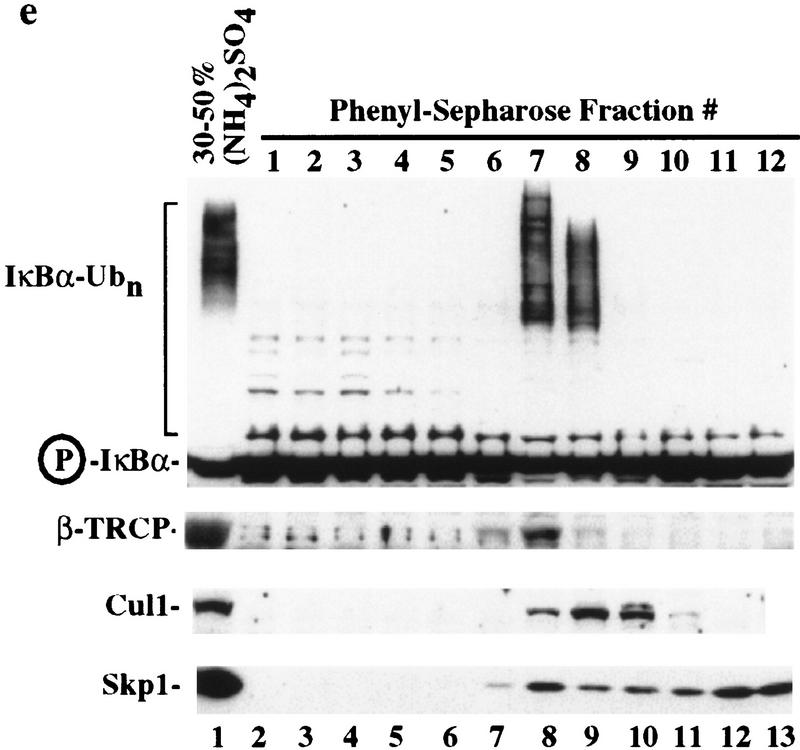
IκBα–ubiquitin ligase activity from human cells cofractionates with β-TRCP. (a) IκBα/p50/p65 complexes were purified to near homogeneity from insect cells as described in Materials and Methods. Proteins were separated by SDS-PAGE and stained with Coomassie blue. (b) Phosphorylation of the IκBα/p50/p65 complex by IκK-β. The IκBα complex (lanes 1,2) or a nonphosphorylatable IκBα mutant (S32/36A) complex (lane 3) was incubated in the presence of ATP and IκK-β as indicated in Materials and Methods. Products were analyzed by immunoblotting with anti-IκBα antibodies to detect a mobility shift accompanying phosphorylation that is absent in the nonphosphorylatable mutant (top) or with antibodies that specifically detect the Ser-32-phosphorylated form of IκBα (bottom). (c) Ubiquitination of IκBα complexes by crude cell lysates was performed as described in Materials and Methods. Phosphorylation leads to a ∼10- to 20-fold increase in ubiquitin conjugates relative to the unphosphorylated complex, whereas no activity is observed with the nonphosphorylatable IκBα complexes. (d) Inhibition of IκBα ubiquitination by phosphorylated IκBα destruction motif peptides but not by nonphosphorylatable destruction motif peptides (p19). Ubiquitination reactions were performed with crude cell extracts and IκK-β phosphorylated IκBα complexes in the presence or absence of phosphorylated or nonphosphorylatable destruction motif peptides. Specific inhibition of ubiquitination was observed with the phosphorylated peptide. (e) Cofractionation of β-TRCP with endogenous IκBα–ubiquitin ligase activity. Crude extracts from THP.1 cells were precipitated with ammonium sulfate. Solubilized proteins containing ubiquitin ligase activity were fractionated using a phenyl–Sepharose column and activity in each fraction determined as described in Materials and Methods. Aliquots of column fractions were assayed for β-TRCP, Cul1, and Skp1 by immunoblotting. Fractions containing β-TRCP, Cul1, and Skp1 (fractions 7,8) contain IκBα–ubiquitin ligase activity.
Biochemical association of endogenous IκBα–ubiquitin ligase activity with β-TRCP
Crude cell lysates from the human monocyte cell line THP.1 contain potent IκBα–ubiquitin ligase activity (Fig. 4c). In the context of an IκBα/NF-κB complex, efficient IκBα ubiquitination by these lysates is dependent on phosphorylation by IκK (Fig. 4c). As reported earlier (Yaron et al. 1997), this IκBα–ubiquitin ligase activity is strongly inhibited by phosphorylated IκBα destruction motif peptides (Fig. 4d, lanes 4,5) but not by nonphosphorylatable destruction box peptides (lanes 2,3). Thus, this assay reiterates the requirements for IκBα ubiquitination observed in vivo. Greater than 95% of the IκBα–ubiquitin ligase activity in these extracts can be precipitated with 30%–50% ammonium sulfate (Fig. 4e, lane 1) and can be further purified by chromatography on a phenyl–Sepharose column (Fig. 4e; see Materials and Methods). Peak fractions of IκBα–ubiquitin ligase activity (Fig. 4e, lanes 8,9) elute at 0.5 m ammonium sulfate.
Having partially purified components of the IκBα–ubiquitin ligase, we examined whether β-TRCP and other SCF components were contained in active fractions from the phenyl–Sepharose column. Skp1 has an extended elution profile, but both Skp1 and Cul1 are contained in the active fractions 7 and 8 (Fig. 4e). Skp1 and Cul1 can interact with multiple F-box proteins, and the identity of the F-box protein in complexes with Cul1 and Skp1 is likely to affect elution properties on phenyl–Sepharose. In contrast with Skp1, β-TRCP levels peak in fraction 7, as determined using affinity-purified carboxy-terminal antibodies, coincident with maximal activity (Fig. 4e, lane 8). β-TRCP was also detected in active fraction 8 (Fig. 4e, lane 9). This fraction contained lower levels of IκBα–ubiquitin ligase activity, as assessed by the extent of conjugation, consistent with the lower levels of β-TRCP. As shown below, under some gel conditions the β-TRCP protein is resolved into a closely spaced doublet of proteins at 58–60 kD. Interestingly, although fraction 6 contains detectable levels of Skp1 and β-TRCP, it lacks detectable Cul1 and IκBα–ubiquitin ligase activity (Fig. 4e, lane 7). Likewise, fraction 9 containing Skp1 and Cul1 but no β-TRCP is also inactive (Fig. 4e, lane 10).
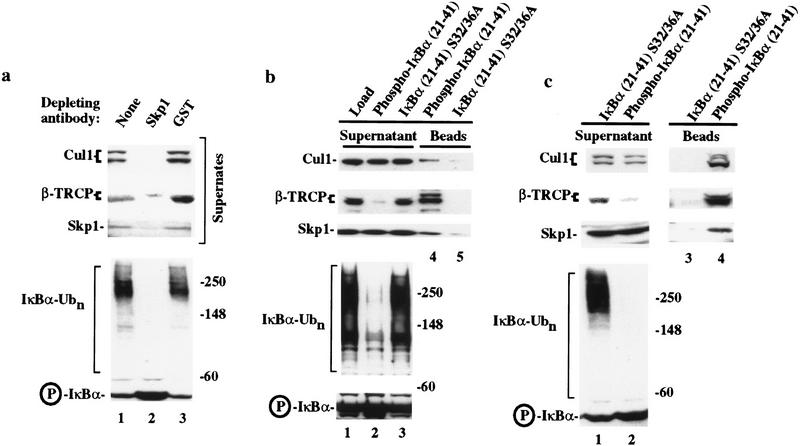
Consistent with a role for Skp1 in the IκBα–ubiquitin ligase, antibodies against Skp1, but not control GST antibodies, deplete IκBα–ubiquitin ligase activity from the active phenyl–Sepharose fractions (Fig. 5a). As expected, Skp1 and Cul1 are largely depleted from these active extracts (lane 2). Importantly, the majority of β-TRCP is also removed by Skp1 antibodies (Fig. 5a, lane 2). β-TRCP migrated as a closely spaced doublet at 58 and 60 kD. The faster migrating form, corresponding to ∼80% of the total, was essentially depleted by anti-Skp1 antibodies (lane 2), when compared with control GST-depleted extracts. The source of the heterogeneity in β-TRCP is not known at present, but it is possible that the more slowly migrating form is not associated with Skp1 or is dislodged from Skp1 by the anti-Skp1 antibodies. We also note that Cul1 migrated as a doublet (Fig. 5a). The slower migrating form is likely to correspond to a form of the protein conjugated to NEDD8, a homolog of Rub1 that is known to be covalently linked to Cdc53 in budding yeast (for review, see Hochstrasser 1998).
Figure 5.
Depletion of IκBα–ubiquitin ligase activity by anti-Skp1 antibodies and destruction motif peptides correlates with removal of β-TRCP. (a) Ubiquitin ligase activity from phenyl–Sepharose fractions 7 and 8 was depleted with antibodies against Skp1 or GST and the supernatant assayed for ubiquitination activity toward phosphorylated IκBα (bottom). The levels of Skp1, Cul1, and β-TRCP in the supernatants from the depleted fractions were determined by immunoblotting (top). (b,c) Endogenous β-TRCP associates with phosphorylated IκBα destruction motif peptides during depletion of IκBα–ubiquitin ligase activity. Crude cell extracts (b) or active fractions from a phenyl–Sepharose column (c) were incubated with beads containing either phosphorylated or nonphosphorylatable IκBα peptides and the supernatants assayed for ubiquitin ligase activity (bottom panels). The levels of Skp1, Cul1, and β-TRCP in the supernatant and associated with destruction motif peptides were determined by immunoblotting (top). β-TRCP is associated with the phosphorylated destruction motif peptides and is substantially depleted from active ubiquitin ligase fractions.
We also found that phospho-IκBα destruction motif peptides (but not the nonphosphorylatable counterparts) were able to deplete ubiquitin ligase activity from both active fractions from the phenyl–Sepharose column (Fig. 5b) and crude cell extracts (Fig. 5c). Skp1, Cul1, and β-TRCP were all associated with the phosphorylated destruction motif beads but not with the nonphosphorylatable destruction motif (Fig. 5, b, lanes 4 and 5, and c, lanes 3 and 4). Although the levels of Skp1 and Cul1 in supernatants were essentially unaffected (Fig. 5, b, lanes 1–3, and c, lanes 1 and 2), the level of β-TRCP in the supernatant from both crude and purified fractions was substantially reduced (∼80% for the crude lysate and >90% for the phenyl–Sepharose fraction) (Fig. 5b,c, lanes 1,2). These data are consistent with β-TRCP-associated Skp1 being a small fraction of the total Skp1/Cul1 complexes present in the cell (see Fig. 1b). Currently available antibodies against β-TRCP were unable to immunodeplete β-TRCP from crude lysates or purified fractions, prohibiting a direct analysis of the effects of removal of β-TRCP on IκBα–ubiquitin ligase activity. Nevertheless, the finding that depletion of SCFβ-TRCP from either crude lysates or purified fraction correlates with loss of ubiquitin ligase activity strongly implicates this SCF complex as being involved in IκBα ubiquitination.
Stimulation of IκBα ubiquitination by an SCFβ-TRCP complex in vitro
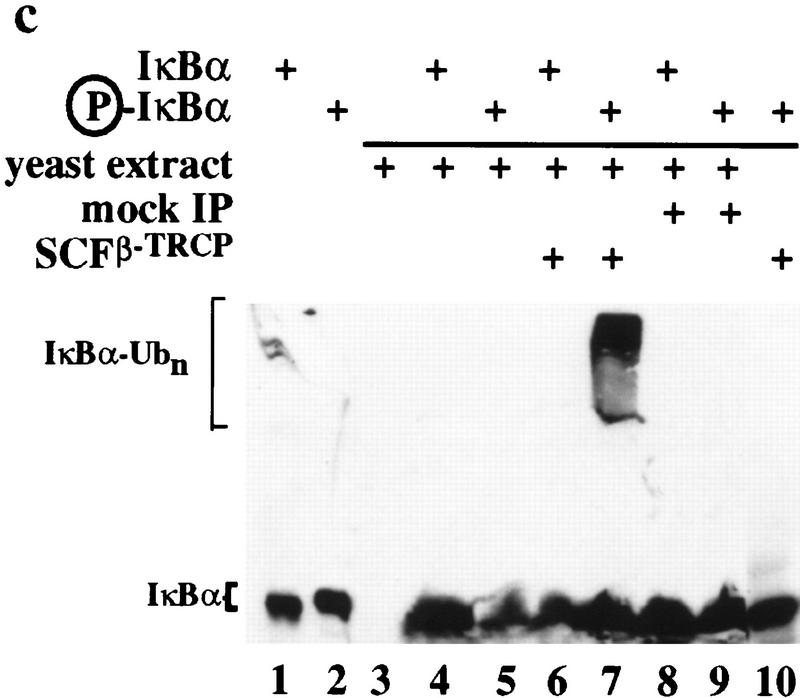
The results described thus far are consistent with a role for SCFβ-TRCP in IκBα ubiquitination. If β-TRCP functions as a specificity factor for ubiquitination of IκBα through an SCF-dependent pathway, it should be possible to confer IκBα ubiquitination activity by introducing β-TRCP into a system that lacks such an activity. Although budding yeast contains a number of E2 enzymes that could potentially support IκBα ubiquitination, its closest homolog to β-TRCP, Met30, does not associate with the IκBα destruction motif (Fig. 1c). We therefore anticipated that yeast extracts shown previously to support SCF-dependent ubiquitination of Cln2 and Sic1 (Deshaies et al. 1995; Skowyra et al. 1997; Verma et al. 1997) would be inactive toward either phosphorylated or unphosphorylated IκBα and this was the case (Fig. 6c, lanes 4,5). However, when these same reaction mixtures were supplemented with Flag-tagged SCFβ-TRCP complexes isolated from 293T cells (Fig. 6a), IκBα ubiquitination was observed (Fig. 6c, lane 7). The activity was dependent upon phosphorylation of IκBα (lane 6) and was absent in reaction mixtures containing anti-Flag immunoprecipitates from mock-transfected cells (lanes 8,9). Moreover, SCFβ-TRCP was unable to stimulate ubiquitination when mixed with E1, ATP, and ubiquitin in the absence of yeast extract (lane 10), indicating that a specific E2 activity is not efficiently immunoprecipitated with the SCFβ-TRCP complex. As expected, the active SCFβ-TRCP complexes associated with phosphorylated IκBα while control immune complex did not (Fig. 6b).
Figure 6.
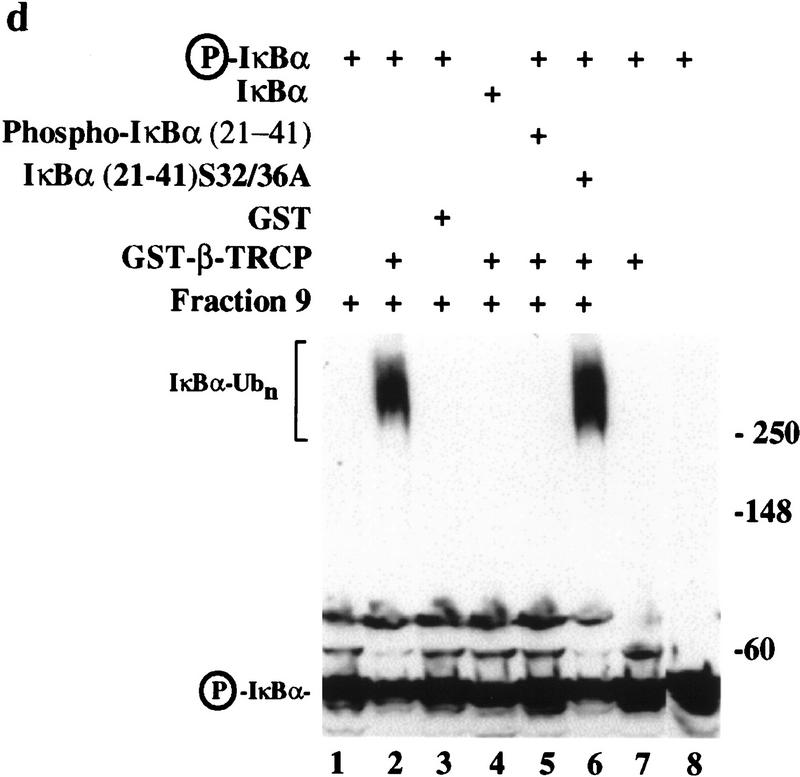
Stimulation of IκBα–ubiquitin ligase activity by SCFβ-TRCP in vitro. (a) Flag-tagged SCFβ-TRCP was prepared after transient transfection in 293T cells by immunoprecipitation along with a Flag immune complex from mock-transfected cells. Immune complexes were analyzed for the presence of Cul1HA, Skp1Myc, and β-TRCPFlag by immunoblotting (lanes 3,4). Crude lysates used for immunoprecipitation are shown as controls (lanes 1,2). (b) β-TRCPFlag immune complexes associate with phosphorylated IκBα in vitro. Immune complexes (10 μl beads) from (a) were incubated with 15 nm phosphorylated IκBα/NF-κB complexes in a total volume of 100 μl. Washed beads were subjected to SDS-PAGE and IκBα determined by immunobloting. The asterisk indicates a sample containing 15% of the input IκBα complex. (c) Stimulation of IκBα–ubiquitin ligase activity by SCFβ-TRCP in vitro. Yeast extracts (supplemented with E1, ubiquitin, and an ATP-regenerating system) were incubated with unphosphorylated or phosphorylated IκBα/NF-κB complexes (25 nm) in the presence of 10 μl of control immune complexes (lanes 8,9) or β-TRCPFlag immune complexes (lanes 6,7). After 90 min, reaction mixtures were separated by SDS-PAGE and IκBα detected by immunoblotting with anti-IκBα antibodies. As controls, untreated IκBα complexes (lanes 1,2), supplemented yeast lysates (lane 3), and an SCFβ-TRCP immune complex reaction mixture containing all components except the yeast extract (lane 10) were also included. (d) Reconstitution of IκBα ubiquitination activity in mammalian extracts by addition of purified GST–β-TRCP. Reaction mixtures, prepared as described in Materials and Methods, contained E1, ubiquitin, ATP, HQ unbound as a source of E2 activity, and other components as indicated (lanes 1–6). Control reactions (lanes 7,8) lacked phenyl–Sepharose fraction 9. After 90 min, products were analyzed by SDS-PAGE and immunoblotting with anti-IκBα antibodies.
In an alternative approach, we tested whether recombinant GST–β-TRCP, purified to near homogeneity from insect cells (data not shown), could support ubiquitination of IκBα in the mammalian ubiquitination system described in Figure 4. As noted above, fraction 9 from the phenyl–Sepharose column contains Cul1 and Skp1 but lacks detectable β-TRCP and IκBα ubiquitination activity (Fig. 6d, lane 1). However, when this fraction was supplemented with GST–β-TRCP, a potent phosphorylation-dependent IκBα–ubiquitin ligase activity was generated (Fig. 6d, lane 2). This activity was not observed when this fraction was supplemented with purified GST protein (Fig. 6d, lane 3). Also, addition of a phosphorylated IκBα destruction motif peptide (but not the unphosphorylated peptide) completely blocked IκBα ubiquitination (Fig. 6d, lanes 5,6). Finally, the activity was not observed when the GST–β-TRCP protein was incubated in the reaction conditions lacking the phenyl–Sepharose fraction, suggesting a requirement for Cul1 and Skp1 (Fig. 6d, lane 7). Taken together, these two assay systems provide compelling evidence that SCFβ-TRCP functions as an IκB–ubiquitin ligase.
Discussion
Activation of NF-κB involves an extensive signal transduction pathway that culminates in the destruction of the NF-κB inhibitor IκBα. Although the protein kinase pathways that control the timing of NF-κB activation have been defined, the molecules responsible for the actual ubiquitination events have not been elucidated. In this work, we provide biochemical evidence that the WD-40-containing F-box protein, β-TRCP, functions as a specificity factor in an SCF complex to promote signal-dependent ubiquitination of IκBα (Fig. 7). A role for β-TRCP in controlling IκBα ubiquitination is supported by the following findings. (1) Destruction of IκBα is known to require IκK-dependent phosphorylation of residues (Ser-32 and Ser-36) located in a destruction motif. β-TRCP and its SCF complex associate with this IκBα destruction motif and with the IκBα/NF-κB complex in a manner that is dependent upon phosphorylation of the IκBα destruction motif. A variety of other F-box proteins, including two other WD-40 containing F-box proteins (Met30 and MD6), failed to associate with either phosphorylated or unphosphorylated IκBα destruction motifs, pointing to the specificity of the interaction with β-TRCP. We believe that the interaction between β-TRCP and the IκBα destruction motif is direct, as peptide beads containing this motif precipitate GST–β-TRCP from insect cell lysates in the absence of other abundant proteins (J. Winston, S. Elledge, and J. Harper, unpubl.). (2) β-TRCP forms a complex with two proteins, Skp1 and Cul1, that have been linked previously to phosphorylation-dependent ubiquitination. β-TRCP is localized in the cytoplasm where IκBα ubiquitination is thought to occur. (3) β-TRCP copurifies with IκBα–ubiquitin ligase activity from tissue culture cells, and these active fractions also contain Cul1 and Skp1. (4) Depletion of β-TRCP with either anti-Skp1 antibodies or phosphorylated destruction motif peptides correlates with loss of IκBα–ubiquitin ligase activity. (5) SCFβ-TRCP complexes stimulated phosphorylation-dependent IκBα–ubiquitin ligase activity when supplemented with E1, ubiquitin, ATP, and a yeast extract. These yeast extracts lack IκBα–ubiquitin ligase despite the presence of multiple SCF complexes (Bai et al. 1996; Patton et al. 1998a,b), providing further evidence of a role for β-TRCP as a specificity factor for IκBα, but provide E2 activities and possibly other components that support IκBα ubiquitination by the β-TRCP complex. (6) Addition of β-TRCP to fractions containing Cul1 and Skp1 but lacking IκBα–ubiquitination activity leads to robust ubiquitination activity that is phosphorylation dependent and inhibited by a phosphorylated IκBα destruction motif peptide. At present, we have been unable to reconstitute IκBα–ubiquitin ligase activity using SCFβ-TRCP complexes isolated from transfected cells and column fractions depleted of β-TRCP by either anti-Skp1 antibodies or phospho-IκBα peptides. This may reflect removal of an essential factor by depletion that is not present in sufficient levels in the transiently expressed SCF complex to support IκBα ubiquitination but are provided in trans by yeast extracts or undepleted mammalian extracts. Taken together, these data provide strong evidence that SCFβ-TRCP functions in IκBα ubiquitination. After submission of this paper, Yaron et al. (1998) reported that β-TRCP is a component of the IκBα–ubiquitin ligase and demonstrated that mutants lacking the F-box stabilize IκBα and block NF-κB activation in vivo. However, no data linking β-TRCP to an SCF-dependent process was presented, and it was suggested that β-TRCP might function independently of Cul1 and Skp1. Our data provide compelling and complementary biochemical evidence that β-TRCP functions in the context of an SCF pathway, a result that has important mechanistic implications and further implicates the SCF pathway in phosphorylation-dependent ubiquitination reactions. Currently, the identity of the E2(s) involved in IκBα ubiquitination in vivo is unknown, as is the nature of the heterogeneity observed with β-TRCP. We note, however, that other F-box proteins including Skp2 are modified by phosphorylation (Lisztwan et al. 1998), and such modifications could potentially play regulatory roles. The methods we have employed offer two general approaches for determining whether a particular ubiquitination process involves an SCF complex: (1) Depletion of active fractions with Skp1 antibodies, and (2) the use of substrates as affinity reagents to examine association with cloned F-box proteins. The expanding number of F-box protein sequences available will greatly facilitate the identification of SCF-dependent processes through these types of approaches.
Figure 7.
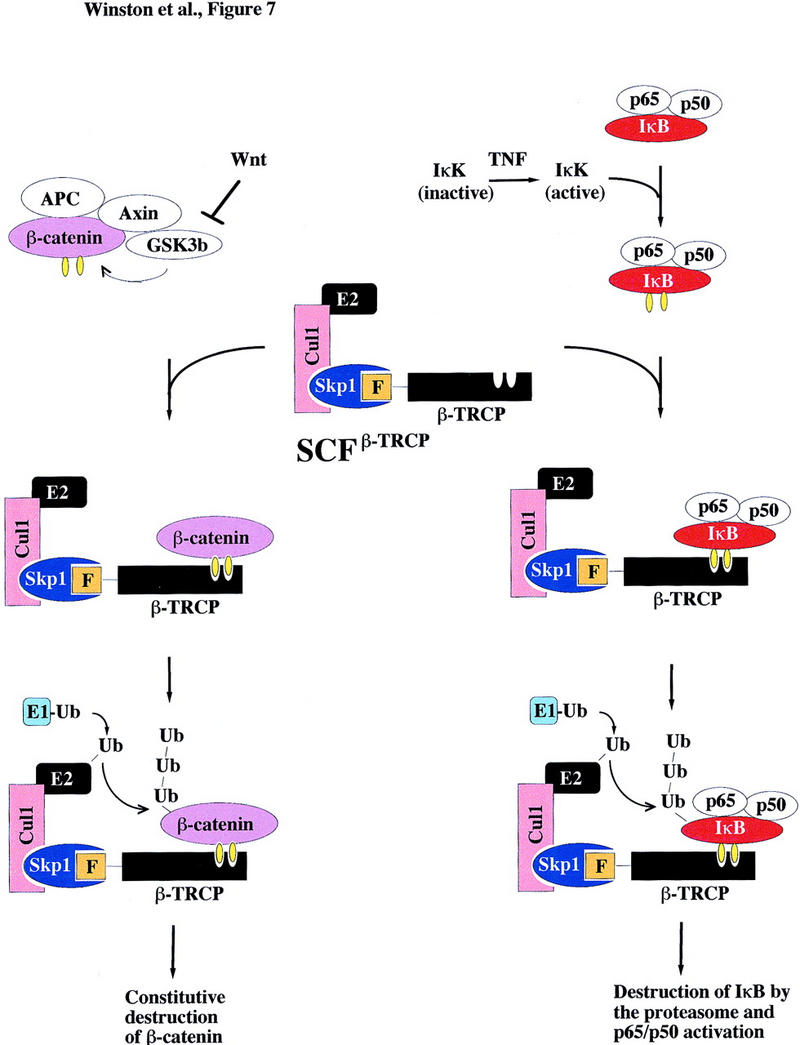
Schematic representation of the proposed pathways controlling ubiquitin-mediated proteolysis of IκBα and βcatenin. β-TRCP, an F-box protein, is a component of an SCF–ubiquitin ligase. In response to appropriate signals (i.e., TNFα), the IκK complex is activated and phosphorylates IκBα in complexes with NF-κB on Ser-32 and Ser-36. This complex is then recognized by β-TRCP in an SCF complex, facilitating ubiquitination by an E1- and E2-dependent mechanism. βCatenin, in complexes with APC, axin, and GSK3β, is phosphorylated on Ser-33 and Ser-37. This phosphorylated β-catenin can then associate with SCFβ-TRCP, resulting in ubiquitination. It is not clear at present whether β-catenin alone or the APC/β-catenin complex is the relevant target. Yellow ovals indicate phosphorylation.
The sequence conservation of the IκBα destruction motif with a region of β-catenin implicated in its turnover, coupled with a genetic requirement for the β-TRCP homolog slimb in turnover of the β-catenin homolog Armadillo (Jiang and Struhl 1998), led us to address whether β-TRCP might interact directly with β-catenin. Phosphorylation of serine residues 33 and 37 was sufficient to allow for a peptide spanning this region to associate with β-TRCP and its SCF complex but not other F-box proteins. β-Catenin is a component of the Wingless/Wnt signaling pathway and functions with Tcf/Lef transcription factors to regulate patterning and other developmental decisions (Peifer 1997). Recent work has revealed that expression of a β-TRCP protein lacking the F-box leads to accumulation of β-catenin and ectopic activation of the Wnt pathway in Xenopus (Marikawa and Elinson 1998) and β-catenin stabilization in mammalian cells (Latres et al. 1999). This, together with our data linking β-TRCP to direct recognition of the phosphorylated β-catenin destruction motif, strongly implicates SCFβ-TRCP as the β-catenin–ubiquitin ligase (Fig. 7). The levels of β-catenin are regulated by the APC (adenomatous polyposis coli) tumor suppressor protein, axin, and the protein kinase GSK3β (Korinek et al. 1997; Morin et al. 1997; Rubinfeld et al. 1997). Formation of an APC/axin/GSK3β/β-catenin complex is thought to be required to allow appropriate phosphorylation of β-catenin by GSK3β (Hart et al. 1998; Ikeda et al. 1998) and in the absence of Wnt signaling, β-catenin levels remain low due to constitutive phosphorylation and ubiquitin-mediated proteolysis. Wnt signaling inactivates GSK3β, leading to increased levels of β-catenin and activation of transcription (Peifer 1997). Mutations in either the APC gene or in β-catenin allow for β-catenin accumulation (Morin et al. 1997; Rubinfeld et al. 1997). Such mutations are found in a large fraction of colon cancers (Morin et al. 1997) and have also been observed in melanoma (Rubinfeld et al. 1997), prostate cancer (Voeller et al. 1998), and experimentally induced cancers (Dashwood et al. 1998). Interestingly, stabilizing mutations in β-catenin are localized to its destruction motif and include mutations in both the phosphoacceptor sites S33 and S37 and other residues in the consensus β-TRCP recognition motif including D32, G34, and I35. Mutations in these residues would be expected to weaken or abolish association of β-catenin with β-TRCP, leading to accumulation of β-catenin.
The role for β-TRCP in β-catenin turnover suggests that it might function as a tumor suppressor. We localized the human β-TRCP gene to 10q24. This region displays genetic abnormalities in a limited number of prostatic, melanocytic, and neural cancers (Parmiter et al. 1988; Lundgren et al. 1992; Rasheed et al. 1992). However, a preliminary analysis of four colon cancers which are wild-type for β-catenin and APC failed to reveal mutations in the β-TRCP protein (Sparks et al. 1998). Given the role for β-TRCP in IκBα ubiquitination, it is conceivable that mutations that inactivate β-TRCP are incompatible with transformation because of loss of survival pathways dependent on NF-κB activation (Beg and Baltimore 1996; Van Antwerp et al. 1996; Wang et al. 1996).
Our results indicate that an SCFβ-TRCP complex functions in two critical transcriptional control pathways, in one case by inactivating an inhibitor of transcription and in the other case by inhibiting an activator of transcription (Fig. 7). Genetic data in Drosophila suggest that the β-TRCP homolog slimb may also regulate the Hedgehog pathway (Jiang and Struhl 1998). In slimb mutants, the Ci transcription factor accumulates inappropriately, although the question of whether this regulation is direct remains to be determined. Moreover, other recent studies have revealed that β-TRCP is coopted by the HIV protein Vpu to facilitate destruction of the CD4 protein (Margottin et al. 1998). Interestingly, Vpu contains a β-TRCP recognition motif very similar to that found in IκBα and β-catenin and phosphorylation is required for it to recruit CD4 to β-TRCP and to bind β-TRCP in the two-hybrid system. Further studies are required to determine whether any of the many proteins containing the DSGφXS motif are also substrates for SCFβ-TRCP. In addition, we note that the anti-inflammatory effects of aspirin are mediated through inhibition of IκK activity (Grilli et al. 1996; Yin et al. 1998), thereby blocking NF-κB activation. Molecules that selectively block recognition of IκBα by β-TRCP may also constitute an alternative therapeutic target for anti-inflammatory agents.
Materials and methods
Plasmid and baculovirus construction
cDNAs encoding mouse and human homologs of Xenopus β-TRCP, human Skp2, and human MD6 were obtained as expressed sequence tags and the sequences determined by automated DNA sequencing. The sequence of human β-TRCP was reported previously. The sequence of mouse β-TRCP was deposited in GenBank (accession no. AF110396). The sources of other novel F-box proteins will be reported elsewhere (J. Winston, D. Koepp, S.J. Elledge, and J.W. Harper, in prep.). Open reading frames were amplified by PCR using Expand high-fidelity polymerase (Boehringer Mannheim) and cloned into the univector derivative pUNI10 as NdeI–SalI fragments. Similarly, human Skp1 was cloned into pUNI10 as NdeI–BamHI fragment. In vitro Cre-recombinase-mediated plasmid fusion (Lui et al. 1998) of univector plasmids with various pHOST recipient plasmids was then used to create plasmids for expression of proteins as amino-terminal Myc3, Flag, or Ha3 fusion proteins under control of the CMV/T7 promoter, and a GST fusion protein for expression in insect cells. Coding sequences for His6-tagged IκBα, IκK-β, p50, and p65 were amplified by PCR and baculoviruses generated using the Bac-to-Bac system (Invitrogen). Plasmids for expression of Cul1HA (Lisztwan et al. 1998) and cyclin F (Bai et al. 1996) were from previous studies. GST fusion proteins were purified from insect cells and eluted from glutathione–Sepharose beads as described previously (Skowyra et al. 1997).
Antibodies
Polyclonal antibodies against human Skp1 and Cul1 were generated in rabbits after injection of GST–Skp1 or GST–Cul1(536–815) produced in bacteria. Antisera were depleted of anti-GST reactivity using immobilized bacterial GST protein and then affinity purified using immobilized GST–Skp1 or GST–Cul1(536–815). Antibodies against the carboxyl terminus of β-TRCP were generated in rabbits using the peptide NETRSPSRTYTYISR. Antibodies were affinity purified using this peptide coupled to Affigel-10 (Bio-Rad). Monoclonal antibodies recognizing Myc, Ha, or Flag epitopes were obtained from BabCO or Sigma. Antibodies against the phosphorylated IκBα destruction motif were from New England Biolabs. All other antibodies were from Santa Cruz Biotechnology.
In vitro binding
Twenty-one (p21, residues 21–42) and 19 (p19, residues 23–41) amino acid peptides containing the IκBα destruction motif were synthesized in both the unphosphorylated and doubly phosphorylated forms where serines 32 and 36 are phosphorylated as described previously (Yaron et al. 1997). Analogous unphosphorylated and doubly phosphorylated peptides overlapping a candidate destruction motif in β-catenin were also synthesized. p21 and β-catenin peptides were coupled to Affigel (Bio-Rad) at 2 mg/ml of resin and p19 peptides were coupled to cyanogen bromide activated Sepharose (Pharmacia) at 10 mg/ml resin. Coupling efficiencies were determined to be >90% by analytical reverse-phase HPLC of reaction supernatants. To examine association of Skp1 containing complexes with destruction motifs, binding reactions were performed in a total volume of 120 μl of 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.35% NP-40, 1 mm DTT, 0.75 mm EDTA, and 5 μg/ml antipain, leupeptin, and aprotinin using 15 μl of peptide beads and the indicated quantity of cell lysate or column fraction. After incubation at 4°C for ∼1 hr, beads were washed three times with 1 ml of binding buffer [20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% NP-40, 1 mm DTT, 1 mm EDTA, and 5 μg/ml antipain, leupeptin, and aprotinin] prior to SDS-PAGE (13.5%) and immunoblotting using the indicated antibodies. Antibody detection was accomplished using ECL (Amersham). To examine association of F-box proteins with peptide beads, [35S]-methionine-labeled in vitro translation products of various F-box proteins were employed. Comparable quantities of translation products (typically 5–10 μl) were diluted to 120 μl with binding buffer prior to incubation with 15 μl of peptide beads (4°C, 1 hr). Beads were washed three times with binding buffer prior to electrophoresis and autoradiography.
In vivo interactions
To examine assembly of SCFβ-TRCP complexes in mammalian cells, 293T cells were transfected with various combinations of plasmids (1 μg each/10-cm dish) expressing Myc, Flag, or Ha-tagged Skp1, β-TRCP (mouse), and Cul1 under control of the CMV promoter using lipofectamine (GIBCO/BRL). The total DNA level was kept constant at 10 μg using empty vector DNA and each transfection contained 100 ng of pCMV–GFP DNA to facilitate determination of transfection efficiency (typically >90%). Forty hours post-transfection, cells were lysed in binding buffer supplemented with 10 mm β-glycerol phosphate, 5 mm NaF, and 1 mm p-nitrophenylphosphate (30 min on ice). After centrifugation (14,000 rpm, 20 min), lysates (1 mg/0.5 ml) were subjected to immunoprecipitation using 10 μl of anti-Myc or anti-HA immobilized on agarose beads, or used in peptide bead binding reactions as described above. Washed immune complexes were separated by SDS-PAGE and immunoblotted using the indicated antibodies. In some experiments, immune complexes from transfected cells were used for ubiquitination reactions (see below) or for binding to IκBα/NF-κB complexes. For binding experiments, immobilized SCF complexes from one 10-cm dish of transfected 293T cells (10 μl of beads) were incubated with 15 nm IκBα/NF-κB complexes in a total volume of 100 μl of binding buffer for 60 min. Beads were washed three times with 600 μl of binding buffer prior to immunoblotting for p50 and IκBα.
In vitro ubiquitination
To generate IκBα/NF-κB complexes as a substrate for ubiquitination, insect cells were coinfected with viruses expressing His6–IκBα, p50, and p65 and complexes purified on nickel–NTA resin. Proteins were eluted with imidazole and the heterotrimeric complex size-selected by gel filtration. To generate phosphorylated IκBα, the IκBα/p50/p65 complex (3 μm) was incubated with IκK-β (500 nm) purified from insect cells and 300 μm ATP, 10 mm MgCl2, 1 mm DTT, 50 mm Tris (pH 7.5) for 2 hr at 25°C. IκBα–ubiquitin ligase activity in THP.1 cell extracts and column fractions was assayed as described previously (Chen et al. 1995) using 25 nm phosphorylated or unphosphorylated IκBα/NF-κB complexes, HQ unbound (33 μg), and 200 nm E1. Proteins were separated by 4%–20% Novex SDS-PAGE, transferred to nitrocellulose, and IκBα revealed with anti-IκBα antibodies. THP.1 cell extracts were prepared by Dounce homogenization (10 strokes) in eight volumes (wt/vol) of buffer containing 50 mm Tris, 1 mm DTT, 1 mm EDTA, 1 mm EGTA, 1× Boehringer Mannheim protease inhibitor cocktail (EDTA-free) and centrifuged at 25,000g for 25 min. The filtered supernatant was fractionated on a POROS HQ column equilibrated in dialysis buffer lacking glycerol to obtain unbound fraction (HQ unbound) or precipitated with ammonium sulfate. The 30%–50% ammonium sulfate cut equivalent to 1 gram of cell paste was solubilized in 3 ml of dialysis buffer (25 mm Tris at pH 7.5, 10% glycerol, 1 mm DTT, 1 mm Benzamidine, 0.4 mm PMSF). The solubilized pellet (2 ml) was adjusted to 1.2 m ammonium sulfate, loaded to phenyl–Sepharose column (Pharmacia 5/5) equilibrated in dialysis buffer containing 1.2 m ammonium sulfate but without glycerol, and bound proteins eluted with a 20 column volume gradient 1.2–0 m ammonium sulfate, collecting two-column volume fractions followed by dialysis. To deplete IκBα–ubiquitin ligase activity from crude extracts (5 mg/ml) or phenyl–Sepharose fractions (0.7 mg/ml), p19 or pp19 beads (5 μl) were incubated with 30 μl of sample for 2 hr. Unbound material was removed for ubiquitination assays or immunoblotting and proteins associated with washed beads were subjected to SDS-PAGE and immunoblotting. Skp1 was immunodepleted by precoupling 30 μl of protein G beads (Pierce) with 1 ml of anti-Skp1 or anti-GST mouse monoclonal IgG1 antibodies (Transduction Laboratories) at 45 μg/ml for 2 hr. Washed beads were incubated with phenyl–Sepharose pools for 3 hr as indicated above.
To assay IκBα ubiquitination activity in yeast extracts, 50 μg of crude yeast extract (Deshaies et al. 1995) was supplemented with 100 ng of human E1, 200 μg of ubiquitin, 2 mm ATP (together with an ATP regenerating system), 10 μm LLNL, and 25 nm of IκBα/NF-κB complexes as substrate in a total volume of 10 μl (50 mm Tris-HCl at pH 7.5, 5 mm MgCl2, 0.6 mm DTT). Reaction mixtures were incubated with SCF immune complexes or control immune complexes from transfected 293T cells (10 μl of beads per assay) for 90 min at 30°C and products analyzed by SDS-PAGE and immunoblotting with antibodies against IκBα. We estimate that the reaction mixture contained 50–100 ng of SCF complexes. To assay IκBα ubiquitination activity in reconstituted mammalian extracts, GST–β-TRCP or GST proteins purified from insect cells (200 ng) were added to 100 μg of phenyl–Sepharose fraction 9 followed by HQ unbound (33 μg), E1 (200 ng), ubiquitin (5 μm), and ATP (2 mm) in a final volume of 15 μl (90 min). In some cases, IκBα peptides were added at 50 μg/ml.
Chromosomal localization and in situ hybridization
Genomic clones (Bacmids 171I9 and 350H4) that hybridize to the human β-TRCP cDNA were obtained from Research Genetics. The chromosomal localization of the human β-TRCP gene was determined by fluorescence in situ hybridization by the Baylor College of Medicine FISH facility. RNA expression analysis by in situ hybridization was performed as described (Parker et al. 1995) using antisense mouse β-TRCP sequences as a probe and sense sequences as a negative control (data not shown).
Acknowledgments
We are extremely grateful to Mark Rolfe at Mitotix for his efforts in coordinating this work. We thank W. Krek for a Cul1 expression plasmid; C. Wong for in situ hybridization; D. Koepp for F-box protein expression constructs; S. Glass for molecular biology; I. Chiu and A. Frederick for p50 cloning; M. Pelletier for IκK reaction optimization; and M. Caligiuri, A. Theodoras, L. Brizuela, G. Draetta, R. Copeland, K. Auger, K. Wee, and D. Skowyra for insightful discussions. This work is supported by grants from the National Institutes of Health [National Institute on Aging (NIA)], the Welch Foundation, and the Baylor Specialized Program of Research Excellence in Prostate Cancer to J.W.H and S.J.E.; and by the Dupont Pharmaceutical Company to Mitotix, Inc. J.W. is supported by a postdoctoral training grant from NIA. S.J.E. is an Investigator with the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jharper@bcm.tmc.edu; FAX (713) 796-9438.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin-proteasome pathway. Proc Natl Acad Sci. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Mathias N, Hofmann K, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulation to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Baltimore D. NF-κB: Ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- Beg AA, Baltimore D. An essential role for NF-κB in preventing TNFα-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS., Jr IκB interacts with the nuclear localization sequences of the subunits of NF-κB: A mechanism for cytoplasmic retention. Genes & Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- Beg AA, Finco TS, Nantermet PV, Baldwin ASJ. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκBα: A mechanism for NF-κB activation. Mol Cell Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Gerstberger S, Carlson L, Fransozo G, Siebenlist U. Control of IκBα proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes & Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- Cohen L, Henzel JW, Baeuerle PA. IKAP is a scaffold protein of the IkappaB kinase complex. Nature. 1998;395:292–296. doi: 10.1038/26254. [DOI] [PubMed] [Google Scholar]
- Dashwood RH, Suzui M, Nakagam H, Sugimura T, Nagao M. High frequency of β-catenin (Ctnnb1) mutations in the colon tumors induced by two heterocyclic amines in the F344 rat. Cancer Res. 1998;58:1127–1129. [PubMed] [Google Scholar]
- Deshaies RJ, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato JA, Mercurio F, Karin M. Phosphorylation of IκBα precedes but is not sufficient for its dissociation from NF-κB. Mol Cell Biol. 1995;15:1302–1311. doi: 10.1128/mcb.15.3.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- Elledge JW, Harper JW. Proteolysis in cell cycle control and cancer. Biochem Biophys Acta. 1998;1377:M61–M70. doi: 10.1016/s0304-419x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Finco TS, Beg AA, Baldwin ASJ. Inducible phosphorylation of IκBα is not sufficient for its dissociation from NF-κB and is inhibited by protease inhibitors. Proc Natl Acad Sci. 1994;91:11884–11888. doi: 10.1073/pnas.91.25.11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore TD, Koedood M, Piffat KA, White DW. Rel/NF-κB/IκB proteins and cancer. Oncogene. 1996;13:1367–1378. [PubMed] [Google Scholar]
- Grilli M, Pizzi M, Memo M, Spano P. Neuroprotection by aspirin and sodium salicylate through blockade of NF-kappa B activation. Science. 1996;274:1383–1385. doi: 10.1126/science.274.5291.1383. [DOI] [PubMed] [Google Scholar]
- Hart MJ, de los Santos R, Albert I, Rubinfeld B, Polakis P. Downregulation of β-catenin by human Axin and its assocation with the APC tumor suppressor, β-catenin and GSK3β. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of the ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- ————— Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- ————— There’s the rub: A novel ubiquitin-like modification linked to cell cycle regulation. Genes & Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the hedgehog and wingless pathways by the F-box/WD40-repeat protein slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Nature. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Latres, E., D.S. Chiaur, and M. Pagano. 1999. The human F box protein β-TRCP associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene (in press). [DOI] [PubMed]
- Lin Y-C, Brown K, Siebenlist U. Activation of NF-κB requires proteolysis of the inhibitor IκB-α: Signal-induced phosphorylation of IκBα alone does not release active NF-κB. Proc Natl Acad Sci. 1995;92:552–556. doi: 10.1073/pnas.92.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): Evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-G, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- Liu Q, Li MZ, Leibman D, Cortez D, Elledge SJ. The univector plasmid fusion system, a method for rapid construction of recombinant DNA molecules without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- Lundgren R, Mandahl N, Heim S, Limon J, Henrikson H, Mitelman F. Cytogenetic analysis of 57 primary prostatic adenocarcinomas. Genes Chromosomes Cancer. 1992;4:16–24. doi: 10.1002/gcc.2870040103. [DOI] [PubMed] [Google Scholar]
- Lyapina SA, Correll CC, Kipreos ET, Deshaies RJ. Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc Natl Acad Sci. 1998;95:7451–7456. doi: 10.1073/pnas.95.13.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD40 protein, h-βTrCP, that interacts with HIV-1 vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- Marikawa Y, Elinson RP. β-TRCP is a negative regulator of Wnt/β-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech Dev. 1998;77:75–80. doi: 10.1016/s0925-4773(98)00134-8. [DOI] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M. IKK-1 and IKK-2: Cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Michel JJ, Xiong Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:435–449. [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Nature. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Parmiter AH, Balaban G, Clark WH, Jr, Nowell PC. Possible involvement of the chromosome region 10q24—q26 in early stages of melanocytic neoplasia. Cancer Genet Cytogenet. 1988;30:313–317. doi: 10.1016/0165-4608(88)90200-2. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes & Dev. 1998a;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Genet. 1998b;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, Olson EN, Harper JW, Elledge SJ. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- Peifer M. β-Catenin as oncogene: The smoking gun. Science. 1997;275:1752–1753. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- Rasheed BK, Fuller GN, Friedman AH, Bigner DD, Bigner SH. Loss of heterozygosity for 10q loci in human gliomas. Genes Chromosomes Cancer. 1992;5:75–82. doi: 10.1002/gcc.2870050111. [DOI] [PubMed] [Google Scholar]
- Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- Rolfe M, Beer-Romero P, Glass S, Eckstein J, Berdo I, Theodoras A, Pagano M, Draetta G. Reconstitution of p53-ubiquitination reactions from purified components: The role of human ubiquitin-conjugating enzyme UBC4 and E6-associated protein (E6AP) Proc Natl Acad Sci. 1995;92:3264–3268. doi: 10.1073/pnas.92.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1793. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Scherer DC, Brockman JA, Chen Z, Maniatis T, Ballard DW. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig K, Tyers M, Elledge SJ, Harper JW. F-box proteins are components of E3 complexes and act as receptors to recruit phosphorylated substrates for ubiquitination. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Vogelstein B, Kinzler K. No mutations of the slimb homolog, beta-TRCP, in colorectal cancer. Neg Obser Gen Oncol. 1998;2:23. [Google Scholar]
- Spevak W, Keiper BD, Stratowa C, Castanon MJ. Saccharomyces cerevisiae cdc15 mutants arrested in a late stage in anaphase are rescued by Xenopus cDNAs encoding N-ras or a protein with beta-transducin repeats. Mol Cell Biol. 1993;13:4953–4966. doi: 10.1128/mcb.13.8.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNFα-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- Verma R, Feldman RM, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeller HJ, Truica CI, Gelmann EP. β-Catenin mutations in human prostate cancer. Cancer Res. 1998;58:2520–2523. [PubMed] [Google Scholar]
- Wang C-Y, Mayo MW, Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: Potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- Wulczyn FG, Krappmann D, Scheidereit C. Signal-dependent degradation of IkappaBalpha is mediated by an inducible destruction box that can be transferred to NF-kappaB, bcl-3, or p53. Nucleic Acids Res. 1998;26:1724–1730. doi: 10.1093/nar/26.7.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, Kirk HE, Kay RJ, Israel A. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- Yaron A, Gonen H, Alkalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning AM, Ciechanover A, Ben-Neriah Y. Inhibition of NF-κB cellular function via specific targeting of the IκB-ubiquitin ligase. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A, Hatzubal A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IkBa-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Yin MJ, Yamamoto Y, Gaynor RB. The anti-inflammatory agents aspirin and salicylate inhibit the activity of I(kappa)B kinase-beta. Nature. 1998;396:77–80. doi: 10.1038/23948. [DOI] [PubMed] [Google Scholar]
- Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- Zandi E, Chen Y, Karin M. Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: Discrimination between free and NF-kappaB-bound substrate. Science. 1998;281:1360–1363. doi: 10.1126/science.281.5381.1360. [DOI] [PubMed] [Google Scholar]