Abstract
Sister chromatid cohesion is crucial for chromosome segregation during mitosis. Loss of cohesion very possibly triggers sister separation at the metaphase → anaphase transition. This process depends on the destruction of anaphase inhibitory proteins like Pds1p (Cut2p), which is thought to liberate a sister-separating protein Esp1p (Cut1p). By looking for mutants that separate sister centromeres in the presence of Pds1p, this and a previous study have identified six proteins essential for establishing or maintaining sister chromatid cohesion. Four of these proteins, Scc1p, Scc3p, Smc1p, and Smc3p, are subunits of a ‘Cohesin’ complex that binds chromosomes from late G1 until the onset of anaphase. The fifth protein, Scc2p, is not a stoichiometric Cohesin subunit but it is required for Cohesin’s association with chromosomes. The sixth protein, Eco1p(Ctf7p), is not a Cohesin subunit. It is necessary for the establishment of cohesion during DNA replication but not for its maintenance during G2 and M phases.
Keywords: ECO1, Cohesin complex, SCC2, SCC3, establishment of cohesion, sister chromatid separation
Sister chromatids move away from each other at the metaphase → anaphase transition because their kinetochores attach to microtubules emanating from opposite spindle poles (Rieder and Salmon 1998). Chromosomes are not mere passengers during this process. During the early stages of mitosis, the tendency of microtubules to pull sister chromatids apart is counteracted by cohesion that holds them together. Therefore, cohesion between sisters generates the tension by which cells align sister chromatids on the metaphase plate (Nicklas 1988). Cohesion between sisters appears to exist along their entire length during metaphase; that is, it is not confined to centromeres (Selig et al. 1992; Guacci et al. 1994). Sister chromatid cohesion is an essential aspect of mitosis. Were sisters to separate before spindle formation, it is difficult to imagine how cells could distinguish sisters from chromatids that were merely homologous. Cohesion also prevents chromosomes from falling apart as a result of double strand breaks and it presumably facilitates their recombination-mediated repair. A sudden loss of cohesion, rather than an increase in the traction exerted by microtubules attached to kinetochores, is thought to trigger sister separation during anaphase (Miyazaki and Orr-Weaver 1994).
Until recently, little was known about the nature of sister chromatid cohesion or about the mechanism by which it is destroyed. Recent work has shown that sister separation depends on the destruction, by ubiquitin-mediated proteolysis, of anaphase inhibitors like budding yeast Pds1p and fission yeast Cut2p (Securins), which bind to and inhibit the Esp1/Cut1 class of sister separating proteins (Separins) (Funabiki et al. 1996; Ciosk et al. 1998). The ubiquitin protein ligase that mediates Securin proteolysis is a large multisubunit complex called anaphase promoting complex (APC) (Irniger et al. 1995; Cohen-Fix et al. 1996; Funabiki et al. 1996; Zachariae et al. 1998). In Saccharomyces cerevisiae, Esp1p is necessary for the disappearance from chromosomes of Scc1p, a protein that is required for sister chromatid cohesion (Michaelis et al. 1997; Ciosk et al. 1998). It has therefore been suggested that Esp1p’s role is to destroy the bridges, or links, that hold sister chromatids together.
Scc1p (also known as Mcd1p) is one of four proteins currently known to be required for sister chromatid cohesion in mitotic cells (Guacci et al. 1997; Michaelis et al. 1997). The others are Scc2p and two members of the Smc family, Smc1p and Smc3p (Michaelis et al. 1997). Proteins belonging to the Smc family also exist in bacteria, which suggests that Smc proteins are modulators of chromosome structure that must have existed in the common ancestor of all living organisms. Members of the Smc protein family have been implicated in chromosome condensation, dosage compensation, and in DNA repair (Jessberger et al. 1998). Two pieces of evidence suggest that Scc1p binds to chromosomes as part of a Cohesin complex with Smc1p and Smc3p. First, Scc1p’s association with chromosomes depends on Smc1p (Michaelis et al. 1997). Second, the Xenopus homolog of Scc1p is found in a soluble multisubunit complex that contains Smc1p, Smc3p, and two other proteins (Losada et al. 1998). It is not yet known whether Cohesin participates in the links that actually hold sister chromatids together. It is known, however, that establishment of cohesion depends on the presence of Scc1p during DNA replication, which suggests that links might be produced only when sister chromatids are in close proximity immediately after passage of replication forks (Uhlmann and Nasmyth 1998). Mis4p (Scc2p’s homolog in Schizosaccharomyces pombe) must also be active during S phase (Furuya et al. 1998).
Because our previous search for mutants defective in sister chromatid cohesion unearthed only four different genes (Michaelis et al. 1997), we have improved and repeated our screen and isolated 29 new mutants that separate sister chromatids without first having destroyed the anaphase inhibitor Pds1p. By this means we have identified two new genes required for sister chromatid cohesion, which we call SCC3 (sister chromatid Cohesion) and ECO1 (Establishment of Cohesion). We also isolated many new alleles of scc1, scc2, and smc3. We show here that Scc1p, Scc3p, Smc1p, and Smc3p are subunits of a Cohesin complex that binds chromosomes from late G1 until the metaphase → anaphase transition. We also show that Scc2p, although not a stoichiometric Cohesin subunit, is nevertheless required for the association of Cohesin with chromosomes. Finally, we show that Eco1p is neither a Cohesin subunit nor required for Cohesin’s association with chromosomes. It is required for the establishment of cohesion during DNA replication but not for maintaining cohesion during G2 and M phases. We suggest that Eco1p might catalyze the formation of links between sisters as they emerge from replication forks. Homologs of all six cohesion proteins exist in most, if not all eukaryotes, suggesting that the mechanisms by which sister cohesion is established in yeast will prove to be conserved in humans.
Results
SCC3 and ECO1: two new genes required for sister chromatid cohesion
To isolate new mutants defective in sister chromatid cohesion, we used a modified version of the screen described by Michaelis et al. (1997). Because sister chromatid cohesion is required for faithful segregation of sister chromatids, partial defects in cohesion should cause cells to lose chromosomes at a high frequency, whereas a complete defect should be lethal and cause sister separation even in the presence of Pds1p. Therefore, we screened for temperature-sensitive mutants that lose a supernumerary chromosome with high frequency at 25°C, cannot proliferate at 37°C, and separate sister chromatids even when Pds1p proteolysis is blocked by a mutation in an APC subunit. The parental strain contained the ochre-suppressible ade2-1 mutation, which causes cells to accumulate a red pigment, and a supernumerary marker chromosome carrying SUP11, which suppresses ade2-1 (Spencer et al. 1990). The parental strain, therefore, forms white colonies, whereas mutants with high rates of chromosome loss form red–white-sectored colonies. To inactivate the APC conditionally in this strain, we deleted CDC26 (which encodes an APC subunit that is only essential at 37°C) and integrated a functional copy along with CAN1 at the lys2 locus. The integration created direct repeats of LYS2 DNA-flanking CAN1 and CDC26 genes, which enabled us to loop out the CDC26 gene from the genome upon selection against the CAN1 gene and thereby create strains with a thermolabile APC. Sister chromatid separation was followed by visualizing tetracycline repressor–GFP (green fluorescent protein) fusion proteins bound to tandem repeats of tet operators integrated close to the centromere of chromosome V (Michaelis et al. 1997). Fewer than 5% of Δcdc26 cells separate sisters upon shift to 37°C but known cohesion mutations, such as scc1-73, increase this to 65%.
Of 1675 ‘sectoring’ mutants generated by EMS mutagenesis, 360 were temperature sensitive for growth, and of these 96 separated sisters in ⩾25% of the cells at 37°C upon deletion of CDC26. Tetrad analyses of progeny from crosses between mutant and parental strains showed that chromosome loss at 25°C, lethality at 37°C, and sister chromatid cohesion defects in the absence of CDC26 at 37°C were tightly linked in 29 mutant strains. Complementation analysis showed that the 29 mutations were all recessive and were located in six different genes. Most of the mutations were in genes previously known to be involved in sister chromatid cohesion. Six mutants failed to complement scc1-73, 12 failed to complement scc2-4, 3 failed to complement smc3-42, and 3 failed to complement a pds1 deletion strain. The remaining two genes were cloned by rescuing the temperature-sensitive lethality of mutants with plasmids containing a library of yeast genomic DNA fragments. In both cases, the mutants were rescued by a single ORF, which was shown subsequently to be tightly linked to the mutations causing temperature sensitive lethality. Four mutations were located in a gene called IRR1, which was known to have an essential but not understood function (Kurlandzka et al. 1995). Because of its role in sister chromatid cohesion, we renamed this gene SCC3. It encodes a protein with a predicted molecular mass of 133 kD. S. pombe contains at least two genes that encode homologous proteins: a closely related protein whose function is unknown and a less related protein called Rec11p, which is required for recombination during meiosis (DeVeaux and Smith 1994). The amino-terminal half of Scc3p is 18%–25% identical to members of the SA (stromal antigen) protein family (Carramolino et al. 1997) from Caenorhabditis elegans, Drosophila melanogaster, and from mammals (Fig. 1A,B).
Figure 1.
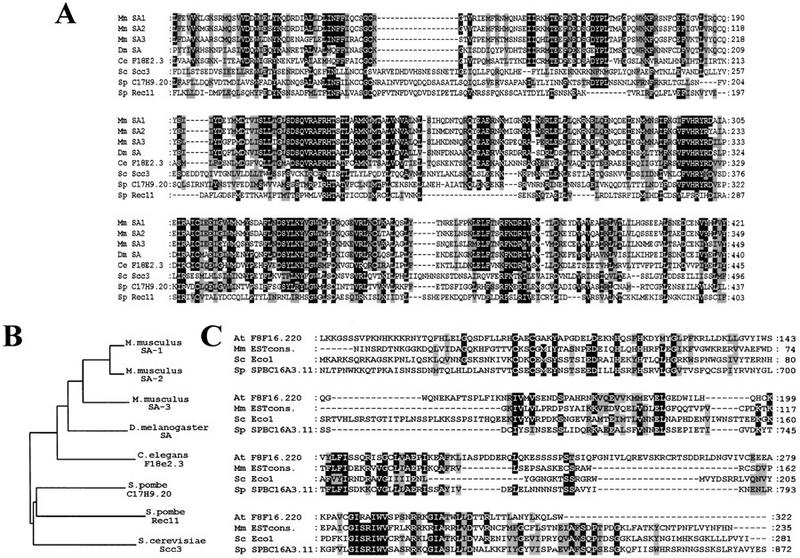
Scc3p and Eco1p are conserved among eukaryotes. (A) Sequence alignment of the amino-terminal half of S. cerevisiae (Sc) Scc3p and its homologs from mouse (Mm SA-1, Mm SA-2, Mm SA-3), D. melanogaster (Dm SA), C. elegans (Ce F18e2.3), and S. pombe (Sp C17H9.2, Sp Rec11). Identical residues in proteins are shown on a black background. Conservative amino acid substitutions are shown on a gray background. Because the human SA1 and SA2 are almost identical to the corresponding mouse proteins, we did not include the human proteins in this alignment. (B) Phylogenetic tree of the Scc3p homologs based on the above aligned sequences. (C) Sequence alignment of S. cerevisiae Eco1p (Sc) and its homologs from A. thaliana (At F8F16.220), mouse (Mm ESTcons.), fission yeast (Sp SPBC16A3.11).
The second gene was a hypothetical ORF located on chromosome VI. Because of its role in establishing cohesion between sister chromatids (see below), we called this gene ECO1. The predicted Eco1 protein sequence shares homology over its entire length with putative proteins from S. pombe, Arabidopsis thaliana, Mus musculus, and humans (Fig.1C). Eco1p’s amino-terminal half contains a conserved Kruppel-like C2H2 zinc finger domain. The S. pombe homolog of Eco1p is ∼650 amino acids longer than the S. cerevisiae protein. Although the amino-terminal 600 amino acid of the fission yeast gene is not homologous to ECO1, it is strongly related to the RAD30 gene in S. cerevisiae. RAD30 functions in a postreplication repair mechanism and it is related to the Escherichia coli, dinB and umuC DNA repair genes (McDonald et al. 1997). We isolated only a single mutant allele of ECO1, in which a conserved glycine at position 211 is replaced by aspartic acid. Tetrad analysis of progeny from a diploid strain heterozygous for an ECO1 deletion showed it to be essential for growth at all temperatures.
Scc3p and Eco1p are required to prevent premature sister chromatid separation
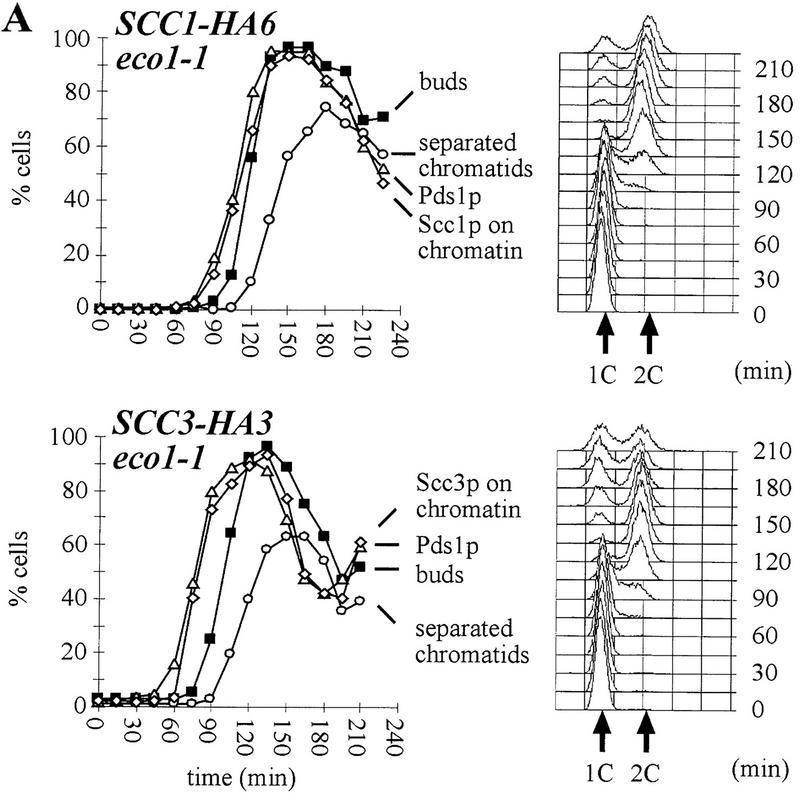
We identified the SCC3 and ECO1 genes because mutations in them affect sister chromatid cohesion during the metaphase arrest attributable to deletion of CDC26. To address whether these genes are required to prevent premature sister separation during normal cell cycles, we followed the separation of GFP-marked chromosome V centromeres (CenV–GFP) in synchronized cultures of eco1-1, scc3-1, and wild-type strains. Small G1 cells obtained by centrifugal elutriation were incubated at 37°C in the presence or absence of nocodazole. Samples were collected every 15 min and were analyzed for DNA content, budding, separation of sister CenV sequences, and the presence of Pds1p (Fig. 2A). The timing of sister separation in wild-type and mutant cells was compared as described by Michaelis et al. (1997). Whereas wild-type cells separate CenV sequences 45 min after budding and replication, eco1-1 and scc3-1 mutants do so after 30 min Small budded wild-type cells never separate sister chromatids but eco1-1 and scc3-1 mutant cells frequently do so (Fig. 2B). In wild-type, sister CenV sequences never separate before the disappearance of Pds1p from nuclei. Both degradation of Pds1p and cytokinesis are delayed in eco1 and scc3 mutants and sister separation largely occurs in the presence of Pds1p (Fig. 2). The delayed Pds1p destruction might be attributable to surveillance mechanisms that monitor either integrity of the genome or the mitotic spindle (Hoyt et al. 1991; Li and Murray 1991).
Figure 2.
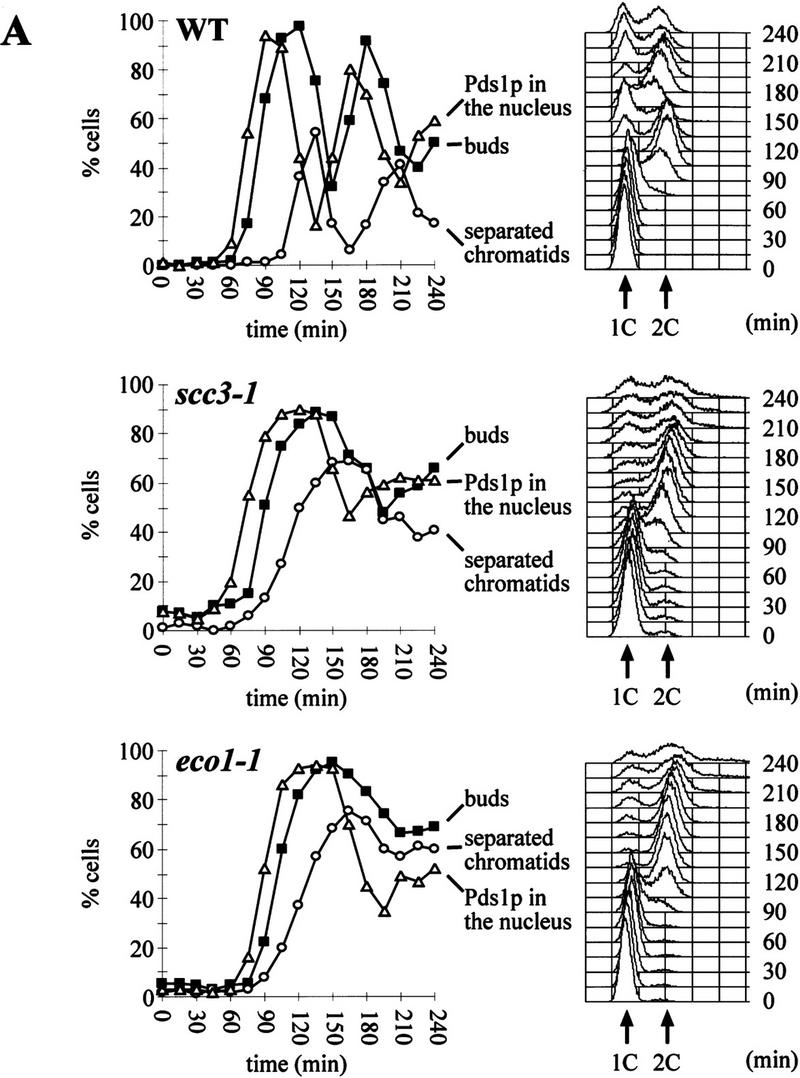
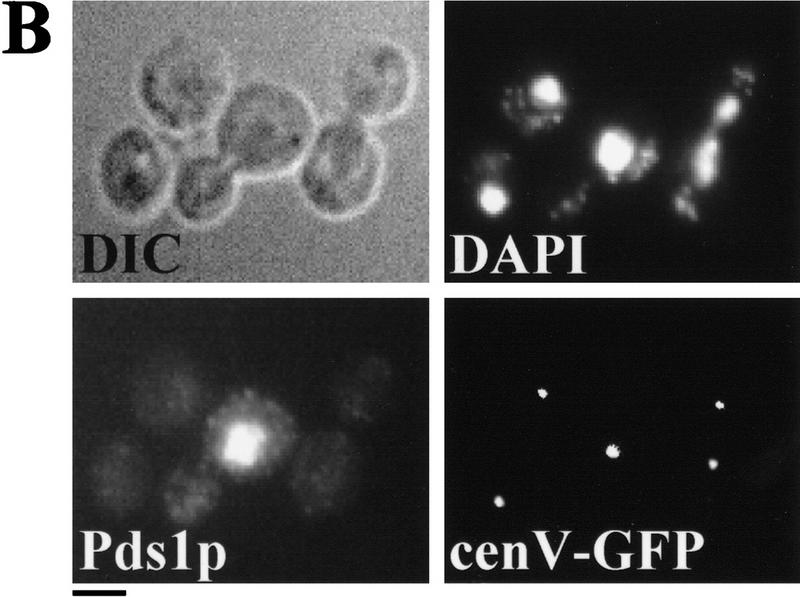
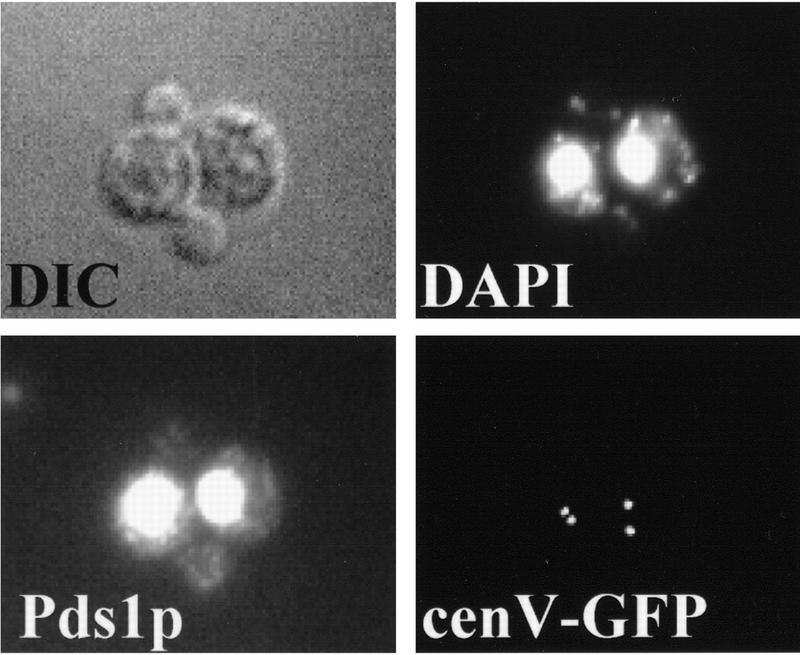
Sister chromatids separate prematurely in scc3-1 and eco1-1 mutants. Small G1 cells of wild-type (K7479), scc3-1 (K7515), and eco1-1 (K7542) strains expressing Pds1–myc18p and with CenV marked by GFP, were isolated by centrifugal elutriation and incubated at 37°C. (A, left) The fraction of budded cells (█), cells that have separated their cenV–GFP ‘dots’ (○) and cells with Pds1p in their nucleus (▵). (A, right) DNA content measured by flow cytometry (FACS). In wild-type cells, degradation of Pds1p always precedes sister chromatid separation. In scc3-1 and eco1-1 mutant cells sister chromatids are separated even in the presence of Pds1p. (B) Immunofluorescence of wild-type (top) and eco1-1 (bottom) cells. DNA was stained by DAPI. Pds1p was detected by antibody to the myc epitope. CenV was visualized by GFP. Bar, 4 μm.
Separated CenV sequences are rarely seen in the same half of budded wild-type cells because sister chromatids move rapidly to opposite poles once cohesion between sisters has been destroyed. In contrast, sister centromeres were frequently found separated but close to each other in the same half of eco1-1 and scc3-1 mutant cells at a time when they should have segregated to opposite poles. Furthermore, when mutant cells eventually undergo cytokinesis at 37°C 20%–30% of unbudded progeny contained either two or no CenV–GFP dots. In conclusion, eco1 and scc3 mutants not only separate sister CenV sequences prematurely but also mis-segregate them with a very high frequency, which would explain their lethality at 37°C.
Sister chromatid separation, cytokinesis, and DNA rereplication were all blocked by nocodazole in wild-type. Nocodazole also blocked cytokinesis and rereplication in eco1 and scc3 mutant cells, but despite this, sister centromeres separated in 50%–60% of the cells within 60 min of their having budded at 37°C (data not shown).
Premature sister separation in eco1 and scc3 mutants is independent of Esp1p
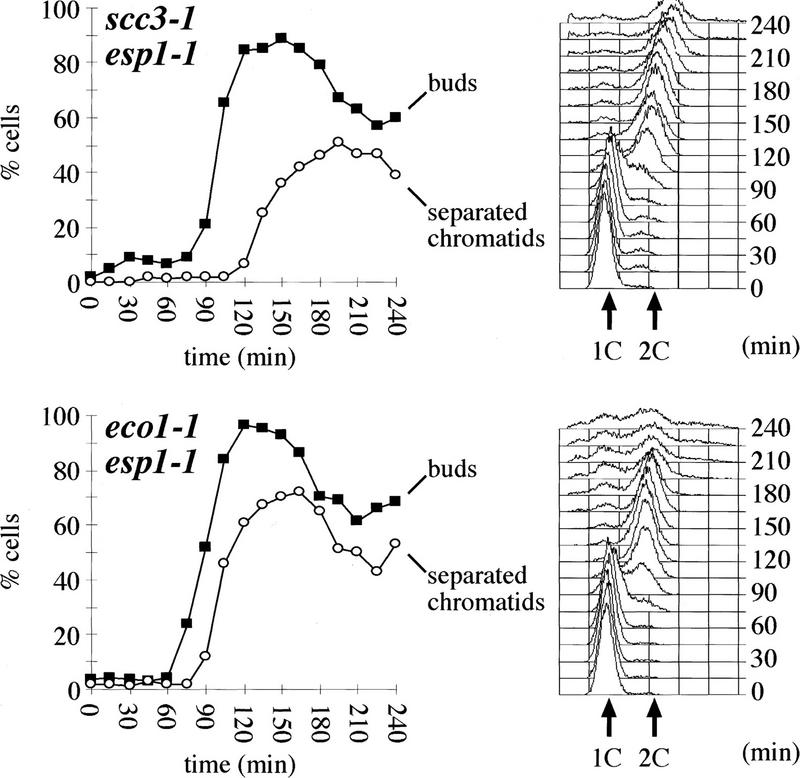
Sister separation in the presence of Pds1p in scc3-1 and eco1-1 mutant cells could be attributable either to resistance of Esp1p to Pds1 inhibition in the mutants or to separation in the absence of Esp1 activity. To distinguish between these two possibilities, we compared the kinetics of sister separation in esp1-1 single mutants with that in scc3-1 esp1-1 and eco1-1 esp1-1 double mutants at 37°C (Fig. 3). Whereas esp1-1 cells separate sisters very inefficiently (Ciosk et al. 1998), both double mutants separated sisters with similar kinetics to eco1 and scc3 single mutants. Thus, premature sister separation in eco1 and scc3 mutants is independent of Esp1 function. This suggests that Scc3p and Eco1p, unlike Pds1p, are not regulators of Esp1p. They are likely to be directly involved in sister chromatid cohesion.
Figure 3.
Scc3p and Eco1p are required to prevent sister chromatid separation in esp1-1 mutants. Small unbudded cells of scc3-1 esp1-1 (K7546) and of eco1-1 esp1-1 (K7544) strains containing cenV–GFP were inoculated into YEPD medium at 37°C. Samples were taken every 15 min and the fraction of cells with buds (█) and cells with separated sister chromatids (○) were determined. The control experiment had been published by Ciosk et al. (1998).
Scc3p and Eco1p are nuclear proteins that associate with chromatin
To detect Scc3p and Eco1p, we tagged the endogenous genes either with 18 Myc epitopes or with 3 HA epitopes at their carboxyl termini. Using Western blotting, we measured the level of Eco1–myc18p as unbudded G1 cells isolated by elutriation progressed through the cell cycle. Its abundance fluctuated only modestly, accumulating to maximal levels during S phase (data not shown). In situ immunofluorescence showed that Eco1–myc18p accumulated in the nuclei and chromosome spreads showed that Eco1–myc18p associated with chromatin (data not shown). We also measured the chromosome binding of Eco1p using cell fractionation (Liang and Stillman 1997). After lysis of spheroplasts with Triton X-100, cell lysates were fractionated by centrifugation into a ‘chromatin’ pellet containing most if not all of the DNA and a supernatant containing ⩾95% of cellular protein. Proteins from both fractions were separated on SDS-PAGE. Western blot analyses showed that most of the Eco1–HA3p was tightly associated with chromatin in cycling cells (Fig. 4A).
Figure 4.
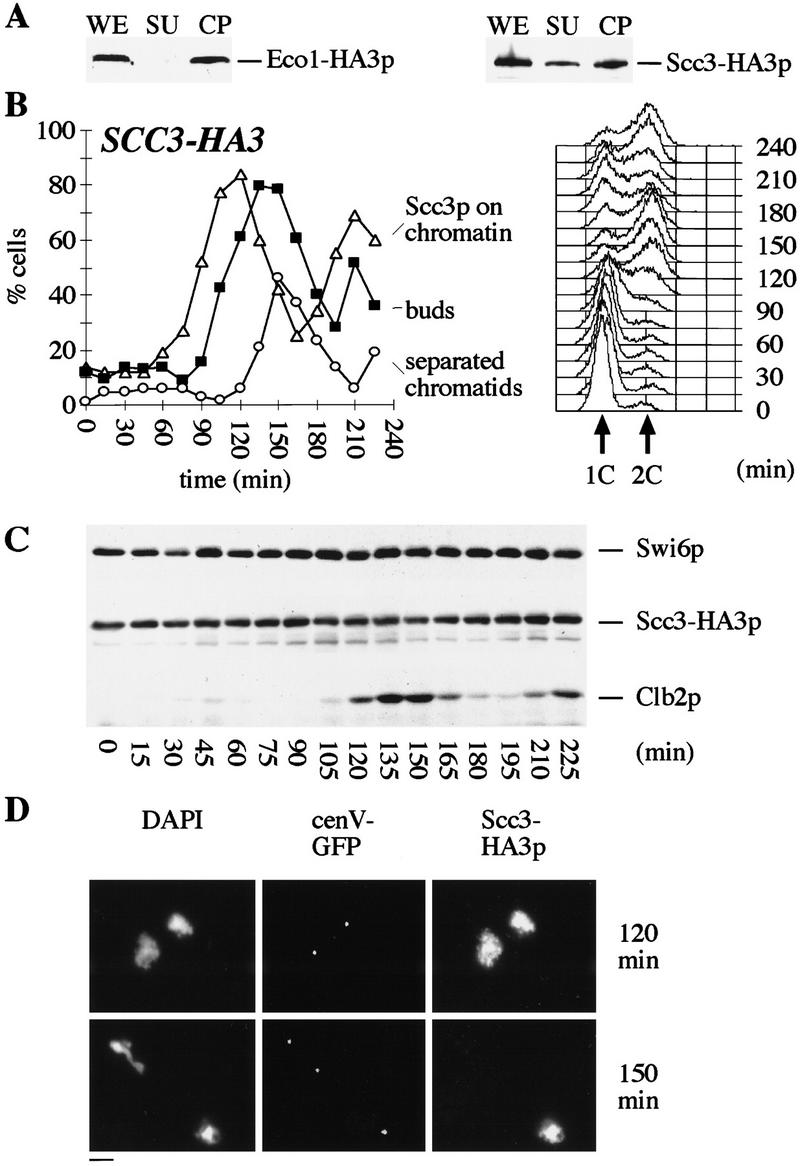
Scc3p and Eco1p are associated with chromatin. (A) Triton X-100 insoluble chromatin pellet was obtained from cycling cultures of ECO1–HA3 and SCC3–HA3 strains as described (Liang and Stillman 1997). Eco1–HA3p and Scc3–HA3p were detected with anti-HA antibody (16B12). (WE) whole cell extract; (SU) Triton X-100 soluble supernatant; (CP) Triton X-100 insoluble chromatin pellet. (B) Small G1 cells expressing Scc3–HA3p (K7461) containing cenV–GFP were incubated at 25°C. DNA content of cells, percentage of budded cells (█), cells with separated sister chromatids (○), and the percentage of cells with Scc3p associated to chromosomes (▵) were determined. (C) Scc3–HA3p, Clb2p, and Swi6p levels were determined by Western blot analysis of protein extracts. (D) Chromosome spreads from 120- and 135-min time points. DNA was stained by DAPI. Scc3–HA3p was detected by antibody to HA epitopes. CenV was visualized by GFP. Scc3p colocalizes with chromatin until the onset of anaphase (top panel). Scc3p association is largely reduced in cells that separated sister chromatids (bottom panel, top left corner). Bar, 4 μm.
Cell fractionation suggested that the bulk of Scc3–HA3p is associated with chromatin in cycling cultures (Fig. 4A). We used chromosome spreading to analyze the association of Scc3–HA3p with chromatin as unbudded G1 cells isolated by elutriation progressed through the cell cycle (Fig. 4B). Indirect immunofluorescence showed that Scc3–HA3p was located within the nuclei throughout the cell cycle (data not shown). Although the abundance of Scc3–HA3p remained roughly constant (Fig. 4C), the amount associated with chromatin varied greatly at different cell cycle stages. Scc3p was largely absent from chromosomes during early G1, but was tightly associated with them from late G1 until metaphase (Fig. 4D). The amount of Scc3p associated with chromosomes dropped dramatically as soon as sister centromeres separated. Thus, there was little or no Scc3p associated with chromosomes in early anaphase cells containing two CenV–GFP dots (Fig. 4D). This pattern of chromatin association resembles that of the cohesin Scc1p.
Scc1p and Scc3p form a stable ‘Cohesin’ complex with Smc1p and Smc3p
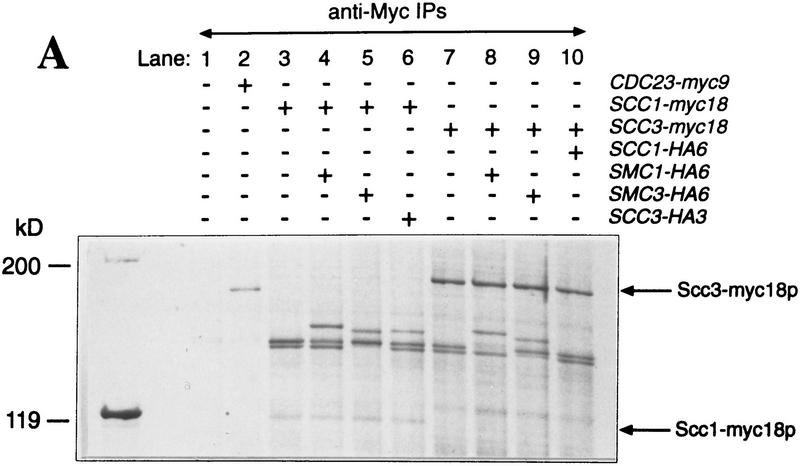
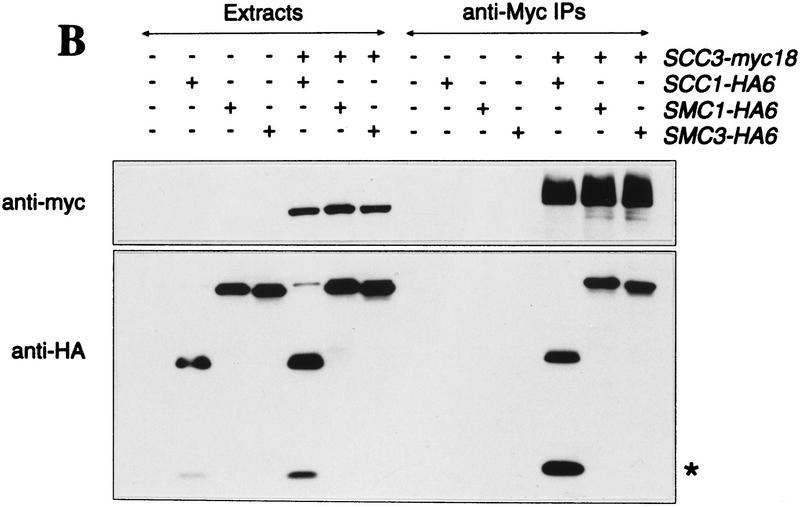
The similarities between Scc1p’s and Scc3p’s cell cycle dependent association with chromatin and the dependence of the former on Smc1p (Michaelis et al. 1997) suggests that these proteins might form a complex together. To investigate this, we constructed haploid strains expressing Myc epitope-tagged versions of either Scc1p or Scc3p in combination with HA epitope-tagged versions of Scc1p, Scc2p, Scc3p, Smc1p, Smc2p, Smc3p, and Eco1p. The Myc-tagged proteins were immunoprecipitated from whole cell extracts, the precipitated proteins were separated by SDS-PAGE, and then detected by silver staining. Scc1–myc18p immunoprecipitates contained at least three additional high molecular weight proteins, which were identified as Smc1p, Smc3p, and Scc3p attributable to changes in their mobility when HA tagged (Fig. 5A, lanes 3–6). Likewise, we detected Smc1p and Smc3p in Scc3–myc18p immunoprecipitates. We could not, however, detect Scc1p, presumably because it is not very agrophilic (Fig. 5A, lanes 7–10). Precipitation of Scc1p, Scc3p, and their associated proteins was myc tag dependent (Fig. 5A, lane 1). Furthermore, none of these proteins were present in Cdc23–myc9p immunopecipitates (Fig. 5A, lane 2). Western blotting of Scc3–myc18p immunoprecipitates confirmed that Smc1–HA6p, Smc3–HA6p, and Scc1–HA6p coprecipitate with Scc3–myc18p (Fig. 5B). A similar analysis showed that Scc1–myc18p associates with Scc3–HA3p, Smc1–HA6p, and Smc3–HA6p (data not shown).
Figure 5.
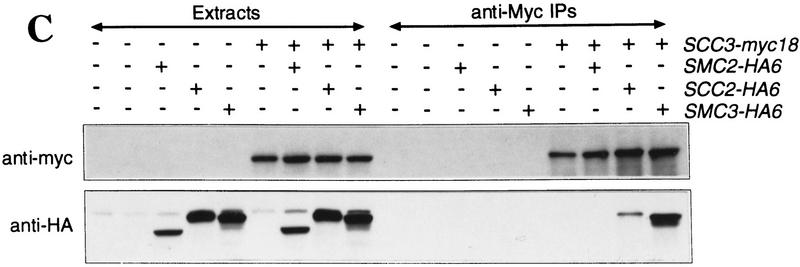
Physical associations between proteins involved in sister chromatid cohesion. Protein extracts were prepared from cells of the indicated genotypes. (+) An epitope-tagged gene; (−) a wild-type allele. Myc-tagged proteins were immunoprecipitated with anti-myc mouse mAb 9E10. Myc-tagged and HA-tagged proteins were detected by immunoblotting, using anti-Myc 9E10 or anti-HA mouse mAb 12CA5, respectively. Protein extracts or immunoprecipitates were separated on 7.5% SDS–polyacrylamide gels and either stained with silver (A) or immunobloted with antibodies (B,C). (A) Scc1/3 and Smc1/3 proteins form a soluble complex. Protein extracts of control cells (no tag, lane 1), cells expressing a myc9-tagged version of Cdc23p (lane 2), and cells expressing myc18-tagged versions of either Scc1p or Scc3p (lanes 3–10), were immunoprecipitated with an antibody specific for the Myc epitope. Proteins that were present in immunoprecipitates were visu alized by staining with silver. Only those proteins that specifically associated with Scc1–myc18p and Scc3–myc18p are shown. Proteins that were specifically present in Scc1–myc18p (lanes 3–6) and Scc3–myc18p precipitates (lanes 7–10) were identified by reducing electrophoretic mobility of candidate proteins by their HA tagging. Scc1–myc18p and Scc3–myc18p are indicated with arrows. Scc1p with no tag is not detectable with silver under these conditions. (B) Scc3–myc18p precipitates Scc1–HA6p, Smc1–HA6p, and Smc3–HA6p. (*) A degradation product of Scc1–HA6p. (C) A small fraction of Scc2–HA6p, but not Smc2–HA6p, is present in Scc3–myc18p precipitate.
We then investigated the association of Scc3–myc18p or Scc1–myc18p with Scc2–HA6p, Eco1–HA3p, and with Smc2–HA6p (Smc2p is implicated in chromosome condensation but not thus far in cohesion). Scc2–HA6p coprecipitated with both Scc3–myc18p (Fig. 5C) and Scc1–myc18p (data not shown), but the stoichiometry of this association was low compared to Smc3–HA6p. No Smc2–HA6p was found in Scc3–myc18p (Fig. 5C) or Scc1–myc18p (data not shown) immunoprecipitates. Finally, Eco1–HA3p was not coprecipitated with either Scc3–myc18p or Scc1–myc18p (data not shown).
These data suggest that Scc1p, Scc3p, Smc1p, and Smc3p form a soluble ‘Cohesin’ complex in extracts from yeast. Scc2p also associates with this complex, but it is not a stoichiometric component. A recent study has described a similar Cohesin complex from Xenopus, which contains homologs of Smc1p, Smc3p, and Scc1p along with two other proteins (Losada et al. 1998). Our data suggest that at least one of these uncharacterized proteins is likely to be the Xenopus homolog of Scc3p. Our finding that Smc2p is not a subunit of the yeast Cohesin complex is consistent with the finding in Xenopus and in fission yeast that Smc2p/Cut14 and Smc4p/Cut3 are subunits of a different complex called Condensin, which is required for chromosome condensation (Saka et al. 1994; Hirano et al. 1997).
Colocalization and interdependent chromatin association of Cohesin subunits
To analyze whether the binding to chromatin of individual Cohesin subunits depends on other members of the complex and on other proteins needed for sister chromatid cohesion, we constructed scc1-73, scc2-4, scc3-1, smc1-259, smc3-42, eco1-1, and wild-type strains expressing HA-tagged versions of either Scc1p or Scc3p. Cells presynchronized in G1 by α-factor treatment were released into nocodazole-containing medium at the restrictive temperature (in this case 35.5°C) and association of Scc1–HA6p or Scc3–HA3p with chromatin was analyzed using two techniques: chromosome spreading and cell fractionation (Fig. 6A,B). A large fraction of Scc1p and Scc3p is found in the chromatin pellet in wild-type cells arrested in nocodazole and both proteins colocalize with DNA in chromosome spreads (Fig. 6C,D). The abundance of Scc3p in chromatin pellets, but not in supernatants, was reduced drastically in scc1-73, scc2-4, smc1-259, and smc3-42 mutant strains. As a consequence, most Scc3p is found in the supernatant in these mutants. In contrast, distribution of Sccp3p was barely, if at all, affected by eco1-1. The amounts of Scc3p associated with chromosomes in spreads was also greatly reduced in scc1-73 (Fig. 6D), scc2-4, smc1-259, and smc3-42 mutants (data not shown).
Figure 6.
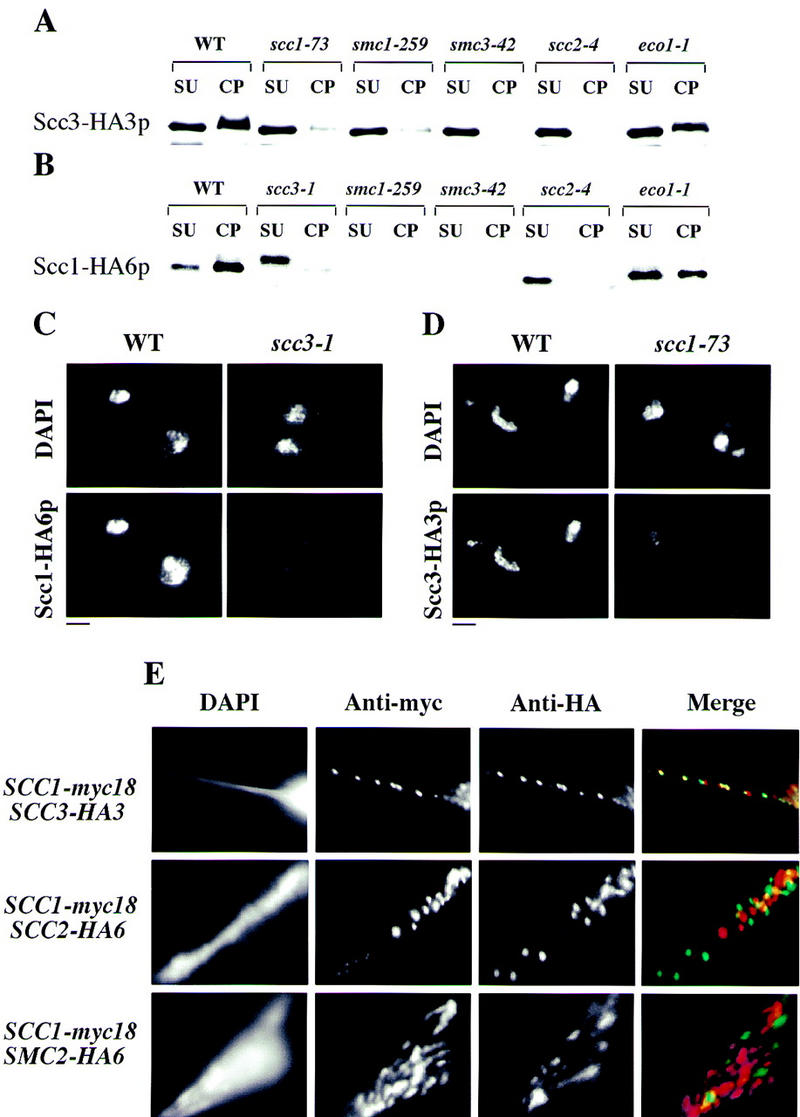
Colocalization of Cohesin subunits and their interdependent chromatin association. Wild-type (K7570), scc1-73 (K7734), scc2-4 (K7689, K7586), scc3-1 (K7548), smc1-259 (K7686, K7589), smc3-42 (K7687, K7583), and eco1-1 (K7586, K7669) strains containing the SCC1 or SCC3 gene tagged with HA epitopes were released from α-factor into nocodazole at 35.5°C. After 2.5 hr, >90% of cells had arrested with large buds and a 2C DNA content. (A,B) The amount of Scc1–HA6p and Scc3–HA3p was detected by Western blot analysis of Triton X-100 insoluble (chromatin pellet, CP) and soluble (supernatant, SU) fractions (Liang et al. 1997). (C) Scc1–HA6p detected in chromosome spreads in wild-type and scc3-1 mutant cells. (D) Scc3–HA3p detected in chromosome spreads in wild-type and scc1-73 mutant cells. Bar, 4 μm. (E) Chromosome spreads were prepared from strains expressing Scc1–myc18p in combination with either Scc2–HA6p, Scc3–HA3p, or Smc2–HA6p. DNA was stained by DAPI. Myc epitopes were detected with anti-myc mouse mAb 9E10 and anti-mouse cy3-conjugated goat antibody (Amersham); HA-tagged proteins were visualized with anti-HA rat mAb 3F10 and anti-rat cy2-conjugated goat antibody (Amersham).
Scc1p’s abundance in both supernatants and chromatin pellets was greatly reduced in both smc1-259 and smc3-42 mutants, whereas its abundance in chromatin pellets decreased while its abundance in supernatants modestly increased in scc3-1 and scc2-4 mutants. We also noticed that Scc1–HA6p migrated more slowly during SDS-PAGE in scc3-1 mutant cells than in wild type. The eco1-1 mutation had only modest effects on Scc1p’s distribution (Fig. 6B). The amount of Scc1–HA6p bound to chromosomes in spreads was also strongly reduced in scc3-1 (Fig. 6C), scc2-1, smc1-259, and smc3-42 mutant strains (data not shown). The reduced abundance of Scc1p in smc1 and smc3 mutants might be attributable to a reduction of its stability when not bound to these Smc proteins. The continued association of Scc1p and Scc3p with chromosomes in eco1-1 mutants was not attributable to this mutation having only a modest effect on sister chromatid cohesion, because 50% or more of metaphase (nocodazole) spreads had two separate CenV–GFP dots despite the presence of both Scc1p and Scc3p on chromosomes (data not shown). We conclude that the association of Scc1p and Scc3p with chromosomes requires Scc1p, Scc2p, Scc3p, Smc1p, and Smc3p, but not Eco1p.
To investigate whether Cohesin’s subunits might also bind to chromatin as a complex, we compared in chromosome spreads the localization of Scc1–myc18p with Scc3–HA3p, Smc3–HA6p, Scc2–HA6p, and Smc2–HA6p. Scc1–myc18p frequently colocalized with Scc3–HA3p (Fig. 6E) and with Smc3–HA6p (data not shown), but it did not, in most cases, colocalize with Scc2–HA6p or with Smc2–HA6p (Fig. 6E).
The colocalization of Scc1p, Scc3p, and Smc3p in chromosome spreads and the interdependence of their association with chromosomes suggests that these cohesin subunits bind chromatin in the form of a complex that resembles that found in soluble extracts. Scc2p neither colocalizes with Cohesin subunits in spreads nor does it appear to be a stoichiometric component of the soluble Cohesin complex. Nevertheless, Scc2p is essential for the efficient association of Cohesin with chromatin, which might be the reason for defective cohesion in scc2 mutants.
Eco1p is not required for Cohesin’s association with and dissociation from chromosomes
Despite being essential for sister chromatid cohesion, Eco1p is neither a Cohesin subunit nor apparently is it even required for Cohesin to bind chromosomes, at least when the cell cycle is arrested with nocodazole. To investigate Eco1p’s role in cells not treated with nocodazole, we measured the chromosomal association of Scc1–HA6p and Scc3–HA3p when unbudded G1 eco1-1 mutant cells isolated by elutriation proceeded through the cell cycle at 37°C (Fig. 7A). As in wild-type cells, Scc1p and Scc3p associated with chromosomes just before S phase and remained associated with them until Pds1p degradation, whereupon both proteins largely disappeared from chromosomes. The disappearance of both proteins from chromatin is delayed relative to wild-type but this is most probably attributable to the delay in Pds1p destruction, which presumably retards Esp1p’s activation. Thus, regulation of Cohesin’s association with and dissociation from chromosomes is unaffected by the eco1-1 mutation. Nevertheless, sister CenV sequences separated prematurely. Thus, 60 min after budding a large fraction of chromosome spreads contained two separate CenV–GFP dots despite being strongly stained for Scc1p and Scc3p (Fig. 7A,B). eco1 mutants are therefore unique in being defective in sister cohesion despite the presence of Cohesin on chromosomes.
Figure 7.
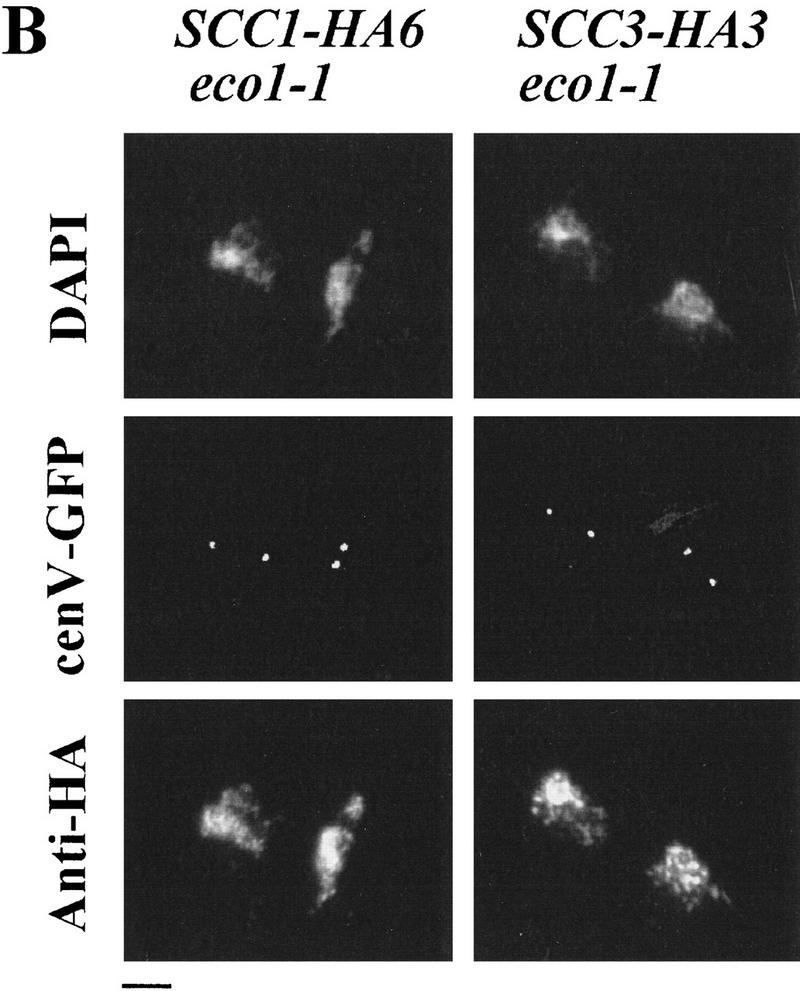
Regulation of Scc1p and Scc3p association with the chromatin is normal in eco1-1 mutant cells. Small unbudded G1 eco1-1 mutant cells expressing Pds1–myc18p and either Scc1–HA6p (K7752) or Scc3–HA3p (K7669) were incubated at 37°C. Samples were taken every 15 min. (A) DNA content of cells and the percentage of budded cells (█), cells with separated sister chromatids (○), cells with Pds1p in their nucleus (▵), and chromosome spreads associated with high amounts of Scc1–HA6p or Scc3–HA3p (⋄). (B) Chromosome spreads of cells taken at 135 min (eco1-1 SCC3–HA3) or at 165 min (eco1-1 SCC1–HA6). DNA was stained by DAPI. CenV was visualized by GFP. Chromatin-associated Scc1–HA6p and Scc3–HA3p were detected by indirect immunofluorescence. Bar, 4 μm.
Eco1p is only required during S phase
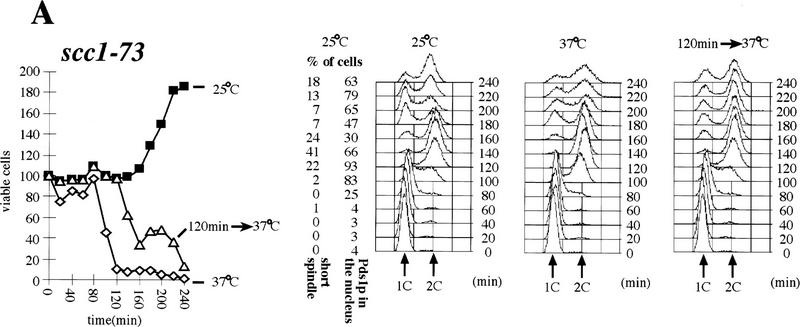
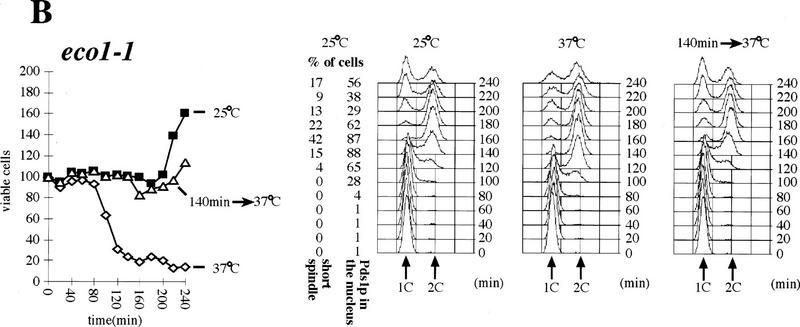
Although Cohesin is able to bind chromosomes from late G1 until M phase, it can only establish cohesion during S phase (Uhlmann and Nasmyth 1998). This suggests that links between sisters might be established only when they are in close proximity during replication. This raises the possibility that there might exist specialized proteins whose role is to establish sister–sister links at the replication fork. Our observation that Eco1p is essential for establishing sister cohesion, but not for Cohesin’s association with chromosomes, is consistent with Eco1p having such a role. If so, Eco1p, like Cohesin, should be required during S phase to establish cohesion, but, unlike Cohesin, it should no longer be required to maintain cohesion during G2. To investigate this, we isolated unbudded G1 eco1-1 and scc1-73 mutant cells by elutriation and split these synchronous cultures into three aliquots. One was incubated at 25°C, another at 37°C, and a third shifted from 25°C to 37°C after most cells had just completed DNA replication (120 min for scc1-73 and 140 min for eco1-1). In each culture, we followed cell viability (by plating cells at 23°C), cellular DNA content, and the fraction of cells whose nuclei contained high levels of Pds1p (Fig. 8A,B). In cultures grown at 37°C, cell viability dropped with kinetics that were similar if not identical to that with which cells completed DNA replication. Thus, restoration of Scc1 activity during G2 cannot compensate for its lack of activity during S phase, as has been found using a SCC1 gene expressed by a galactose-inducible promoter. The same is true for eco1-1. Thus, cell viability depends on activity of both Scc1p and Eco1p during S phase.
Figure 8.
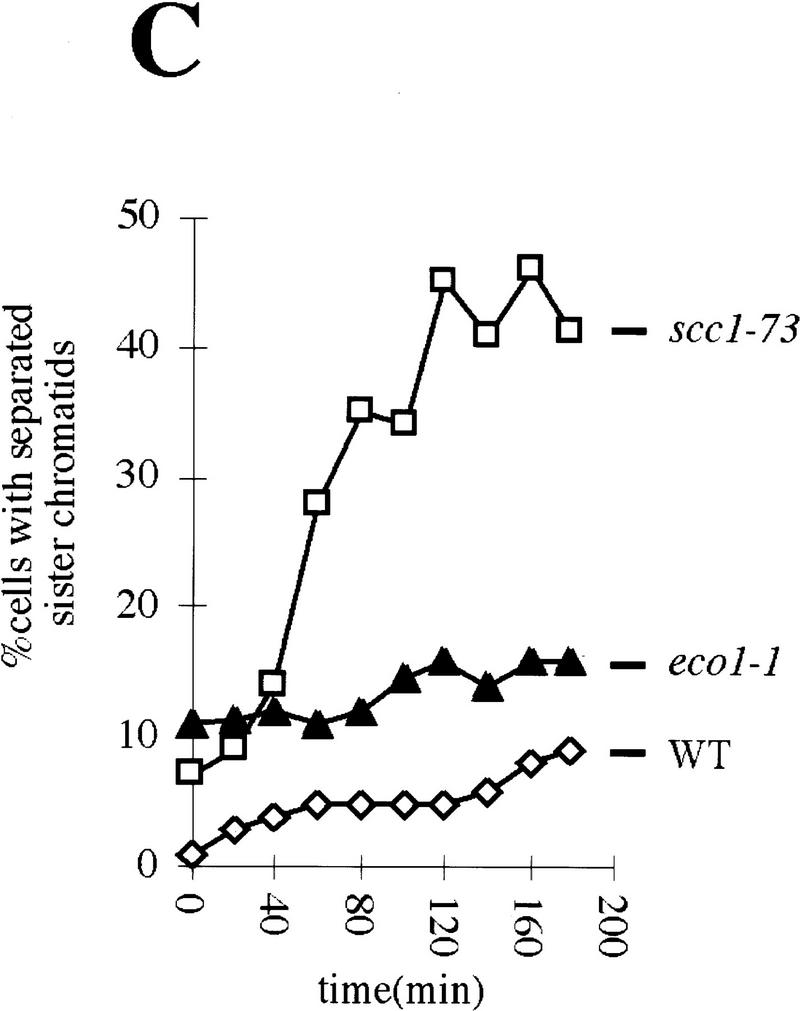
Eco1p is essential during S phase but not during G2 and M phases. (A,B) Small G1 cells of eco1-1 (K7542) and scc1-73 (K6800) strains expressing Pds1–myc18p were inoculated into YEPD medium at 25°C and 37°C (⋄). Aliquots from cultures grown at 25°C (█) were shifted to 37°C at 120 min (scc1-73; ▵) and at 140 min (eco1-1; ▵). DNA contents and the percentages of viable cells were determined in each of these cultures every 20 min. The presence of Pds1p and short bipolar spindles within nuclei were determined by immunofluorescence at each time point from the cultures grown at 25°C. (C) Wild-type (K7101; ⋄), eco1-1 (K7542; ▴), and scc1-73 (K6800; □) strains were released from α-factor arrest into nocodazole-containing medium at 22.5°C. Cultures were shifted to 37°C when >85% of cells had budded (time point 0 min). We followed the separation of CenV–GFP dots for 180 min after the temperature shift. Cells were arrested with 2C DNA content during the entire experiment.
At 25°C, 85% of the scc1-73 cells had completed S phase by 120 min, but only a few had formed bipolar mitotic spindles and almost none had commenced degrading Pds1p. Upon shift to 37°C, 65% of the cells rapidly lost viability (Fig. 8A). This implies that Scc1p is required both during S phase and during G2/M. In the eco1-1 culture (25°C) at 140 min, most but not all cells had completed S phase and few if any had commenced with Pds1p degradation. In contrast, there was only a small drop in viability soon after the culture was shifted to 37°C, which correlated with the small number of G1 cells that underwent replication after the temperature shift to 37°C. This suggests that eco1-1 cells acquire the ability to execute mitosis at 37°C soon after cells complete DNA replication (Fig. 8B). Unlike Scc1p, which is required during both S and G2 phases, Eco1p might be required only during S phase.
To address whether Eco1p and Scc1p are required to maintain sister chromatid cohesion once cells have completed S phase, we released wild-type, eco1-1 and scc1-73 mutant cells from an α-factor-induced G1 arrest into nocodazole at 22.5°C. All three cultures were shifted to 37°C 110 min after release from pheromone, when >85% of the cells had completed replication. Sister chromatid cohesion was monitored subsequently by counting the fraction of cells with one and two CenV–GFP dots (Fig. 8C). At the time of the temperature shift, the number of wild-type cells with two dots was <3% and it increased only very slowly with time; 90% or more of wild-type cells still had only a single GFP dot 180 min after shift to 37°C. In scc1-73 mutants, in contrast, the number of cells with separated CenV sequences increased rapidly, from 6% at the shift to 50% 2 hr later. Remarkably, the pattern in eco1-1 cells resembled that of wild-type cells, but not that of scc1-73 mutants. Although 11% of the cells had two CenV–GFP dots at the time of the shift (the eco1-1 allele is still partly defective at 22.5°C), this number increased with time at the same low rate as in wild-type. We conclude that Scc1p but not Eco1p is required to maintain during M-phase sister cohesion that was established during S phase. Our results suggest that Eco1p activity might be required exclusively during S phase.
Discussion
By screening for temperature-sensitive yeast mutants that lose chromosomes at a high frequency and separate sister centromeres in the absence of APC activity, we have identified six chromosomal proteins (Michaelis et al. 1997; this study), which are essential for preventing premature sister chromatid separation and for ensuring that sisters segregate to opposite spindle poles during mitosis. Homologs of all six of these cohesion proteins exist in a variety of eukaryotes including humans. Four of them, Scc1p (Mcd1p), Scc3p, Smc1p, and Smc3p are subunits of a Cohesin complex that binds to yeast chromosomes in late G1 and disappears from them at the metaphase → anaphase transition. The composition and function of yeast Cohesin resembles that of a similar 14S complex in Xenopus, which is known to contain proteins homologous to Smc1p, Smc3p, and Scc1p (Losada et al. 1998). The fifth protein, Scc2p, is related to Mis4p in fission yeast, to Rad9p from Coprinus, and to proteins encoded by Drosophila and human cDNAs. A small but significant fraction of Scc2p is found associated with Scc1p and Scc3p and is essential for both of these proteins to associate with chromosomes.
The sixth protein, Eco1p (also called Ctf7p) (R.V. Skibbens et al., pers. comm.), is neither a subunit of Cohesin in yeast nor is it required for Cohesin to associate with chromosomes. Furthermore, whereas Cohesin and its subunits are essential to maintain cohesion during G2 and M phase in yeast, Eco1p is only needed to establish cohesion during S phase (Fig. 9A). Similar conclusions as to Eco1/Ctf7’s unique function in establishing cohesion have been drawn independently by R.V. Skibbens et al. (pers. comm.).
Figure 9.
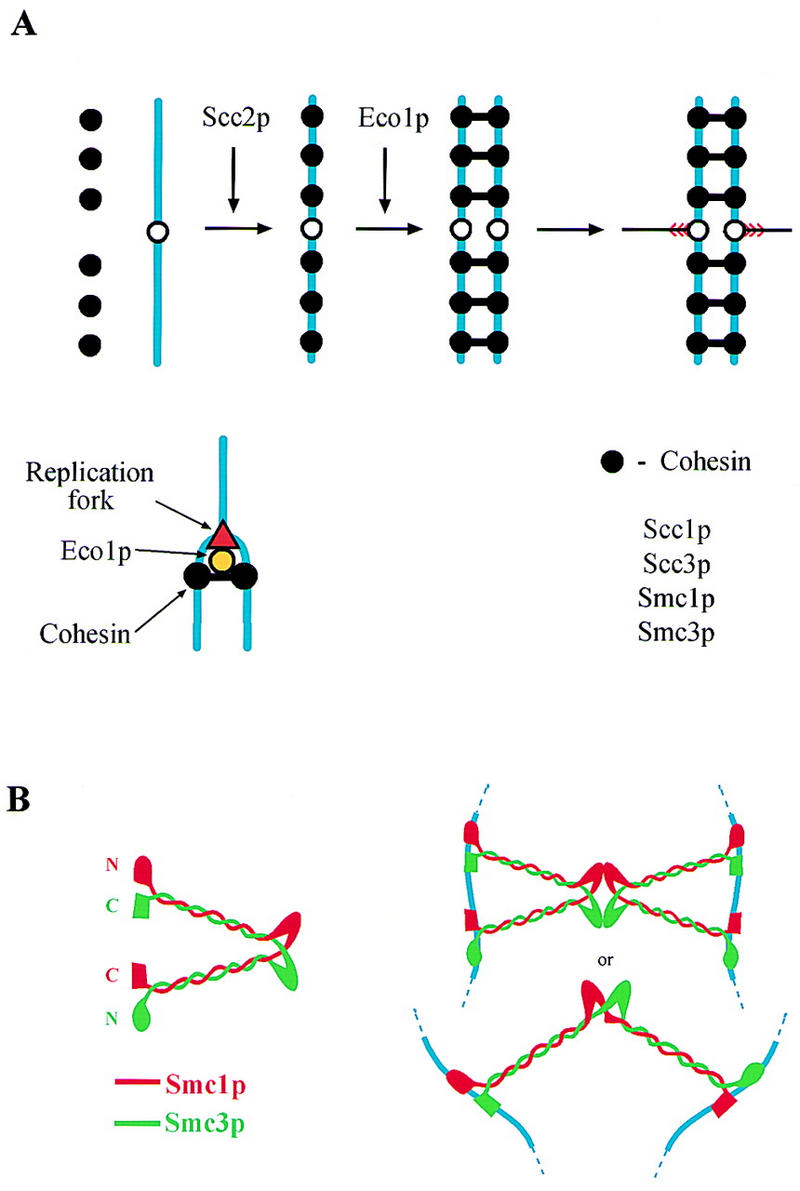
Models for the function of different cohesion proteins. (A) A Cohesin complex composed of Scc1p, Scc3p, Smc1p, and Smc3p is loaded onto chromosomes at the end of G1. Association of Cohesin with chromosomes depends on Scc2p. During S phase, Eco1p, acting at replication forks, catalyzes the formation of links between sister chromatids. These links are portrayed as being composed of Cohesin. It is possible, however, that the links merely need Cohesin for their formation. (B) Two models for how Smc1/3 heterodimers might join sister chromatids.
A Cohesin complex
Previous work showing that Scc1p (Mcd1p), Smc1p, and Smc3p are required for sister chromatid cohesion and that Scc1p’s association with chromosomes depends on Smc1p suggested that these cohesin proteins might bind chromosomes as a multisubunit complex (Michaelis et al. 1997). The demonstration that a small but unquantified fraction of Scc1p bound to Smc1p when overproduced was consistent with this scenario (Guacci et al. 1997). We have now identified a fourth cohesin protein, called Scc3p, and showed that it forms a stable complex, at least when not bound to chromosomes, with Smc1p, Smc3p, and Scc1p. Immunoprecipitates of Scc1p contain Smc1p, Smc3p, and Scc3p in roughly equal amounts, according to silver staining. Surprisingly, all three proteins appear to be more abundant than Scc1p itself. This is probably because Scc1p stains poorly with silver, as Western blot analysis of Scc3–myc18p immunoprecipitates showed that roughly equal amounts of Scc1p, Smc1p, and Smc3p are associated with Scc3p.
The colocalization of Scc1p with Scc3p and Smc3p in chromosome spreads and the interdependence of their association with chromosomes suggests that Scc1p, Scc3p, Smc1p, and Smc3p also bind chromosomes as a multisubunit complex. It is an open question whether there exist other stoichiometric subunits of yeast Cohesin.
Does Cohesin connect sister chromatids?
Although there is no direct evidence that Cohesin actually constitutes the links that hold sisters together during G2, it is nevertheless worth considering how it might do so. Two subunits of yeast Cohesin, Smc1p and Smc3p, belong to the conserved Smc protein family, members of which contain a hinge region that separates two very long stretches of α-helix at the ends of which there are globular amino- and carboxy-terminal domains (Koshland and Strunnikov 1996). A recent electron microscopic analysis of MukBp from E. coli and Smcp from Bacillus subtilis suggests that they form homodimers whose hinge region connects two antiparallel coiled coils, at both ends of which amino- and carboxy-terminal domains are brought together into a composite globular domain with a potential ATPase and DNA-binding activity (Melby et al. 1998).
The finding that both yeast and Xenopus Cohesin contains equimolar amounts of Smc1p and Smc3p suggests that these two proteins might form heterodimers in which α-helical sections of Smc1p coil in an antiparallel fashion around those of Smc3p. If the structure of such a heterodimer were similar to bacterial Smcs, then the globular domain at one end of the heterodimeric V would contain Smc1p’s amino-terminal domain next to Smc3p’s carboxy-terminal domain, whereas the other end would contain Smc3p’s amino-terminal domain next to Smc1p’s carboxy-terminal domain (Fig. 9B). The globular domains of a single Smc1/3 heterodimer in principle could bind sister DNA molecules and thereby form bridges between them. Such bridges might alternatively be produced through the association (possibly mediated by Scc1p or Scc3p) of two different Smc1/3 heterodimers, each of which is bound in a bivalent manner to a single chromatid (Fig. 9B).
Eco1p, a protein needed for establishing cohesion
Cohesin’s binding to chromosomes is necessary but not sufficient for sister chromatid cohesion. Under at least two circumstances, for example, when Eco1p is inactive or Scc1p is only synthesized after S phase (Uhlmann and Nasmyth 1998), Cohesin still binds to chromosomes in normal amounts without producing cohesion. This suggests that functional sister–sister connections (possibly involving Cohesin) are only produced during DNA replication and that Eco1p might be involved in their production. Eco1p is much less abundant than Cohesin subunits. Furthermore, it neither associates with soluble Cohesin nor is it required for Cohesin’s association with chromosomes. Eco1-1 mutant cells divide without loss of viability when shifted to 37°C during G2 and maintain cohesion between sister CenV sequences when cells that had been arrested in nocodazole at 22.5°C are shifted to 37°C. Both properties imply that Eco1p is needed to establish cohesion during S phase but is not necessary to maintain cohesion during G2 and M phases. Therefore, we propose that the proximity of sister chromatids immediately after the passage of replication fork is not sufficient for establishing stable links between them but this is instead an active process catalyzed by Eco1p (Fig. 9A). Establishment of sister chromatid cohesion is quite distinct from the loading of Cohesin onto chromosomes, which can occur during most but not all phases of the cell cycle and does not need Eco1p.
We cannot strictly exclude the possibility that the maintenance of sister cohesion in eco1-1 mutants during G2 or M phase is attributable to the mutant gene product becoming thermoresistant during DNA replication. However, several arguments can be raised against this interpretation. The cohesion defect of eco1-1 mutants is more severe than that of any other cohesin mutant that we have characterized and yet sister chromatids remain tightly associated for up to 3 hr when nocodazole-arrested cells are shifted to 37°C. Crucially, other eco1 alleles (ctf7–203) seem to behave in a similar manner, implying that the behavior of eco1-1 is not allele specific (R.V. Skibbens et al. pers. comm.). Finally, as Eco1p is present throughout the cell cycle, it is unlikely that thermolabile eco1 alleles are simply temperature-sensitive for synthesis.
The S. pombe homolog of Eco1p is a much larger protein whose amino-terminal half is homologous to Rad30p from S. cerevisiae and to dinBp and umuCp from E. coli, which function in postreplicative DNA repair (McDonald et al. 1997). This suggests that Eco1p might associate with Rad30p in vivo and, therefore, that it could have a role in DNA repair as well as sister chromatid cohesion.
Regulation of Cohesin binding to chromosomes
Cohesin’s association with chromosomes is presumably necessary for sister chromatid cohesion, whereas the disappearance from chromosomes of at least two of its subunits (Scc1p and Scc3p) at the metaphase to anaphase transition suggests that Cohesin’s removal is necessary for loss of cohesion. Regulation of Cohesin’s association with chromosomes is, therefore, a vital aspect of the eukaryotic cell cycle.
Our observation that neither Scc1p nor Scc3p is stably bound to chromosomes in scc2 mutants suggests that Scc2p is required for the stable association of Cohesin with chromosomes (Fig. 9A). The lack of Cohesin on chromosomes in scc2 mutants could explain their defective sister chromatid cohesion. Scc2p’s homolog in S. pombe, a protein called Mis4p, is also required for sister chromatid cohesion and it has been suggested that Mis4p might act independently of Cohesin (Furuya et al. 1998). Our observations suggests otherwise. A key question is whether Scc2p is required for the formation of Cohesin complexes or only for their association with chromosomes. The finding that Scc1p is much less abundant in smc1 or smc3 mutants than wild-type or even scc2 mutants and the implication that it may not be stable unless complexed with Smc proteins suggests that the soluble Scc1p protein found in scc2 mutants is complexed with Smc1p and Smc3p. In which case, scc2 mutants may be primarily defective in Cohesin binding to chromosomes. We detected a weak association between Scc2p and soluble Cohesin complexes. A transient interaction between Cohesin and Scc2p would be consistent with Scc2p having a role in loading Cohesin on chromatin. Alternatively chromatin-bound Scc2p might be required for the formation of structures that are recognized and bound by Cohesin.
At least two subunits of Cohesin, Scc1p and Scc3p, disappear suddenly from yeast chromosomes at the onset of anaphase in a process that depends on liberation of Esp1p from its inhibitor Pds1p (Ciosk et al. 1998). This suggests that Esp1p induces a change in yeast Cohesin, which by destroying cohesion between sisters, would allow the segregation of sister kinetochores to opposite spindle poles. It is conceivable that Scc2p is involved in regulating the amount of Cohesin associated with chromosomes not only during G2 but also during mitosis. A reduction in Scc2p’s activity might contribute to Cohesin dissociation from chromatin at the onset of anaphase.
Other roles for cohesins?
Cohesins may well have important roles in processes other than sister cohesion. The bovine orthologs of Smc1p and Smc3p are components of a mammalian recombination complex RC-1 (Jessberger et al. 1996). Furthermore, Scc1p’s homolog in S. pombe, a protein called Rad21p, is necessary for double strand break repair (Birkenbihl and Subramani 1995), whereas Scc2p’s homologs in Coprinus (Rad9p) and in fission yeast (Mis4p) are involved in DNA repair and meiosis (Zolan et al. 1992; Furuya et al. 1998). Cohesins holding sister chromatids together maintain proximity between homologous sequences that might be a prerequisite for recombination and recombination-mediated DNA repair. Alternatively, cohesin proteins might be required for recombination and DNA repair independently from their role in sister chromatid cohesion.
Three different classes of cohesion proteins
In summary, the six cohesion proteins that we have identified by genetic analysis fall into three groups. Scc1p, Scc3p, Smc1p, and Smc3p are subunits of a Cohesin complex that possibly resides at the sites that link sister chromatids together during G2. Scc2p is necessary for Cohesin’s association with chromosomes, whereas Eco1p mediates the formation of links between sisters during replication. These three types of cohesion proteins appear to be conserved between yeast and humans. Whether and how Cohesin mediates the connections between sisters remains to be investigated.
Materials and methods
Yeast strains and growth conditions
Cells were grown in YEP (Rose et al. 1990) media supplemented with either 2% raffinose (YEPRaf) or 2% glucose. Canavanine plates contained 0.0066% canavanine and all amino acids except arginine. Elutriation was performed as described previously (Schwob and Nasmyth 1993). Small G1 cells were inoculated into YEPD medium at 25°C or 37°C. Cells were arrested with α-factor or with nocodazole as previously described (Irniger et al. 1995).
All strains were derivatives of W303. The centromeric region of chromosome V was visualized by GFP (Michaelis et al. 1997). SCC1, SCC2, SCC3, SMC1, SMC2, SMC3, ECO1, and PDS1 genes were tagged at their carboxyl terminus with multiple Myc or HA epitopes by homologous recombination at their original chromosomal loci, using a PCR-based method (W. Zachariae and K. Nasmyth, unpubl.). Cells were transformed with a cassette containing multiple copies of myc or HA sequences and a marker gene. The cassette was amplified by PCR with target gene-specific primers. The tagged strains all proliferated normally at 37°C, demonstrating that the epitope-tagged proteins were functional.
To obtain a strain in which APC can be inactivated conditionally, the CDC26 gene was disrupted by a PCR targeting method as described before (Wach et al. 1994). To integrate a functional CDC26 gene to the lys2 locus, a YDp-K (Berben et al. 1991) based integrative vector was constructed that contained the functional CDC26 gene at the PstI site and the CAN1 gene at the SmaI site. The plasmid was cut at the XhoI site in the LYS2 gene and was transformed into Δcdc26 cells. After integration, CDC26 and CAN1 genes were flanked by direct repeats of homologous sequences that allowed us to loop out the CDC26 and CAN1 genes upon selection against the CAN1 gene. The above strain proliferated normally at 37°C showing that the CDC26 gene integrated to the lys2 locus is functional.
Genetics
A W303 strain with CenV–GFP, with a supernumerary chromosome carrying SUP11 and with the indolgenous CDC26 gene disrupted and functional CDC26 and CAN1 genes integrated to the lys2 locus (K7074), was mutagenized with ethyl methanesulfonate to 30%–50% survival. Approximately 9 × 105 colonies were plated as previously described (Spencer et al. 1990). Cells were grown at 23°C for 7–10 days. Red–white sectoring colonies (1675) were screened for temperature-sensitive growth. The temperature-sensitive mutants (360) were streaked on canavanine-containing plates to delete CDC26 from the genome. Versions of mutants lacking CDC26 were released from stationary phase (2-day-old patches on YEPD plates) into YEPD medium at 37°C and the ratio of cells with separated sister chromatids (cenV–GFP) was determined.
Complementation analysis and cloning of mutants that separated sisters in the presence of Pds1p showed that we had isolated mutations in six different genes: SCC1, SCC2, SMC3, PDS1, SCC3 (IRR1), and ECO1. SCC3 (IRR1) and ECO1 genes were identified by complementing the temperature-sensitive growth defect of the corresponding mutants. A YCplac vector-based genomic library was transformed into scc3-1 and eco1-1 mutants and transformants were selected at 37°C. ECO1 was replaced by S. pombe his5+ in the wild-type diploid K842 strain as described previously (Shirayama et al. 1998). The heterozygous Δeco1 diploid strain was sporulated and tetrad analysis showed that none of the Δeco1 spores were viable. On the basis of tetrad analysis of spores derived from diploids containing the scc3-1 allele and a functional HA-tagged allele of SCC3 gene or eco1-1 allele and a functional HA-tagged allele of ECO1, we concluded that the temperature-sensitive alleles are linked to the corresponding tagged ORFs.
Immunoprecipitations
For silver staining (Fig. 5A), the Cohesin complex was purified by one-step immunoprecipitation from an unfractionated whole cell extract prepared from Δpep4 cells (0.5 × 1010) using the procedure of Zachariae et al. (1998). For immunoprecipitation-immunoblotting experiments (Fig. 5B,C), cells (1.5 × 109) were broken in 0.4 ml of buffer B70. Extracts (0.33 ml, 6 mg) were incubated with 0.1 ml of protein A–Sepharose and then with 0.033 ml of protein A–Sepharose carrying the anti-myc antibody 9E10. Preparation of extracts and immunoprecipitations were carried out essentially as described previously (Zachariae et al. 1998). In each case, half of the final immunoprecipitate was loaded per lane. In immunoprecipitation–immunoblotting experiments, 100 mg of protein from whole-cell extract was loaded per lane. Extracts and immunoprecipitates were separated on 1-mm thick, 7.5% SDS–polyacrylamide gels and analyzed by immunoblotting.
Cohesion protein chromatin-binding assay
Wild-type (K7570), scc1-73 (K7734), scc2-4 (K7689, K7586), scc3-1 (K7548), smc1-259 (K7686, K7589), smc3-42 (K7687, K7583), and eco1-1 (K7586, K7669) strains containing SCC1 or SCC3 genes tagged with HA epitopes, were released from α-factor arrest into YEPD medium containing 15 μg/ml nocodazole and 1% DMSO at nonpermissive temperature (35.5°C). After 2.5 hr all cells had undergone S phase and were arrested in an M phase-like state. Cells were harvested by centrifugation and a crude Triton X-100 insoluble chromatin preparation was obtained as described (Liang and Stillman 1997) Aliquots of the Triton X-100-soluble supernatant and of the chromatin pellet reflecting the same cell equivalent were loaded for 8% SDS-PAGE. Western blotting was performed with the monoclonal antibody 16B12 (Boehringer) against the HA epitope to detect Scc1–HA6p or Scc3–HA3p in the chromatin fractions.
Viability assay
The viability of cells was measured by diluting aliquots of eco1-1 and scc1-73 cultures at the indicated time points and plating constant volume of dilutions on YEPD plates at 23°C. Number of colonies was counted after 3 days.
Other techniques
Flow cytometric DNA quantification was performed as described (Epstein and Cross 1992). Chromosome spreading was performed as described previously (Michaelis et al. 1997). Protein extracts, Western blot analysis, in situ immunofluorescence, antibody dilutions were performed as described previously (Piatti et al. 1996). On chromosome spreads HA-tagged proteins were detected using 3F10 antibody (Boehringer) and with a rhodamine-conjugated secondary antibody. Colocalization of the Myc- and HA-tagged proteins was assessed using two sets of antibodies: anti-Myc mouse mAb 9E10 with anti-mouse cy3-conjugated goat antibody (Amersham), and anti-HA rat mAb 3F10 with anti-rat cy2-conjugated goat antibody (Amersham). The timing of sister chromatid separation relative to DNA replication and budding was calculated as described (Michaelis et al. 1997).
Acknowledgments
We are grateful to Robert V. Skibbens and Doug Koshland for sharing unpublished results; in particular, their observation that ctf7 mutants might be involved in establishing cohesion. We thank Christine Michaelis and Dunja Knapp for advice on techniques used during the genetic screen; Wolfgang Zachariae, Jan Michael Peters for sharing their biochemical expertise; Tina Meinl, Fedor Severin, Isabel Gonzales, Tomoyuki Tanaka for technical help; Wolfgang Zachariae and Masaki Shirayama for stimulating discussions; Jan Michael Peters and Rosemary Clyne for their critical comments on the manuscript; and H. Tkadletz for helping with figures. F.U. acknowledges support through an European Molecular Biology Organization long-term fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Nasmyth@NT.IMP.UNIVIE.AC.CAT; FAX 43 1 798 71 53.
References
- Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. The YDp plasmids: A uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Subramani S. The rad21 gene product of Schizosacharomyces pombe is a nuclear, cell cycle regulated phosphoprotein. J Cell Biol. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- Carramolino L, Lee B, Zaballos A, Peled A, Barthelemy I, Shav-Tal Y, Prieto I, Carmi P, Gothelf Y, Gonzalez de Buitrago G, Aracil M, Marquez G, Barbero J, D Z. SA-1, a nuclear protein encoded by one member of a novel gene family: Molecular cloning and detection in hemopoietic organs. Gene. 1997;195:151–159. doi: 10.1016/s0378-1119(97)00121-2. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An Esp1/Pds1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters J-M, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- DeVeaux LC, Smith GR. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes & Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: A novel B cyclin from budding yeast with a role in S phase. Genes & Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996a;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature. 1996b;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion protein molecule required in S phase and not destroyed in the G1 phase. Genes & Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E, and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Trotis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jessberger R, Riwar B, Baechtold H, Akhmedov TA. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- Jessberger, R., C. Frei, and S.M. Gasser. 1998. Chromosome dynamics: The SMC protein family. Curr. Opin. Genet. Dev. 8. [DOI] [PubMed]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Ann Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Kurlandzka A, Rytka J, Gromadka R, Murawski M. A new essential gene located on Saccharomyces cerevisiae chromosome IX. Yeast. 1995;11:885–890. doi: 10.1002/yea.320110910. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes & Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J, Levine A, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homolog of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintainance of chromosomes (SMC) and MukB proteins: Long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker J, Diffley JFX, Nasmyth K. Activation of S phase promoting Cdks in late G1 defines a ‘point of no return’ after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes & Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–317. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes & Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Selig S, Okumura K, Ward D, Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/Fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10. [DOI] [PubMed]
- Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJR, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase promoting complex from yeast: Identification of a subunit related to Cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- Zolan M, Crittenden J, Heyler N, Seitz L. Efficient isolation and mapping of rad genes of the fungus Coprinus cinereus using chromosome-specific libraries. Nucleic Acids Res. 1992;20:3993–3999. doi: 10.1093/nar/20.15.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]